The role of biosensors in the detection of emerging infectious diseases
Bobby
Pejcic†
ab,
Roland De
Marco
*a and
Gordon
Parkinson‡
ab
aNanochemistry Research Institute, Department of Applied Chemistry, Curtin University of Technology, GPO Box U 1987, Perth, WA, 6845, Australia. E-mail: R.DeMarco@curtin.edu.au; Fax: +61 8 9266 2300
bAustralian Biosecurity Cooperative Research Centre, Perth, WA, Australia
First published on 11th July 2006
Abstract
Global biosecurity threats such as the spread of emerging infectious diseases (i.e., avian influenza, SARS, Hendra, Nipah, etc.) and bioterrorism have generated significant interest in recent years. There is considerable effort directed towards understanding and negating the proliferation of infectious diseases. Biosensors are an attractive tool which have the potential to detect the outbreak of a virus and/or disease. Although there is a host of technologies available, either commercially or in the scientific literature, the development of biosensors for the detection of emerging infectious diseases (EIDs) is still in its infancy. There is no doubt that the glucose biosensor, the gene chip, the protein chip, etc. have all played and are still playing a significant role in monitoring various biomolecules. Can biosensors play an important role for the detection of emerging infectious diseases? What does the future hold and which biosensor technology platform is suitable for the real-time detection of infectious diseases? These and many other questions will be addressed in this review. The purpose of this review is to present an overview of biosensors particularly in relation to EIDs. It provides a synopsis of the various types of biosensor technologies that have been used to detect EIDs, and describes some of the technologies behind them in terms of transduction and bioreceptor principles.
 Bobby Pejcic. | Bobby Pejcic received his PhD in Chemistry from Curtin University of Technology Australia in 2004. He is currently a Postdoctoral Research Fellow with the Commonwealth Scientific Industrial Research Organization (CSIRO), Petroleum Division, in Perth Australia. His research interests relate to the surface and interface analysis of electrochemical systems and materials. He is also interested in the application of chemical sensors to real world problems. |
 Roland De Marco. | Roland De Marco received his PhD in Chemical Physics from La Trobe University Australia in 1992. In 1995, he joined Curtin University of Technology, and was promoted to Associate Professor in 2002. Currently he is also Head of the Department of Applied Chemistry at Curtin University of Technology. His current research interests include: electrochemical surface and interface analysis; electrochemistry; and electroanalytical chemistry. |
 G. Parkinson. | After completing a Bachelor of Science (Hons) in Chemistry at University College of London, Gordon Parkinson studied his PhD in Solid State Chemistry at the University of Wales. Following 10 years as a Senior Research Chemist at British Petroleum Research Centre, London, he joined Curtin University of Technology in 1994. He was the founding Director of the Curtin Nanochemistry Research Institute. In 2005, he joined Alcoa World Alumina as Manager, Transformational Technology. His research interests include nanochemistry, solid-fluid interface, crystallization, biomimicry and innovation. |
1 Introduction
There is great deal of interest in monitoring and controlling the spread of emerging infectious diseases (EID) in the international community.1–7 EIDs can be broadly classified as: (a) new, previously unrecognised diseases (e.g., severe acute respiratory syndrome); (b) known diseases which have increased in incidence, virulence or geographic range over the past several decades (e.g., foot-and-mouth disease); and (c) diseases which threaten to increase in the near future (e.g., avian influenza).8 A complex interplay of factors such as greater global movement of people and animals, demographic shifts, ecological changes, climate changes, changes in animal husbandry practices, etc. have led to the emergence of an increasing number of new diseases.9,10 Clearly, there are many challenges facing organizations and nations concerned with controlling the proliferation of EIDs. Disease surveillance and diagnosis are integral and crucial components of all public health services. An adequate detection system is an essential step in helping to eliminate or minimize the spread of a virus outbreak before the economic, human, and environmental repercussions become devastating.Because of the threat posed by EIDs, the speedy detection of a virus or antigen has a major impact on the success of strategies for disease zoning, control or eradication. Disease surveillance and detection strategies come in various shapes and forms ranging from syndromic to remote sensing.1 The intention of surveillance is to improve data collection and management, and provide improved decision-making tools. Early warning systems are crucial for disease emergency vigilance. The collection of data and diagnostic specimens particularly from free-living populations of animals can be at times difficult to achieve because of the vast size of territories, the sparse human population, and lack of physical infrastructure. More importantly, the problem with existing surveillance programs is the long time delay between sample collection in remote areas and transportation to laboratories, which can be a significant distance away from the site of collection. Depending on the type of EID and the weather conditions, it may only take several hours for the disease to spread through an animal and/or human population. Searching for a rapid, simple and sensitive method is of considerable interest. One of the problems with EID detection is actually determining whether infection has occurred. The difficulty arises because the initial symptoms after infection can be difficult to distinguish from other infections. Consequently, there is a need to develop rapid and reliable tools/systems for the detection of EIDs in the community. One attractive tool that is capable of providing immediate information on a disease outbreak is the biosensor. It is well established that biosensors play a significant role in medicine/clinical analysis,11–13 food/water analysis,14–16 environmental monitoring,17,18 and agriculture.19,20 In principle, biosensors offer the possibility of real-time monitoring, and the deployment of these devices in the field would provide a means for rapid virus detection. An ideal disease detection and surveillance system would involve various processes as shown in Fig. 1.
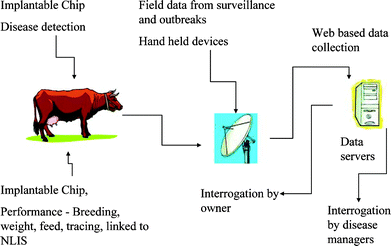 | ||
| Fig. 1 An ideal approach to real-time disease detection and surveillance. | ||
Generally, biosensors are distinguished from one another by the nature of the process and according to their biochemical or biological component, e.g., biocatalytic (i.e., enzyme), immunological (i.e., antibody) and nucleic acid (i.e., DNA). A number of biosensors have been developed for EIDs; however, all of them essentially comprise a biological recognition element or bioreceptor, which interacts with the analyte and responds in some manner that can be detected by a transducer (refer to Fig. 2). The biological recognition element or bioreceptor is a crucial component and its function is to impart selectivity so that the sensor responds only to a particular analyte or biomolecule of interest, hence avoiding interferences from other substances. The transducer is the other component of the biosensor, which also plays an important role in terms of converting the biorecognition event into an electrical signal, and common transducers employed for EID detection include electrochemical, optical, and piezoelectric platforms. An ideal biosensor is one that is reagentless; however, in most studies reported in the literature a co-substrate is normally used. The addition of an enzyme, redox species, etc. is mainly designed to help with the transduction process by facilitating the generation of a detectable product. In fact, detection strategies can be divided into two categories: nonlabeled or label-free types, which are based on the direct measurement of a phenomena occurring during the biochemical reactions on a transducer surface; and labeled, which relies on the detection of a specific label. Research into ‘label-free’ biosensors continues to grow;21–23 however ‘labeled’ ones are more common and are extremely successful in a multitude of platforms.
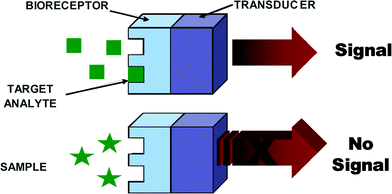 | ||
| Fig. 2 A schematic diagram of a typical biorecognition element. | ||
Since well over 6000 articles have been published in the area of biosensors just from the years 1996 to 2006 alone (English journals), this manuscript is not meant to be a comprehensive review of the field, but rather a critical review, presenting a selection of the most significant technologies and advances in relation to EID monitoring. One of the major challenges when reviewing the literature is the unclear distinction between a biosensor and a bioanalytical instrument. The term ‘biosensor’ has been loosely applied in the literature, noting that on many occasions it has been used to describe an analytical device that incorporates several additional separation steps, which is independent from detection.§ It is not the intention of this review to critically evaluate the various types of biosensors nor will it present a summary of recommendations for effective biosensing. Rather, the objective of this review is to address the development of biosensors for the detection of EIDs. In addition, it will discuss some recent biosensor advances and problems that need to be resolved before they make the journey into the market place. The emphasis will be on the development towards hand-held/implantable analytical devices rather than laboratory instruments. This review covers papers that have been published over the last decade on EID detection and has been structured into four main sections: electrochemical biosensors, optical biosensors, piezoelectric biosensors, and biosensor outlook.
