Open Access Article
Open Access ArticleEx vivo spectroscopic characterisation of the biological activity of pancreatic cyst fluid†
Outmane
Bouzerda
 ab,
Laura E.
Kane
ab,
Laura E.
Kane
 c,
Gregory S.
Mellotte
d,
Barbara M.
Ryan
d,
Stephen G.
Maher
c,
Olivier
Piot‡
c,
Gregory S.
Mellotte
d,
Barbara M.
Ryan
d,
Stephen G.
Maher
c,
Olivier
Piot‡
 ae and
Aidan D.
Meade‡
ae and
Aidan D.
Meade‡
 *b
*b
aBioSpectroscopie Translationnelle (BioSpecT) EA7506, Université de Reims Champagne-Ardenne, Reims, France
bSchool of Physics & Clinical & Optometric Sciences, Technological University Dublin, Kevin Street, Dublin D08 NF82, Ireland. E-mail: aidan.meade@tudublin.ie
cDepartment of Surgery, Trinity St James's Cancer Institute, Trinity Translational Medicine Institute, Trinity College Dublin, Dublin D08 W9RT, Ireland
dDepartment of Gastroenterology, Trinity College Dublin, Tallaght University Hospital, Dublin 24, Ireland
ePlateforme d'Imagerie Cellulaire ou Tissulaire (PICT), Université de Reims Champagne-Ardenne, Reims, France
First published on 13th June 2025
Abstract
Pancreatic cystic lesions (PCLs) are fluid-filled sacs often identified incidentally during abdominal imaging for unrelated pancreatic indications. While most PCLs are non-cancerous (benign) with no potential progression to pancreatic cancer (PC), some PCLs may undergo malignant transformation and are therefore more likely to progress into invasive cancer. The main challenge lies in separating PCLs that are entirely benign from those that are premalignant, as imaging tools and clinical guidelines remain inadequate. Understanding how these PCLs arise and develop is crucial for the proper management of patients with these lesions. Well-established PDAC cell lines and pancreatic cyst fluid (PCF), the fluid contained within PCLs, represent an invaluable tool for investigating the biological processes driving PCL formation and their eventual malignant transformation. Such information could greatly improve patient risk stratification, as well as avoid unnecessary follow-up and treatment for those who do not need it. Novel approaches based on vibrational spectroscopy may represent a useful adjunct to clinical tests of this sample type, offering a mode for objective assessment as well as providing potential in vivo applications. In the present ex vivo investigation, the first of its kind in this field, a focus was brought to developing a cell line-based model with vibrational spectroscopy for discriminating the response of PDAC cell lines to exposure to PCF. We demonstrate that this approach provides a robust assay which may have potential, with further development, to provide models which are predictive of the trajectory of disease progression in precision medicine.
1 Introduction
Due to its poor prognosis, pancreatic cancer (PC) is one of the world's most aggressive cancers, with an incidence rate close to its rate of mortality.1 In 2024, PC had the lowest 5-year survival rate of any malignancy, at just 13% according to the American Cancer Society.2 Indeed, PC is expected to overtake colorectal cancer as the 2nd cause of cancer-related death in next two decades.3 However, its poor survival rate is mainly due to a typically late-stage diagnosis as well as the lack of specific symptoms. The two main forms of PC are pancreatic ductal adenocarcinoma (PDAC) (accounting for 85–90% of disease incidence), and pancreatic neuroendocrine tumor (PNET), which is less common (having an incidence rate less than 5%).4,5Pancreatic cystic lesions (PCLs) are fluid-filled cavities that can be found either on or within the pancreas, and have several subtypes, including pseudocysts (non-neoplastic cysts), intraductal papillary mucinous neoplasms, mucinous cystic neoplasms, serous cystic neoplasms, and other rare cystic lesions.6 Since these PCLs can be either benign or precancerous, they possess variable risk of progressing to PC in any given patient.6,7 The increased identification of PCLs by cross-sectional imaging over the past few years has made their appropriate management