2 EID detection methodologies
Table 1 summarises the main analytical features of a wide range of biosensors, noting that a majority of the existing technologies used for detecting EIDs rely on antibodies as the recognition molecule. Antibodies are the critical part of an immunosensor, since their quality contributes to the sensitivity and specificity.25 As it is beyond the scope of this review, the reader is referred to other articles for a more detailed description of immunosensors.11,26,27 An outline of the key immunological procedures employed for the identification of EIDs can be found in the review by Peruski and Peruski.28 However, it is important to note that the success of an immunosensor depends heavily on which antibodies, labels, and reagents are used in the assay.28,29¶ Once antibodies of the desired specificity and affinity have been developed, they can be incorporated in a wide range of transducer platforms. DNA is an alternative recognition molecule that has also received some attention for the detection of EIDs. A number of excellent reviews have been published elsewhere on sensors based on nucleic acid interaction, noting that the detection of specific DNA sequences provides the fundamental basis for monitoring a wide variety of genetic diseases and viral infections.30–36 Despite the great deal of work in this area the application of a DNA biosensor for EID detection is still in its infancy.| Transducer | Analyte | Biosensor format | Detection limit | Ref. |
|---|---|---|---|---|
| a NA = not available. | ||||
| Amperometric | Newcastle disease | Enzyme-label immunoassay | 11.1 ng ml−1 | 45 |
| Forest–Spring encephalitis | Sandwich gold-label immunoassay | 10−7 mg ml−1 | 46 | |
| Japanese B encephalitis | Fe2+/3+ probe & label-free immunoassay | 6 × 10−9 1g pfu ml−1 | 47 | |
| Hepatitis B | Methylene blue probe, PCR & DNA | NA | 48 | |
| Hepatitis B | Osmium complex probe, PCR & DNA | NA | 49 | |
| Potentiometric | Hepatitis B | Enzyme label immunoassay | ∼50 fM | 51 |
| Japanese B encephalitis | Immunoassay | 6 × 10−9 1g pfu ml−1 | 52 | |
| Light-addressable potentiometric | Newcastle disease | Sandwich enzyme-label immunoassay | 2 ng ml−1 | 53, 54 |
| Venezuelan equine encephalitis | Sandwich enzyme-label immunoassay | 30 ng ml−1 | 56 | |
| Impedance spectroscopy | Hepatitis B | Immunoassay | 8 ng ml−1 | 60 |
| Hepatitis B | Immunoassay | 50 ng l−1 | 61 | |
| Conductometric | Bovine viral diarrhoea | Sandwich immunoassay | Varied between 102–104 CCID ml−1 | 68, 69 |
| Fiber-optic evanescent wave | Newcastle disease | Fluorescein-label sandwich immunoassay | 10 ng ml−1 | 93 |
| Surface plasmon resonance | Foot-and-mouth disease | Immunoassay | NA | 98–100 |
| Hepatitis A | Immunoassay | NA | 101 | |
| Severe acute respiratory syndrome | Peptide binding | NA | 102 | |
| Fluorescence | Various toxins | Immunoassay | ∼ng ml−1 | 109, 110 |
| Dengue | Immunoassay | NA | 111 | |
| Dengue | Nucleic acid | ∼Picomolar | 112, 113 | |
| Piezoelectric | Foot-and-mouth disease | Immunoassay | NA | 115 |
| Foot-and-mouth disease | Immunoassay | NA | 116 | |
| Hepatitis B | Nucleic acid | ∼0.01 μg ml−1 | 117 | |
| Severe acute respiratory syndrome | Immunoassay | 0.6 mg ml−1 | 118 | |
| Dengue | Immunoassay | ∼μg ml−1 | 119–121 | |
One of the major requirements in developing a biosensor for EIDs is the need for a sensitive analytical device that can easily go down to very low detection levels without significant changes in selectivity. Many infectious diseases will spread rapidly through a community before any symptoms are identified1 and a biosensor that can easily detect low levels of antigen at the onset of infection will be invaluable. In addition, a biosensor that is relatively cheap, robust, responds rapidly and provides high-throughput is highly desired for field applications. However, the type of biosensor used for EIDs will in many cases depend on the properties of the analyte (i.e., size, structure, concentration, etc.) and the matrix (i.e., air, liquid) in which the analyte is found. In fact, the sensitivity of the biosensor is highly dependent on the surface preparation/structure and a great deal of effort has been focussed on understanding and tailoring the interfacial properties.37,38|| The most commonly used biosensing methodologies for detecting EIDs will be described in detail.
2.1 Electrochemical biosensors
More than half of the biosensors used for the detection of pathogens and viruses are based on electrochemical transducers.39 Furthermore, a number of reports have demonstrated the importance of electrochemical biosensors in clinical and environmental analysis26,27,40,41 This may not be surprising considering that electrochemical transduction possesses the following advantages: low cost; high sensitivity; independence from solution turbidity; easily miniaturized/well suited to microfabrication; low power requirements; and relatively simple instrumentation.42 These characteristics make electrochemical detection methods highly attractive for field monitoring of infectious diseases and biological warfare agents. Several electrochemical approaches (i.e., amperometric, potentiometric, impedance) have been used to monitor the changes that occur during EID detection. The method used in many cases depends on the type of change (i.e., redox, ionic, conductivity, etc.) along with the properties of the analyte and matrix. For instance, biosensors that involve amperometric detection usually employ an electroactive marker, noting that the antibody–antigen and DNA hybridization reactions do not generate a significant signal on their own. For a discussion of the electron transfer mechanisms that commonly occur with amperometric-based biosensors, we refer readers to the review by Habermuller et al.43 However, the relative merits of each electrochemical transducer will be explored in terms of EID detection requirements.Without a doubt the amperometric sensor for glucose is one of the most studied and successful of all biosensors.44 This has led to the development of a vast array of immunosensors and DNA biosensors that are based on this form of detection. Indeed, the amperometric approach has become a popular choice for the detection of EIDs and biological warfare agents. Recently, Yu and coworkers45 developed an amperometric immunosensor for the assay of Newcastle disease in citrate buffer solutions. The Newcastle disease antigen was immobilized onto a graphite paste matrix, which was prepared according to two different methods. An enzyme labeled (i.e., horseradish peroxidase) antibody was used to catalyze the oxidation of hydrogen peroxide on a tetramethylbenzidine substrate thereby generating a current response that was directly related to the virus concentration. It was shown that the sensor response depends on the Newcastle disease antigen loading, immunosensor preparation conditions, tetramethylbenzidine concentration, and incubation time. Similarly, work by Brainina et al.46 revealed that amperometric detection is an effective and sensitive approach for the diagnosis of Forest–Spring encephalitis. Detection was achieved using a sandwich approach, which involved immobilizing the antigen onto a graphite electrode followed by binding with antibody and a gold-labeled protein A. The sensor was shown to respond to Forest–Spring encephalitis virus over a wide concentration range (i.e., 10−7 to 10−2 mg ml−1). More importantly, the analysis was carried out in real blood serum samples and the analytical results were in excellent agreement compared to the standard ELISA method.46 In many cases, detection with amperometric biosensors is achieved by using an enzyme label in order to improve the sensitivity. However, the group of Yuan47 developed a label-free amperometric immunosensor for Japanese B encephalitis vaccine. The immunosensor was fabricated by immobilizing the antiserum of Japanese B encephalitis on a gold nanoparticle/o-phenylenediamine polymer layer that was deposited onto a Prussian blue coated platinum electrode.47 The response was evaluated as a function of Japanese B encephalitis vaccine concentration, temperature, pH, incubation time, and gold nanoparticle–polymer layer thickness. It was reported that the immunosensor responded to Japanese B encephalitis vaccine in the concentration range 1.1 × 10−8 to 1.9 × 10−6 plaque forming unit ml−1.47 It is evident that the above biosensor studies are based on immunoassay principles; however, there also appears to be some interest in the development of amperometric sensors for DNA detection. Various groups have shown that this approach is particularly suitable for the detection of Hepatitis B virus, noting that a polymerase chain reaction step was also incorporated in the assay.48,49 Despite the analytical success, the main drawback in using DNA biosensors for EID detection is the long assay times (∼4 h) and the difficulty in establishing if a particular disease is infectious. To add further complications, this genetic material is packaged inside an envelope and the quantity is usually very small and not very stable.
The development of potentiometric sensors for the detection of EIDs has also received a great deal of attention. This group of sensors are known to have the longest history and the largest number of applications.50 However, most of the work reported in the literature using potentiometric-sensing principles employs the indirect approach for the detection of virus and biological warfare agents. This involves measuring a change in either the pH, redox potential or changes in the ionic concentration of an elemental species, which occur during a biorecognition event. A common strategy that has been employed is the use of enzymes to catalyze the consumption or production of protons and/or charged elemental species. The approach developed by Purvis and coworkers51 involved the formation of an enzyme labeled (i.e., horseradish peroxidase) immuno-complex at the surface of a polypyrrole-coated gold electrode. The detection of Hepatitis B virus in blood/serum was achieved by a secondary reaction that produced charged products (i.e., changes in the redox state, pH and/or ionic strength), and the potential shift was measured with respect to a silver/silver chloride reference electrode.51 Furthermore, a comparative study was undertaken and the results agreed favourably with the standard ELISA method. Reports suggest that the potentiometric-based immunosensor is rapid (<15 min), stable (i.e., 4 months), reproducible (CV <5% at 0.1 ng ml−1), and sensitive (∼50 fM).51 Others have shown that a potentiometric immunosensor based on a modified platinum electrode can be used to detect Japanese B encephalitis antigen.52 Zhang et al.52 coated the platinum electrode with a colloidal gold nanoparticle/conductive polymer film followed by the immobilization of the Japanese B encephalitis antibody.52 The shift in potential was related to the antigen concentration in solution, and the potentiometric response mechanism was explained in terms of a change in the density of electron charges. Despite the promising analytical results of the potentiometric biosensor, further work still needs to be undertaken in order to clarify the response mechanism in blood.**
Some reports suggest that potentiometric transducers cannot provide the required sensitivity for the detection of antibody–antigen reactions.26 However, a light addressable potentiometric sensor (LAPS) based on a field effect transistor (FET) technology has proved to be highly successful for the immunoassay of various viruses and pathogens. A LAPS device consists of n-type silicon doped with phosphorus and an insulating layer in contact with the aqueous solution. An alternating photocurrent is generated when a light source flashes rapidly, such as a light emitting diode (LED), and the FET is used to detect changes in the potential at the silicon-insulator surface. The group of Lee53,54 developed a LAPS biosensor to detect Newcastle disease virus, and it was shown that the sensor responds linearly to the virus over a wide concentration range. The assay procedure involved a filtration capture step in which the sandwich immuno-complex was passed through a biotin-embedded nitrocellulose membrane and immobilized onto the membrane via the biotin-streptavidin interaction. An enzyme-immunoassay LAPS approach was also used by Uithoven et al.55 who showed that the potentiometric detection platform can rapidly monitor (<15 min) biological warfare (BW) agents in the field. In this process, a BW agent forms an immunocomplex with both a fluorescein-labelled antibody and a biotin-streptavidin-labelled antibody.55 The fluorescein-labelled immunocomplex undergoes a further complexation reaction with an anti-fluorescein urease conjugated antibody, and the enzymatic breakdown of urea causes a change in pH, which is detected potentiometrically.55 More importantly, the biosensor employs an eight-channel instrument, which can assay up to eight BW agents simultaneously.55 Others have used LAPS to identify the virus Venezuelan equine encephalitis in cultured cells.56 In this study, an immunofiltration enzyme assay was used in conjunction with the LAPS device, and a limit of detection of ∼30 ng ml−1 was achieved.56
Impedance spectroscopy is another electrochemical method that has also received some interest for the detection of EIDs. It is well known that during a biorecognition event several physicochemical processes (i.e., changes in the electric double layer and charge transfer) transpire at and near the surface of a biosensor, and by applying a controlled AC electrical stimulus over a selected range of frequencies the variations in the sensor surface properties (i.e., interfacial capacitance, charge transfer resistance) can be linked to the biochemical changes, and the reader is referred to several fundamental reviews which have been published recently on the topic.57–59 An interesting trend that has started to emerge is the development and application of EIS for the detection of viruses.†† Indeed, the group of Yuan60 used it to detect hepatitis B antigen by observing a change in the electron transfer resistance of a redox probe before and after antigen–antibody interaction. The hepatitis B antibody was immobilized onto a platinum electrode modified with colloidal gold and polyvinyl butyral. EIS measurements were performed at an electrode potential of +220 mV and it was demonstrated that the sensor responds linearly to the hepatitis B antigen over the concentration range of 20–160 ng ml−1, and a limit of detection of 8 ng ml−1 was obtained.60 However, Wang et al.61 were able to improve the detection limit for hepatitis B virus (i.e., 50 ng l−1) and extend the linear analytical range (i.e., 0.5–200 μg l−1) by also using colloidal gold nanoparticles. The immunosensor was fabricated by forming a combined self-assembled monolayer of 4-aminothiophenol and colloidal gold nanoparticles. The impedance-based sensor was optimized by investigating the effect of various surface layers, incubation time, and antigen concentration on the response mechanism. It was revealed that the electron transfer resistance increases with elevated antigen concentrations. Unfortunately, no explanation was provided by the authors on the mechanism responsible for the observed change in impedance.
Many of the studies reported in the literature still employ a redox probe (i.e.,[Fe(CN)6]4−/[Fe(CN)6]3−) to assist with detection. It is evident that EIS is a powerful detection method in clean buffered solutions;57–59 however, it remains to be seen if this method works well for monitoring EIDs in a complex matrix such as blood. Signals arising from non-specific adsorption processes are still a problem and a great deal of effort has been devoted to controlling the surface structure.64 Similarly, a number of challenges still exist in relation to sensitivity, and further work needs to be done in this area before this technology can play a future role in the real-time detection of EID. Recent work by the group of Higson and coworkers65 has demonstrated that the fabrication of a microelectrode array may help improve the sensitivity and it will be interesting to see if this approach works for EIDs.
There has been a great deal of effort to understand the relationship between conductivity and antibody–antigen interaction. Subsequently, conductometric-based biosensors have been used to measure the changes in conductance before and after a biorecognition event, noting that during a biorecognition event the ionic concentration/composition changes and this leads to a change in the solution electrical conductivity. Early reports suggested that the sensitivity of this technique is generally inferior compared to other electrochemical methods; however, recent studies have shown that it is capable of rapidly detecting (<10 min) various food borne pathogens down to very low levels.66,67 The group of Alocilja68,69 showed that this approach can be extended and used to detect bovine viral diarrhoea virus (BVDV) over a wide concentration range. The biosensor was fabricated using a conductive polyaniline label in the sandwich immunoassay scheme, and it was demonstrated that polyaniline improves the sensitivity of the biosensor by forming a conductive molecular bridge between the two electrodes. More importantly, it was shown that the sensor can be used to detect BVDV in artificial blood serum samples.68 Regrettably, the authors have failed to validate their BVDV biosensor against a standard immunoassay method. There is no doubt that the conductometric biosensor platform is a sensitive technique; however, biosensor validation studies still need to be performed before it is routinely used for the detection of EIDs.
Gold and carbon are the most common materials used to carry current/charge during an electrochemical event.43,70 Similarly, there has been considerable interest in the development of biosensors that use conductive polymers (e.g. polyaniline, polypyrrole) as an electrochemical transducer.66,71,72 The growth in the use of conductive polymers has primarily been stimulated by improved response characteristics such as increased sensitivity, stability, and reproducibility. It is well known that the response of polymer-based sensors is greatly dependent on the mode of polymerization, the monomer concentration, and the counterions used during polymerization.51,73 By varying these parameters, it allows the surface properties of the biosensor to be modified, and this feature can be exploited in order to optimize the transduction signal. Likewise, the development of nanomaterials as electrodes for electrochemical-based detectors represents an exciting area of research. The growth in nanomaterials has the potential to revolutionize new developments in EID detection. The ability of carbon nanotubes to promote electron-transfer reactions is well documented.74,75 Electrodes modified with carbon-nanotubes have been recently reported by Wang and coworkers for monitoring various biomolecules.76–78 Although they have shown promising results in DNA- and enzyme-based biosensors,76–79 little work has been done using carbon-nanotubes for monitoring EIDs. Obviously, the next step that needs to be taken is to see how these materials perform for the detection of EIDs, particularly in a complex matrix such as blood.
Electrochemical sensors offer many opportunities for the detection of EIDs. In particular, the amperometric-based biosensor appears to be showing promising signs in terms of sensitivity and selectivity compared to other electrochemical methods. Despite the remarkable sensitivity, rapid response, miniaturization capability, and low cost there are still problems with long-term stability and selectivity in blood. In fact, many of the amperometric biosensor studies involving EID detection have been undertaken in well-defined buffer solutions, and it is not known if this technology is selective enough to detect EIDs directly in blood. Obviously, further work would be required to demonstrate that the electrochemical biosensors do not suffer from biofouling problems. Such problems can be partly addressed by covering the transducer or sensor surface with an appropriate selective film that rejects undesirable components.80 However, this approach may not work very well when analyzing large molecules (i.e., viruses), which need to diffuse through the selective layer to reach the sensor surface. Notwithstanding, the use of disposable screen printed electrodes appears to be a realistic strategy for EID detection.‡‡ Consequently, screen printed electrodes have attracted a great deal of attention recently as a platform in DNA, immuno and enzyme-based biosensors.51,72,77,81–87 The technology is particularly attractive for the mass production of cheap disposable electrodes. On the other hand, this strategy may be somewhat limited to applications that do not require long-term real-time monitoring.