of greater importance. Several sets of clinical guidelines are employed globally for the surveillance and management of PCLs, indicating a lack of consensus as to the clinical management of these patients.8 These guidelines differ on several cut-offs and criteria, and have been shown to perform poorly when it comes to risk stratification of patients.8 An accurate risk stratification method is hence essential to correctly identify patients with low-risk PCLs from those at increased risk of PC development and thus guide their clinical management and specify their treatment. Moreover, the diagnostic approaches currently adopted to discriminate between PCLs and assess their risk of malignancy fall short, underscoring an urgent need for further advanced diagnostics modalities.9,10
In the past two decades, vibrational spectroscopy (VS), including Raman and FTIR spectroscopy, when combined with appropriate machine learning and chemometrics approaches, has shown promise for the detection of a variety of cancers such as lung,11 breast,12 prostate,13 and colorectal cancer.14 These non-invasive, sensitive, objective and reproducible techniques allow rapid label-free molecular profiling of biological samples which may be translated into clinically useful diagnostic information.15 Previous work has demonstrated the capability of VS for diagnosis of PC.16 Recent work has coupled Raman spectroscopy (RS) with KNN (K-Nearest Neighbours) and SVM (Support Vector Machine) algorithms to differentiate between tumour repopulating cells and control cells from the PDAC cell line Mia Paca-2 with high accuracy.17 Other research using surface enhanced Raman spectroscopy (SERS) combined with principal component differential function analysis (PC-DFA) investigated a diagnostic model using exosomes derived from normal and PDAC cell lines for the detection of PC.18 Further research has revealed the link between cholesterol esterification (CE) and metastasis in PC using label-free RS,19 where results demonstrated that inhibiting CE is capable of preventing PDAC cells from growing and propagating (metastasis) using a murine model. This research emphasizes the role of RS as a powerful tool to identify biomolecular variations in PC tissues and cell lines.
Szymoński et al., recently reported in two proof-of-concept studies,20,21 that FTIR spectroscopy and Raman hyperspectral imaging (RHM) combined with data science and machine learning approaches including PCA (Principal Component Analysis), HCA (hierarchical cluster analysis), NMF (non-negative matrix factorization), T-SNE (T-distributed stochastic neighbour embedding), and CNN (convolutional neural network), can effectively discriminate between PC subtypes, including PDAC, intraductal papillary mucinous carcinoma, and ampulla of Vater. The main outcomes of these experiments highlighted notable variations in the β-sheet secondary structure composition of proteins, as well as the DNA methylation level among the three PC subtypes. Another study revealed the potential of FTIR hyperspectral imaging to detect distinctive biochemical alterations between disease subtypes, including PDAC and PNET, from healthy and dysplastic tissues.22 Sala et al., also used Attenuated Total Reflection (ATR)-FTIR spectroscopy for detecting PC using a blood-based biopsy.23
The primary goal of this study was to determine whether the exposure of reporter PDAC cell lines of differential phenotypes to pancreatic cyst fluid (PCF) produces biochemical alterations which are observable spectroscopically ex vivo. A secondary goal was to compare the ability of FTIR and Raman spectroscopy, together with linear and non-linear dimensionality reduction approaches (respectively, PCA and UMAP (Uniform Manifold Approximation and Projection)) to discriminate the response across PDAC cell phenotypes when exposed to PCF.
Our in vitro results provide a first demonstration of the potential of spectroscopic technologies as complementary diagnostic tools, opening up a new path for prediction of the trajectory of PC using this approach.