2.2 Optical biosensors
When light (usually monochromatic) is passed through a sample, several things can transpire. The light can either be reflected back or it can be transmitted through the sample. The process that occurs will depend on the wavelength of light, the angle of incidence, the sample composition (i.e., the type and concentration of molecules, etc.) and sample thickness. By exploiting the energy from the electromagnetic spectrum it can be used to provide information about the changes in the local environment surrounding the analyte. Optical biosensors, which are sometimes referred to as ‘optodes’, have received considerable interest on the detection of viruses and/or pathogens.88 Various modes of optical measurement exist (i.e., absorption; reflection; fluorescence; chemiluminescence; and phosphorescence);89 however, biosensors based on surface plasmon resonance and fluorescence principles are the most common and promising methods for EID detection. Similarly, recent progress in fiber-optic technology suggests that optical biosensors may become a powerful tool in the imminent future for the real-time and remote detection of EIDs.90–92 In practice, fiber optics can be coupled with all optical techniques, thus increasing their versatility. A number of articles have appeared in the literature reviewing the use of optical fibers,90–92 and it was concluded that the detection limits of optical fiber-based biosensors are comparable to sophisticated large bench-top instruments. Lee and Thompson93 were able to detect the Newcastle disease (ND) virus down to 10 ng ml−1 using a fiber-optic evanescent wave biosensor. A polyclonal antibody was immobilized covalently onto an aminosilane-coated quartz fiber, and detection was achieved using fluorescein-labelled anti-ND in a sandwich format.93 In its simplest form of measurement, the optical fibers are employed as waveguides to transport light to and from a solution to be analyzed, noting that total internal reflection is the underlying mechanism. One of the major advantages of using optical biosensors in conjunction with optical fibers is that it permits sample analysis to be done over long distances and this has important implications for field monitoring. However, the main drawback apart from being relatively expensive is that optical fibers may suffer from miniaturization problems.§§ Notwithstanding, the application of optical fiber-based nanosensors has become an area of significant interest and various methods have been developed to alleviate the problems arising from miniaturization.94Surface plasmon resonance (SPR) is a form of reflectance spectroscopy that has been widely used in biosensor development. It has been demonstrated that SPR plays a significant role in relation to immunogenicity, food analysis, proteomics, drug discovery, and DNA analysis.95–97 This method is particularly attractive for direct label-free detection and a number of papers have shown that the SPR-based biosensor is a powerful tool for EID monitoring. Studies by the group of Andreu98–100 revealed that SPR is a particularly useful tool for screening the virus of foot-and-mouth disease (FMD). It was used to study the effects of combining multiple amino acid replacements within the sequence of the antigenic GH loop of FMD virus.100 The influence of amino acid substitutions within A15 was also determined, and it was shown that SPR can rapidly screen antigens while simultaneously providing kinetic data on the antigen–antibody interaction.98 Validation studies revealed that the results are in agreement with the relative antigenicities as determined by competition ELISA.98 By contrast, Gomara et al.101 developed an SPR-based immunosensor, which uses synthetic peptides for the detection of the hepatitis A virus in human serum. More importantly, it is reported that the sensitivity of SPR is comparable to ELISA.101 Recently, Chen et al.102 used a SPR biosensor to study the coronavirus of severe acute respiratory syndrome (SARS). It was shown that SPR was able to verify that the N-terminal deleted proteinase dimer adopts a state different from that of the full-length proteinase dimer.102
SPR is one of very few techniques that are able to provide non-invasive, real-time kinetic data on association and dissociation rates, along with equilibrium binding constants for receptor or ligand systems.¶¶ Recently, Myszka and coworkers103 demonstrated that it is possible to rapidly screen many antibodies from hybridoma culture samples using the same SPR sensor surface. This is an important requirement when high sample throughputs are needed such as establishing the type of infection in a group of sentinel animals. In fact, various workers have shown that a portable hand-held SPR-based biosensor (Spreeta™) can be deployed for continuous monitoring of toxins.104,105 However, some reports suggest that the Spreeta™ technology is not as sensitive as the standard ELISA method.106 One of the challenges that limit the application of SPR for real-time measurements in blood is that the signal is very sensitive to non-specific physical binding on the surface.39 Careful engineering of the surface may in some cases minimize undesirable adsorption processes; however, the long-term stability of the surface layer is likely to fail when in direct contact with blood. While SPR is an interesting and appealing transduction method, one must note that the measuring range is very limited. It appears that the technique struggles to detect biochemical compounds or biomolecules, which have a molecular weight of less than 5000 daltons. In spite of these pitfalls, there seems to be a general push to develop a hand-held SPR device that can be used to directly detect pathogens in the environment. The development of a technology which can be taken into the field and provide real time monitoring is a step in the right direction in tackling the proliferation of EIDs.
Fluorescence measurements are of particular interest in biosensor systems due to their high sensitivity.26 In most fluorescent-based immunoassays, fluorochromes are used to label the biomolecules and generate the fluorescent signal, noting that neither antigens nor antibodies exhibit any fluorescence properties. Recently, Walt and coworkers107 reviewed the impact of fluorescence-based nucleic acid detection and microarrays on the biological sciences. Biosensor arrays employing fluorescence-based detection of biohazards (i.e., viral, bacterial, toxins, etc.) have undergone a revolution in terms of miniaturization and automation.108 The group of Ligler109,110 showed that a fluorescence-based multianalyte immunosensor array can be used to simultaneously detect various microorganisms and toxins. Furthermore, it is reported that the technology exhibits comparable sensitivity to the standard ELISA method.109 The biosensor consists of a patterned array of biological recognition elements (i.e., antibodies, receptors) immobilized on the surface of a planar waveguide, and a fluorescence assay is performed on the patterned surface, which yields an array of fluorescent spots. Signal transduction is achieved by using a diode laser for fluorescence excitation and a CCD camera to capture the image. Other workers have shown that fluorescence is an amenable and sensitive approach for the detection of dengue virus. Renard et al.111 developed a fluorescent-based immunosensor, whereas the group of Baeumner112,113 fabricated a nucleic acid microfluidic biosensor that employs liposome signal amplification, and showed that detection down to the picomolar range can be achieved. Despite the improved sensitivity and reduced analysis time, further developmental work still needs to be performed in a complex sample such as blood and serum, if these devices are to be routinely used for the detection of infectious diseases in the field. Another drawback of fluorescence technology is that it requires relatively expensive reagents, and often gives rise to a reaction that is time consuming. Notwithstanding, the capability to perform real time measurements is an area of significant interest for the detection of EIDs.108
2.3 Piezoelectric biosensors
Piezoelectric detection works on the principle that frequency variations of an oscillating quartz crystal correspond to changes in mass as a result of a biochemical reaction/biorecognition event (i.e., antibody–antigen interaction, DNA hybridization).16,114 There are two types of piezoelectric sensors which have been used to detect EIDs: (a) bulk wave or quartz crystal microbalance (QCM) and (b) surface acoustic wave. Some reports advocate that the surface acoustic wave format is more sensitive compared to bulk wave.16 Others have evaluated the detection limit of various biosensor platforms and concluded that the piezoelectric method is inferior compared to electrochemical and optical detectors.114 Notwithstanding, a number of articles have recently appeared in the literature, which use the piezoelectric sensor approach to detect a wide range of viruses. The group of Rickert115 reported on a quartz crystal microbalance for FMD virus. A synthetic peptide with the amino acid sequence 135–154 of the capsid protein VP-1 of FMD virus was covalently linked to the gold surface via a self-assembled monolayer of alkane thiol, and the frequency change was followed with time after exposure to antibody.115 Similarly, Gajendragad and coworkers116 developed a piezoelectric immunosensor for FMD and reported a shelf life of 18 weeks after storing the antibody-coated crystal at room temperature. More importantly, it was demonstrated that the biosensor can detect the FMD virus in clinical samples, and the results compared well with ELISA.116 By contrast, Zhou et al.117 successfully used a piezoelectric-based DNA biosensor for the detection of hepatitis B virus. A nucleic acid probe was immobilized onto the gold electrode via a polyethyleneimine–glutaraldehyde cross-linking process. Despite the sensor responding linearly to hepatitis B over a limited concentration range (i.e., 0.02–0.14 μg ml−1), it was demonstrated that the crystal can be regenerated/activated by