2 Materials and methods
2.1 Cell culture and preparation to PCF treatment
Pancreatic cell lines HPNE, BxPC-3 and PANC-1 were purchased from ATCC, while H6c7 cell line was purchased from Kerafast Inc. (Boston, MA, USA). PDAC cell lines (BxPC-3, PANC-1) were maintained in an RPMI-1640 medium (Biosciences, San Jose, CA, USA) supplemented with 10% (v/v) foetal bovine serum (Biosciences) and 1% (v/v) penicillin/streptomycin (Lonza Group, Switzerland). H6c7-normal cells were maintained in keratinocyte serum free medium with L-glutamine, 5 ng mL−1 human recombinant EGF and 50 mg mL−1 bovine pituitary extract (kit) (Bioscience, Dublin, Ireland), supplemented with 50 U mL−1 streptomycin. The base medium for HPNE-intermediate cells is 75% Dulbecco's modified Eagle's medium (DMEM) without glucose with additional 2 mM L-glutamine (ThermoFisher, UK), 1,5 g L−1 sodium bicarbonate, and 25% medium M3 base (Incell Corp., San Antonio, TX, USA). The growth of this cell line was optimized in an alternative growth medium: DMEM – high glucose supplemented with 10% (v/v) FBS, 10 ng mL−1 human recombinant EGF (Bioscience, Ireland) and 50 U mL−1 streptomycin (Lonza Group, Switzerland). All cells were maintained in humidified incubators at 37 °C, with 5% CO2. With no apparent contamination, regular mycoplasma testing was carried out using the MycoAlert mycoplasma detection kit (Biosciences).All cells were seeded in 100 μL of serum-protein free media, in a flat-bottom 96-well plate at a density of 45000 cells per well. Cells were either treated with 5% (v/v) PCF or 5% (v/v) fresh serum-protein free media for 24 h. For H6c7-normal and HPNE-intermediate cell lines, PCF exposure appeared to be significantly cytotoxic, and cell concentration was reduced to such an extent that it was not possible to acquire spectra from a population of these cells. The remainder of the experiment focused therefore on analysis of the two PDAC cell lines. General biological and molecular details of the two PDAC cell lines properties are summarized in Table 1.
| Cell line | BxPC-3 | PANC-1 |
|---|---|---|
| KRAS: v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog. | ||
| Age/gender | 61-Year-old female | 56-Year-old male |
| Cell morphology | Epithelial | Mesenchymal |
| Metastasis | Non-metastatic | Metastatic |
| Differentiation | Moderate to poor | Poor |
| KRAS statue | Wild type | mutated |
After the PCF-exposure period BxPC-3 and PANC-1 cells were then trypsinized, fixed in 4% paraformaldehyde, and washed with dH20. Fixed cells were maintained at 4 °C and suspended within 200 μL of dH20. To enable their spectroscopic analysis, 100 μL of each sample was deposited on CaF2 slides using a Shandon Cytospin® 4 Cytocentrifuge (MM France) at 700 rpm for 7 min, which permits absorption of residual liquid by the sample chamber filter and then left to dry at room temperature. Details of the experimental workflow are illustrated in Fig. S1 in the ESI.†
2.2 Patient sample collection
A total of 20 samples per cell line were prepared in this experiment, including 5 controls (untreated cells) and 15 PCF-treated samples (treated cells), with no technical replicated were included, only biological replicates. PCF samples were collected prospectively by EUS-FNA (endoscopic ultrasound-guided fine-needle aspiration) from 15 patients with PCLs at one of the three institutions in Dublin, Ireland: Tallaght University Hospital, St Vincent's University Hospital, and St James's Hospital. This work was performed in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. Patients provided informed consent for sample and data acquisition, and the study received full ethical approval from Tallaght University Hospital Joint Research Ethics Committee Review Board (ID: 0319-264). The Research Ethics Committee of TU Dublin also provided approval for conduct of the study (REC-20-64).2.3 FTIR microspectroscopy
After visual inspection, cell aggregates were selected, and their infrared spectral images were acquired in transmission mode in the region between 4000–800 cm−1 using a Spotlight 400 FTIR imaging system (PerkinElmer, France), which was purged with CO2-free dry air and equipped with a liquid nitrogen-cooled MCT (mercury–cadmium–telluride) detector. FTIR images were acquired at a spatial resolution of 6.25 μm2 and a spectral resolution of 4 cm−1, with each spectrum acquired using 128 scans.A background spectrum corresponding to the CaF2 window, and the atmospheric environment was recorded with 240 accumulations and automatically subtracted from each spectrum within the PerkinElmer SpectrumIMAGE software.