exposing the surface to a hydroxide and acid solution followed by an ethanol wash.117 Zuo et al.118 fabricated an immunosensor for severe acute respiratory syndrome (SARS), and detected a frequency shift that was proportional to the antigen concentration in the range 0.6–4 μg ml−1. The piezoelectric crystal was coated with horse polyclonal antibody induced by a SARS-associated coronavirus, and the detection of the antigen was achieved by spraying it in the form of an aerosol via ultrasonic oscillation.118 By contrast, the groups of Tai and Wu119–121 used the QCM biosensor to monitor dengue virus. More importantly, Tai et al.120 used a 15-mer peptide template and developed a novel molecular imprinting approach to fabricate their QCM biosensor. It was demonstrated that molecular imprinting may be a promising method for alleviating problems arising from non-specific interactions.The QCM transduction method is an attractive option for real-time monitoring of EIDs. It is apparent from the aforementioned studies that most workers have overcome many of the challenges, which face this technique. In addition, the sensitivity and reliability of the piezoelectric biosensor is comparable to the conventional ELISA method.122 Despite the promising analytical performance of the piezoelectric sensor further work still needs to be undertaken to evaluate the stability of the sensor surface in biological fluids. Problems such as crystal regeneration, relatively long incubation times, non-specific binding of proteins or other biomaterials, and loss of material coating after washing are well known limitations of this technique,16,26,114 which still require further attention before this biosensor technology platform is used routinely for EID detection.||||
3 Biosensor outlook
Biosensors have become an integral aspect of modern life, as they have allowed us to monitor our environment and surroundings, so that we can control and manipulate it in a way that ensures our survival. What does the future hold for biosensors for the detection of EIDs? Before we can answer this question, it is important to ask what is the rate of growth of activity involving biosensors? The number of scientific publications published and patents issued worldwide each year gives a general guide of the growth rate. Between 1984 and 1990, there were approximately 300 scientific publications and 227 patents on biosensors. From 1991 to 1997, roughly 3000 articles were published and over 376 patents were filed. A survey of the period 1998 to 2004 has revealed that well over 6000 articles have been published and about 1100 patents have been issued and/or are pending. The number of papers published and patents issued per year is an important indicator of research activity, and the present growth rate suggests that the future looks very bright indeed. Similarly, there are some indications that biosensors may play an important role in the biosecurity and/or military sectors in the near future..15*** The future research outlook for EID detection using biosensors looks positive, despite reports suggesting that very little market growth or progress has occurred over the past few years.124In spite of all the research, there is a long way to go before biosensors can be used to detect EIDs in real-time. Biofouling of the sensor membrane is still a major obstacle that has limited the widespread application of biosensors.125 It has been shown that the accurate long-term usage of implanted sensors is dictated by fibrosis formation that develops around the sensor and subsequently inhibits the influx of analyte to the detector. If a biosensor is to be employed for real-time monitoring of EIDs in blood, serum or other biological fluids, it must be able to withstand the rigours of long-term exposure. A number of methods have been developed to overcome biofouling problems.126 None of these approaches appear to completely eliminate membrane biofouling, although there are some promising signs in prolonging sensor functionality. Recent efforts have been directed towards developing new ways of improving the biocompatibility of biosensors. Wang et al.80 demonstrated that depositing heparin onto the surface of an amperometric biosensor can significantly improve sensor biocompatibility. Work by Ward et al.127 has shown that vascularization of the foreign body capsule which surrounds a subcutaneous biosensor improves the life of the sensor. Polymers have also received a lot of attention in relation to chemical modification of electrode and sensor surfaces. Brown and Lowry128 examined various Nafion® coating procedures, and demonstrated that ascorbic and uric acid interferences can be removed during in vivo measurements in the brain. Recently, Higson and coworkers129 have shown that biosensors coated with poly(ethylene glycol)-calix[4]resorcinarenetetrathiol may play an important role in suppressing electrode biofouling and/or passivation problems. By contrast, Meyerhoff and coworkers130–133 have tackled the biocompatibility problem by incorporating nitric oxide (NO) releasing polymers [i.e., polyurethane, poly(vinyl chloride) and polydimethylsiloxane doped with diazeniumdiolate functional groups] within the sensor membrane that prevent thrombogenesis. Despite extensive research efforts, the use of biosensors for real-time detection of EIDs has not gained widespread clinical acceptance.
Bioreceptor stability and activity are an important criterion for the development of a robust and a long-lasting biosensor. Irrespective of the type of biosensor, immobilization of the bioreceptor onto a solid support plays a major role in determining the overall performance of the device. For a biosensor to be successful, it is somewhat necessary that the bioreceptor remains attached irreversibly to the transducer.††† While self-assembled monolayers formed on macroscopic gold surfaces have been studied in depth,37,38,134 it is not known if this approach generates a surface that allows long-term measurements of EIDs. However, recent work has shown that enzymes can be attached to porous silicon using an organic linker, and still retain their biomolecular activity.135,136 Others have shown that scanning probe nanolithography can be used in conjunction with chemoselective protein-to-surface linkers to create templates for fabricating virus arrays.137 If a biosensor is to be developed for long-term, real-time measurements of EID in a biological fluid/environmental sample then the surface of the sensor must be durable and selective for the target analyte. Another challenge facing immunosensors is that they are not completely reversible, so that only a single immunoassay can be performed. This can be a pitfall for applications that may require real-time monitoring; however, a great deal of effort has been directed towards the development of renewable antibody surfaces103 and ‘reagentless’ or ‘label-free’ sensing.21,62
The literature contains a vast description of a wide range of biosensors that exploit enzymes, nucleic acids, and antibodies in conjunction with electrochemical, optical, and piezoelectric transducers. An important trend in biosensors is the development of miniaturized devices such as the lab-on-a-chip.12‡‡‡ Equally, the development of microscale separation devices, particularly micromachined capillary electrophoresis (CE) chips, has witnessed an explosive growth in recent years.138 Such miniaturized devices provide the capability to shrink conventional ‘bench-top’ separation systems with major advantages of speed, cost, portability, and solvent and/or sample consumption. Subsequently, the integration of micro-electrochemical systems for local detection, implantable or portable devices, and measurement in small volumes opens the way for applications in EID monitoring. Advancements in nanotechnology will further assist with the development of miniaturized and/or hand-held biosensors. In fact, optical fiber-based nanosensors depend on the construction of nanometre-sized optical fibers. Consequently, the application of atomic force microscopy is becoming an important research tool for characterizing ligand-receptor interactions, and this technique will certainly foster the development of innovative nanosensors. Similarly, the increasing growth of the telecommunications industry will support the development of new and improved optical methods. In fact, fiber optic sensors are beginning to generate intense interest for in vivo monitoring, and this paves the way for new and important advances in real-time clinical monitoring of EIDs.
The type of device selected will depend on a number of issues such as: What is the sensitivity and/or detection limit of the biosensor? Although, many of the biosensors described in this review can be used to detect EIDs at very low levels, the sensor sensitivity will also be system dependent (i.e., the size, type, and structure of the antigen) rather than just transducer dependent. What is the cost? Is the biosensor versatile? Is it preferred to monitor continuously, or to collect samples and periodically test them? If the system stays in the field for weeks or months, then maintaining the biological component of the biosensor can be rather difficult. In fact, most existing biosensors (i.e., immunosensors, DNA biosensors) cannot withstand the rigours of long-term implantation. Another problem that needs to be addressed is the biosensor response time. There are relatively few biosensors around that can accurately assay a biological sample in less than several minutes. Most devices have an analysis time that range from 15 minutes to several hours. Clearly, a biosensor device that is rapid, robust, and durable will be highly desirable for field applications. Very few of the biosensors investigated in this review satisfy the aforementioned criteria; however, much more validation work still needs to be done before they are accepted and embraced by the medical and/or scientific community for real-time monitoring. Detailed data and evaluation protocols on performance characteristics of biosensors, as well as comparisons with established methods, are missing in most cases.
4 Conclusions
There is no doubt that biosensors have recently undergone significant improvements in terms of their achievable selectivity and detection limits. Although there are many hand-held or field biosensors available, most of these technologies have not been fully realized for the detection of EIDs. The development of biosensors for the detection of infectious diseases is still in its infancy. Many technical and/or scientific challenges still exist irrespective of the type of biosensor platform. It has been shown in this review that the biosensor response depends on many experimental factors such as antigen/antibody loading, surface preparation/immobilization conditions, incubation time, temperature, type of biological fluid, etc. These factors determine the success of a biosensor for a particular EID and therefore need to be carefully controlled/optimized if reliable measurements are going to be made. In addition, problems such as matrix interference, specific adsorption of blood components onto the sensor surface and drift are common for all biosensor devices. Currently, most biosensors show excellent results in pristine laboratory samples; however, very few are sufficiently robust to be employed directly for the real-time detection of EID in biological samples (i.e., blood). Similarly, a majority of the biosensors developed for EID detection have so far undergone limited clinical evaluation, and further work needs to be done before they make their way into the market place. The key issue that needs to be addressed in the future is the increasing demand for sensor biocompatibility that will allow biomolecules to be monitored in real time. A great deal of research has been undertaken on the development of new materials (i.e., carbon nanotubes, polymers, etc.), which may overcome some of the challenges and limitations facing existing biosensor technologies. Designers of biosensor devices need to be aware of the specific needs for a particular analytical device. It is a must for the designer to look at the special demands of clinical chemistry prior to making new developments. The significant technological advances made over the past decade are certain to facilitate the application of biosensors for the detection of EIDs.Acknowledgements
The financial support of the Australian Biosecurity Cooperative Research Centre (AB-CRC) for Emerging Infectious Disease, and the Australian Research Council (ARC) are gratefully acknowledged. One of us (BP) is particularly grateful to Curtin University for a Curtin Strategic Research Grant.References
- S. Binder, A. M. Levitt, J. J. Sacks and J. M. Hughes, Science, 1999, 284(5418), 1311 CrossRef CAS.
- H. Field, P. Young, J. M. Yob, J. Mills, L. Hall and J. Mackenzie, Microb. Infect., 2001, 3(4), 307 Search PubMed.
- G. Crameri, L. F. Wang, C. Morrissy, J. White and B. T. Eaton, J. Virol. Methods, 2002, 99, 41 CrossRef CAS.
- B. J. Shiell, D. R. Gardner, G. Crameri, B. T. Eaton and W. P. Michalski, Virus Res., 2003, 92, 55 CrossRef CAS.
- W. J. Bellini, B. H. Harcourt, N. Bowden and P. A. Rota, J. Neurovirol., 2005, 11(5), 481 CrossRef.
- F. R. R. Teles, D. M. F. Prazeres and J. L. Lirna-Filho, Rev. Med. Virol., 2005, 15(5), 287 CrossRef CAS.
- D. L. Wu, C. C. Tu, C. Xin, H. Xuan, Q. W. Meng, Y. G. Liu, Y. D. Yu, Y. T. Guan, Y. Jiang, X. N. Yin, G. Crameri, M. P. Wang, C. W. Li, S. W. Liu, M. Liao, L. Feng, H. Xiang, J. F. Sun, J. D. Chen, Y. W. Sun, S. L. Gu, N. H. Liu, D. X. Fu, B. T. Eaton, L. F. Wang and X. G. Kong, J. Virol., 2005, 79(4), 2620 CrossRef CAS.
- http://www1.abcrc.org.au/pages/programs.aspx?ProgramID=26., accessed January 2006.
- S. T. Nichol, J. Arikawa and Y. Kawaoka, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(23), 12411 CrossRef CAS.
- A. Fenton and A. B. Pedersen, Emerging Infect. Dis., 2005, 11(12), 1815 Search PubMed.
- P. B. Luppa, L. J. Sokoll and D. W. Chan, Clin. Chim. Acta, 2001, 314(1–2), 1 CrossRef CAS.
- P. D'Orazio, Clin. Chim. Acta, 2003, 334, 41 CrossRef CAS.
- S. Andreescu and O. A. Sadik, Pure Appl. Chem., 2004, 76(4), 861 CrossRef CAS.
- M. C. Hennion and D. Barcelo, Anal. Chim. Acta, 1998, 362, 3 CrossRef CAS.
- E. C. Alocilja and S. M. Radke, Biosens. Bioelectron., 2003, 18, 841 CrossRef CAS.
- P. Leonard, S. Hearty, J. Brennan, L. Dunne, J. Quinn, T. Chakraborty and R. O'Kennedy, Enzyme Microb. Technol., 2003, 32, 3 CrossRef CAS.
- S. Rodriguez-Mozaz, M. P. Marco, M. J. L. de Alda and D. Barcelo, Pure Appl. Chem., 2004, 76(4), 723 CrossRef CAS.
- S. Rodriguez-Mozaz, M. J. L. de Alda, M. P. Marco and D. Barcelo, Talanta, 2005, 65, 291 CrossRef CAS.
- I. E. Tothill, Comput. Electron. Agric., 2001, 30, 205 CrossRef.
- M. N. Velasco-Garcia and T. Mottram, Biosyst. Eng., 2003, 84(1), 1 Search PubMed.
- M. A. Cooper, Anal. Bioanal. Chem., 2003, 377(5), 834 CrossRef CAS.
- S. Grant, F. Davis, J. A. Pritchard, K. A. Law, S. P. J. Higson and T. D. Gibson, Anal. Chim. Acta, 2003, 495, 21 CrossRef CAS.
- S. Q. Hu, J. W. Xie, Q. H. Xu, K. T. Rong, G. L. Shen and R. Q. Yu, Talanta, 2003, 61(6), 769–777 CrossRef CAS.
- D. R. Thevenot, K. Toth, R. A. Durst and G. S. Wilson, Pure Appl. Chem., 1999, 71(12), 2333 CrossRef CAS.
- B. Hock, Anal. Chim. Acta, 1997, 347, 177 CrossRef CAS.
- R. I. Stefan, J. F. van Staden and H. Y. Aboul-Enein, Fresenius' J. Anal. Chem., 2000, 366, 659 CrossRef CAS.
- A. Warsinke, A. Benkert and F. W. Scheller, Fresenius' J. Anal. Chem., 2000, 366, 622 CrossRef CAS.
- A. H. Peruski and L. F. Peruski, Clin. Diagnost. Lab. Immunol., 2003, 10(4), 506 Search PubMed.
- D. S. Hage, Anal. Chem., 1995, 67(12), 455R CAS.
- J. Wang, G. Rivas, X. Cai, E. Palecek, P. Nielsen, H. Shiraishi, N. Dontha, D. Luo, C. Parrado, M. Chicharro, P. A. M. Farias, F. S. Valera, D. H. Grant, M. Ozsoz and M. N. Flair, Anal. Chim. Acta, 1997, 347, 1 CrossRef CAS.
- M. Yang, M. E. McGovern and M. Thompson, Anal. Chim. Acta, 1997, 346, 259 CrossRef CAS.
- T. Vo-Dinh and B. Cullum, Fresenius' J. Anal. Chem., 2000, 366, 540 CrossRef CAS.
- M. Fojta, Electroanalysis, 2002, 14(21), 1449 CrossRef CAS.
- J. J. Gooding, Electroanalysis, 2002, 14(17), 1149 CrossRef CAS.
- E. Palecek, Talanta, 2002, 56, 809 CrossRef CAS.
- M. Campas and I. Katakis, Trends Anal. Chem., 2004, 23(1), 49 CrossRef CAS.
- N. K. Chaki and K. Vijayamohanan, Biosens. Bioelectron., 2002, 17, 1 CrossRef CAS.
- J. J. Gooding, F. Mearns, W. Yang and J. Liu, Electroanalysis, 2003, 15(2), 81 CrossRef CAS.
- D. Meadows, Adv. Drug Delivery Rev., 1996, 21(3), 179 CrossRef CAS.
- J. Shah and E. Wilkins, Electroanalysis, 2003, 15(3), 157 CrossRef.
- M. Diaz-Gonzalez, M. B. Gonzalez-Garcia and A. Costa-Garcia, Electroanalysis, 2005, 17(21), 1901 CrossRef CAS.
- G. G. Guilbault, M. Pravda, M. Kreuzer and C. K. O'Sullivan, Anal. Lett., 2004, 37(8), 1481 CrossRef CAS.
- K. Habermuller, M. Mosbach and W. Schuhmann, Fresenius' J. Anal. Chem., 2000, 366, 560 CrossRef.
- A. P. T. Turner, B. Chen and S.A. Piletsky, Clin. Chem., 1999, 45(9), 1596 CAS.
- J. L. Gong, F. C. Gong, G. M. Zeng, G. L. Shen and R. Q. Yu, Anal. Lett., 2003, 36(2), 287 CrossRef CAS.
- K. Brainina, A. Kozitsina and J. Beikin, Anal. Bioanal. Chem., 2003, 376(4), 481 CAS.
- R. Yuan, L. Y. Zhang, Q. F. Li, Y. Q. Chai and S. R. Cao, Anal. Chim. Acta, 2005, 531(1), 1 CrossRef CAS.
- B. Meric, K. Kerman, D. Ozkan, P. Kara, S. Erensoy, U. S. Akarca, M. Mascini and M. Ozsoz, Talanta, 2002, 56, 837 CrossRef CAS.
- H. X. Ju, Y. K. Ye, J. H. Zhao and Y. L. Zhu, Anal. Biochem., 2003, 313, 255 CrossRef CAS.
- E. Bakker, D. Diamond, A. Lewenstam and E. Pretsch, Anal. Chim. Acta, 1999, 393(1–3), 11 CrossRef CAS.
- D. Purvis, O. Leonardova, D. Farmakovsky and V. Cherkasov, Biosens. Bioelectron., 2003, 18(11), 1385 CrossRef CAS.
- L. Zhang, R. Yuan, X. Huang, Y. Chai and S. Cao, Electrochem. Commun., 2004, 6, 1222 CrossRef CAS.
- W. E. Lee, G. H. Thompson, J. G. Hall, E. R. Fulton and J. P. Wong, J. Immunol. Methods, 1993, 166(1), 123 CrossRef CAS.
- W. E. Lee, G. H. Thompson, J. G. Hall and D. E. Bader, Biosens. Bioelectron., 2000, 14(10–11), 795 CrossRef CAS.
- K. A. Uithoven, J. C. Schmidt and M. E. Ballman, Biosens. Bioelectron., 2000, 14, 761 CrossRef CAS.
- W. G. Hu, G. H. Thompson, A. Z. Alvi, L. P. Nagata, M. R. Suresh and E. R. Fulton, J. Immunol. Methods, 2004, 289(1–2), 27 CrossRef CAS.
- E. Katz and I. Willner, Electroanalysis, 2003, 15(11), 913 CrossRef CAS.
- J. G. Guan, Y. Q. Miao and Q. J. Zhang, J. Biosci. Bioeng., 2004, 97(4), 219 Search PubMed.
- I. O. K'Owino and O. A. Sadik, Electroanalysis, 2005, 17(23), 2101 CrossRef.
- D. P. Tang, R. Yuan, Y. Q. Chai, J. Y. Dai, X. Zhong and Y. Liu, Bioelectrochemistry, 2004, 65(1), 15 CrossRef CAS.
- M. Wang, L. Wang, G. Wang, X. Ji, Y. Bai, T. Li, S. Gong and J. Li, Biosens. Bioelectron., 2004, 19, 575 CrossRef CAS.
- G. Lillie, P. Payne and P. Vadgama, Sens. Actuators, B, 2001, 78, 249 CrossRef.
- G. Farace, G. Lillie, T. Hianik, P. Payne and P. Vadgama, Bioelectrochemistry, 2002, 55, 1 CrossRef CAS.
- S. J. Ding, B. W. Chang, C. C. Wu, M. F. Lai and H. C. Chang, Anal. Chim. Acta, 2005, 554(1–2), 43 CrossRef CAS.
- A. C. Barton, S. D. Collyer, F. Davis, D. D. Gornall, K. A. Law, E. C. D. Lawrence, D. W. Mills, S. Myler, J. A. Pritchard, M. Thompson and S. P. J. Higson, Biosens. Bioelectron., 2004, 20, 328 CrossRef CAS.
- Z. Muhammad-Tahir and E. C. Alocilja, Biosens. Bioelectron., 2003, 18, 813 CrossRef CAS.
- Z. Muhammad-Tahir and E. C. Alocilja, Biosyst. Eng., 2004, 88(2), 145 Search PubMed.
- Z. Muhammad-Tahir, E. C. Alocilja and D. L. Grooms, IEEE Sens. J., 2005, 5(4), 757 Search PubMed.
- Z. Muhammad-Tahir, E. C. Alocilja and D. L. Grooms, Biosens. Bioelectron., 2005, 20(8), 1690 CrossRef.
- F. Lucarelli, G. Marrazza, A. P. F. Turner and M. Mascini, Biosens. Bioelectron., 2004, 19, 515 CrossRef CAS.
- A. Sargent, T. Loi, S. Gal and O. A. Sadik, J. Electroanal. Chem., 1999, 470, 144 CrossRef CAS.
- A. J. Killard, L. Micheli, K. Grennan, M. Franek, V. Kolar, D. Moscone, I. Palchetti and M. R. Smyth, Anal. Chim. Acta, 2001, 427, 173 CrossRef CAS.
- J. N. Barisci, C. Conn and G. G. Wallace, Trends Polym. Sci., 1996, 4(9), 307 CAS.
- C. E. Banks and R. G. Compton, Analyst, 2006, 131(1), 15 RSC.
- J. J. Gooding, Electrochim. Acta, 2005, 50, 3049 CrossRef CAS.
- R. P. Deo and J. Wang, Electrochem. Commun., 2004, 6, 284 CrossRef CAS.
- Y. Lin, F. Lu and J. Wang, Electroanalysis, 2004, 16(1–2), 145 CrossRef CAS.
- J. Wang, G. Liu and R. M. Jan, J. Am. Chem. Soc., 2004, 126, 3010 CrossRef CAS.
- M. Guo, J. Chen, L. Nie and S. Yao, Electrochim. Acta, 2004, 49(16), 2637 CrossRef CAS.
- J. Wang, L. Chen, S. B. Hocevar and B. Ogorevc, Analyst, 2000, 125, 1431 RSC.
- J. C. Schmidt, Field Anal. Chem. Technol., 1998, 2(6), 351 Search PubMed.
- S. Laschi, M. Franek and M. Mascini, Electroanalysis, 2000, 12(16), 1293 CrossRef CAS.
- G. Chiti, G. Marrazza and M. Mascini, Anal. Chim. Acta, 2001, 427, 155 CrossRef CAS.
- F. Lucarelli, I. Palchetti, G. Marrazza and M. Mascini, Talanta, 2002, 56, 949 CrossRef CAS.
- F. Lucarelli, A. Kicela, I. Palchetti, G. Marrazza and M. Mascini, Bioelectrochemistry, 2002, 58, 113 CrossRef CAS.
- F. Lucarelli, G. Marrazza, I. Palchetti, S. Cesaretti and M. Mascini, Anal. Chim. Acta, 2002, 469, 93 CrossRef CAS.
- S. Susmel, G. G. Guilbault and C. K. O'Sullivan, Biosens. Bioelectron., 2003, 18, 881 CrossRef CAS.
- G. A. Canziani, W. Zhang, D. Cines, A. Rux, S. Willis, G. Cohen, R. Eisenberg and I. Chaiken, Methods, 1999, 19, 253 Search PubMed.
- A. F. Collings and F. Caruso, Rep. Prog. Phys., 1997, 60(11), 1397 CrossRef CAS.
- E. D. S. Kerslake and C. G. Wilson, Adv. Drug Delivery Rev., 1996, 21, 205 CrossRef CAS.
- D. Uttamchandani and S. McCulloch, Adv. Drug Delivery Rev., 1996, 21, 239 CrossRef CAS.
- D. J. Monk and D. R. Walt, Anal. Bioanal. Chem., 2004, 379, 931 CrossRef CAS.
- W. E. Lee and H. G. Thompson, Can. J. Chem., 1996, 74(5), 707 CrossRef CAS.
- B. M. Cullum and T. Vo-Dinh, Trends Biotechnol., 2000, 18(9), 388 CrossRef CAS.
- R. Karlsson, J. Mol. Recognit., 2004, 17, 151 CrossRef CAS.
- N. Bianchi, C. Rutigliano, M. Tomassetti, G. Feriotto, F. Zorzato and R. Gambari, Clin. Diagnost. Virol., 1997, 8, 199 Search PubMed.
- K. Kukanskis, J. Elkind, J. Melendez, T. Murphy, G. Miller and H. Garner, Anal. Biochem., 1999, 274, 7 CrossRef CAS.
- P. Gomes, E. Giralt and D. Andreu, Vaccine, 1999, 18(3–4), 362 CrossRef CAS.
- P. Gomes, E. Giralt and D. Andreu, Mol. Immunol., 2000, 37(16), 975 CAS.
- P. Gomes, E. Giralt, W. Ochoa, N. Verdaguer and D. Andreu, J. Pept. Res., 2002, 59(5), 221 CrossRef CAS.
- M. J. Gomara, G. Ercilla, M. A. Alsina and I. Haro, J. Immunol. Methods, 2000, 246, 13 CrossRef CAS.
- S. Chen, L. L. Chen, J. Z. Tan, J. Chen, L. Du, T. Sun, J. H. Shen, K. X. Chen, H. L. Jiang and X. Shen, J. Biol. Chem., 2005, 280(1), 164 CAS.
- G. A. Canziani, S. Klakamp and D. G. Myszka, Anal. Biochem., 2004, 325(2), 301 CrossRef CAS.
- A. N. Naimushin, S. D. Soelberg, D. K. Nguyen, L. Dunlap, D. Bartholomew, J. Elkind, J. Melendez and C. E. Furlong, Biosens. Bioelectron., 2002, 17, 573 CrossRef CAS.
- T. M. Chinowsly, J. G. Quinn, D. U. Bartholomew, R. Kaiser and J. L. Elkind, Sens. Actuators, B, 2003, 91, 266 CrossRef.
- B. D. Spangler, E. A. Wilkinson, J. T. Murphy and B. J. Tyler, Anal. Chim. Acta, 2001, 444, 149 CrossRef CAS.
- J. R. Epstein, I. Biran and D. R. Walt, Anal. Chim. Acta, 2002, 469, 3 CrossRef CAS.
- K. E. Sapsford, Y. S. Shubin, J. B. Delehanty, J. P. Golden, C. R. Taitt, L. C. Shriver-Lake and F. S. Ligler, J. Appl. Microbiol., 2004, 96(1), 47 CrossRef CAS.
- C. A. Rowe-Taitt, J. P. Golden, M. J. Feldstein, J. J. Cras, K. E. Hoffman and F. S. Ligler, Biosens. Bioelectron., 2000, 14, 785 CrossRef CAS.
- C. A. Rowe-Taitt, G. P. Anderson, B. M. Lingerfelt, M. J. Feldstein and F. S. Ligler, Anal. Chem., 2002, 74, 6114 CrossRef CAS.
- M. Renard, L. Belkadi and H. Bedouelle, J. Mol. Biol., 2003, 326(1), 167 CrossRef CAS.
- S. Kwakye and A. Baeumner, Anal. Bioanal. Chem., 2003, 376(7), 1062 CAS.
- N. V. Zaytseva, V. N. Goral, R. A. Montagna and A. J. Baeumner, Lab Chip, 2005, 5(8), 805 RSC.
- D. Ivnitski, I. Abdel-Hamid, P. Atanasov and E. Wilkins, Biosens. Bioelectron., 1999, 14, 599 CrossRef CAS.
- J. Rickert, T. Weiss, W. Kraas, G. Jung and W. Gopel, Biosens. Bioelectron., 1996, 11(6–7), 591 CrossRef CAS.
- M. R. Gajendragad, K. N. Y. Kamath, P. Y. Anil, K. Prabhudas and C. Natarajan, Vet. Microbiol., 2001, 78(4), 319 CrossRef CAS.
- X. Zhou, L. Liu, M. Hu, L. Wang and J. Hu, J. Pharm. Biomed. Anal., 2002, 27, 341 CrossRef CAS.
- B. L. Zuo, S. M. Li, Z. Guo, J. F. Zhang and C. Z. Chen, Anal. Chem., 2004, 76(13), 3536 CrossRef CAS.
- C. C. Su, T. Z. Wu, L. K. Chen, H. H. Yang and D. F. Tai, Anal. Chim. Acta, 2003, 479(2), 117 CrossRef CAS.
- D. F. Tai, C. Y. Lin, T. Z. Wu and L. K. Chen, Anal. Chem., 2005, 77(16), 5140 CrossRef CAS.
- T. Z. Wu, C. C. Su, L. K. Chen, H. H. Yang, D. F. Tai and K. C. Peng, Biosens. Bioelectron., 2005, 21(5), 689 CrossRef CAS.
- Y. Liu, W. Zhang, X. Yu, H. Zhang, R. Zhao, D. H. Shangguan, Y. Li, B. Shen and G. Liu, Sens. Actuators, B, 2004, 99, 416 CrossRef.
- K. R. Rogers and M. Mascini, Field Anal. Chem. Technol., 1998, 2(6), 317 Search PubMed.
- U.S. & Worldwide: Biosensor Market, R&D, Applications and Commercial Implication, Fuji-Keizai USA, Inc., New York, 2004 Search PubMed.
- N. Wisniewski, F. Moussy and W. M. Reichert, Fresenius' J. Anal. Chem., 2000, 366, 611 CrossRef CAS.
- N. Wisniewski and M. Reichert, Colloids Surf., B, 2000, 18, 197 CrossRef CAS.
- K. W. Ward, M. J. Quinn, M. D. Wood, K. L. Tiekotter, S. Pidikiti and J. A. Gallagher, Biosens. Bioelectron., 2003, 19, 155 CrossRef.
- F. O. Brown and J. P. Lowry, Analyst, 2003, 128, 700 RSC.
- S. Collyer, F. Davis, A. Lucke, C. J. M. Stirling and S. P. J. Higson, Electroanalysis, 2004, 16(14), 275 CrossRef CAS.
- M. C. Frost, S. M. Rudich, H. Zhang, M. A. Maraschio and M. E. Meyerhoff, Anal. Chem., 2002, 74, 5942 CrossRef CAS.
- M. H. Schoenfisch, H. Zhang, M. C. Frost and M. E. Meyerhoff, Anal. Chem., 2002, 74, 5937 CrossRef CAS.
- M. M. Batchelor, S. L. Reoma, P. S. Fleser, V. K. Nuthakki, R. E. Callahan, C. J. Shanley, J. K. Politis, J. Elmore, S. I. Merz and M. E. Meyerhoff, J. Med. Chem., 2003, 46(24), 5153 CrossRef CAS.
- M. C. Frost, M. M. Batchelor, Y. Lee, H. Zhang, Y. Kang, B. Oh, G. S. Wilson, R. Gifford, S. M. Rudich and M. E. Meyerhoff, Microchem. J., 2003, 74, 277 CAS.
- T. Peng, Q. Cheng and Q. Cheng, Electroanalysis, 2002, 14(6), 455 CrossRef CAS.
- B. R. Hart, S. E. Letant, S. R. Kane, M. Z. Hadi, S. J. Shields and J. G. Reynolds, Chem. Commun., 2003, 3, 322 RSC.
- S. E. Letant, B. R. Hart, S. R. Kane, M. Z. Hadi, S. J. Shields and J. G. Reynolds, Adv. Mater., 2004, 16(8), 689 CrossRef CAS.
- C. L. Cheung, J. A. Camarero, B. W. Woods, T. Lin, J. E. Johnson and J. De Yoreo, J. Am. Chem. Soc., 2003, 125(23), 6848 CrossRef CAS.
- J. Fischer, J. Barek and J. Wang, Electroanalysis, 2006, 18(2), 195 CrossRef CAS.
Footnotes |
| † Present address: CSIRO Petroleum, ARRC, PO Box 1130, Bentley, WA, 6102, Australia. |
| ‡ Present address: Alcoa, Perth, WA, Australia. |
| § According to IUPAC recommendations, a biosensor is “a self-contained integrated receptor–transducer device, which is capable of providing selective quantitative or semi-quantitative analytical information using a biological recognition element”.24 |
| ¶ One of the major challenges with detecting EIDs is the availability of specific antibodies, and a great deal of effort has been undertaken in generating acceptable receptors/reagents. |
| || Controlling the surface chemistry and coverage is paramount in ensuring high reactivity, stability, orientation and accessibility as well as minimizing non-specific binding processes. |
| ** There is no doubt that the potentiometric methods are highly sensitive to changes in the chemistry and ionic properties of blood and this could severely limit their application for real-time monitoring of EIDs. |
| †† An important feature of EIS is that it is able to provide ‘reagentless’ or ‘label-free’ sensing,62,63 and this makes it highly attractive for real-time monitoring. |
| ‡‡ It is well known that ‘memory effects’ and/or ‘membrane fouling’, which is sometimes observed with electrochemical-based biosensors, can be alleviated when using disposable screen printed electrode sensors. |
| §§ Unlike electrochemical-based transducers that are concentration sensitive detectors and therefore perform better when miniaturized (i.e., microelectrodes), diminishing the sample volume with mass sensitive optical biosensors reduces the concomitant signal intensity or sensitivity. |
| ¶¶ SPR has the advantage that it can measure complex formation without labelling the reactants, and it can analyse samples from crude preparations. |
| |||| Controlling the physical dimensions and properties of the piezoelectric material is another problem that also needs to be addressed. Some reports suggest that each crystal needs to be calibrated separately, since its frequency depends on the crystal geometry and the immobilization technique.123 |
| *** Since September 11th, 2001, the detection of biohazards in the environment has become an issue of great concern.18 The war on terrorism is driving the need for biosensors to rapidly detect biowarfare agents, and a great deal of research has been directed towards this area. |
| ††† Immobilizing the bioreceptor onto selected sites while still retaining the activity and the binding power of the biological moiety is a significant challenge in biosensor research. |
| ‡‡‡ Miniaturization saves large amounts of expensive or rare biological material, and thus lowers the development and production costs. |
| This journal is © The Royal Society of Chemistry 2006 |