2.4 Raman microspectroscopy
Raman images were captured via an XPlora microspectrometer (HORIBA, France) over the 600–1800 cm−1 fingerprint region using an exposure time of 15 s and a pixel size of 2 μm2. The spectrometer was equipped with a 532 nm laser diode excitation source, delivering an output power of 64 mW at the sample. Spectra were recorded using a 25% laser filter, with laser light focused on the sample using an Olympus MS Plan 100 lens with a numerical aperture of 0.95 and a focal length of 180 mm. The grating system was set at 1200 lines per mm, with a confocal hole of 300 μm, and a slit size of 200 μm.2.5 Preprocessing of FTIR and Raman spectra
Next, the images were corrected for atmospheric contributions, including water vapor and carbon dioxide. Following that, a resonant Mie scattering correction was employed using 10 iterations, 14 principal components of the CRMieSC (Clustered Resonant Mie Scattering Correction) algorithm,25 taking Matrigel as the reference spectrum. To correct remaining distortion of the FTIR spectral baseline, the Concave Rubberband method was employed using 5 iterations. Lastly, the spectra were smoothed using the Savitzky–Golay filter with a 5th polynomial order and a window size of 15, and then vector normalized prior to spectroscopic analysis.
Due to the heterogeneity of the samples which led to diverse spectral distortions, the adjustments were performed separately for each image using the OCTAVVS software v0.1.24.25 An example of the preprocessing workflow is provided in Fig. S2 in the ESI.†
The procedure of preprocessing was carried out mostly within the Orange Data mining software v3.36.1.
2.6 Multivariate statistical analysis
• Uniform manifold approximation and projection (UMAP) unlike PCA, is a non-linear manifold learning technique for dimensionality reduction. In terms of visualization quality, the UMAP preserves a larger portion of the global structure of a dataset due to its faster runtime compared to PCA or T-SNE. Moreover, UMAP may also be used as a dimensionality reduction methodology for data pre-treatment prior to input into machine learning algorithms since it does not impose any limitations on the embedding dimension.27
3 Results
3.1 Characterization of the mean Raman spectra of BxPC-3 and PANC-1 cells
Raman imaging was carried out on fixed PDAC cells, BxPC-3 and PANC-1, to interrogate the capability of Raman spectroscopy in differentiating between BxPC-3 and PANC-1 cell lines, prior to PCF exposure, as well as to highlight the effect of exposing these cells to PCF. The average normalized Raman spectra were compared and highlighted in Fig. 1. The main bands present in each cell type and their corresponding assignments reflecting their biochemical profiles are displayed in Table 1.Visually, the spectral differences between untreated and PCF-treated cells for both cell types can be seen principally in four distinctive regions denoted by the grey shaded areas. The first region (area 1) between 600–700 cm−1 is mainly attributed to DNA and RNA vibration bands and a few amino acids bands, particularly ring breathing vibrations from DNA bases (C–C stretching and twisting vibrations of phenylalanine, tyrosine and guanine) and the C–C twisting mode of phenylalanine and tyrosine linked to some protein signatures. The second region (area 2) between 1200–1360 cm−1 is dominated by protein signatures, notably amide III bands, the spectral signatures originating in the CH3/CH2 twisting mode of lipids, and the primary and secondary structures of proteins such as α-helix, β-sheet, and random coils. The third region (area 3) between 1400–1490 cm−1 is attributed to the CH2 bending mode of lipid and proteins. Lastly, the spectral variations of the fourth region (area 4) are marked by signatures of carotenoids principally attributed to β-carotene present in the two cell types after PCF treatment (Table 2).28
| Raman shift (cm−1) | Peak assignments |
|---|---|
| 620 | C–C twisting mode of phenylalanine (proteins) |
| 642 | C–C twisting mode of tyrosine and phenylalanine (proteins) |
| 670 | Ring breathing vibrations of guanine and tyrosine (DNA/RNA) |
| 723 | C–C symmetric stretch vibration A-DNA (ring breathing mode of DNA/RNA) |
| 751 | C–C symmetric breathing of tryptophan (protein) |
| 782 | Uracil, thymine, cytosine (ring breathing modes in the DNA/RNA) |
| 827 | O–P–O stretching in DNA, tyrosine, proline |
| 852 | Proline, hydroxyproline, tyrosine ring breathing modes (proteins) |
| 957 | Carotenoids, cholesterol (symmetric stretching vibration) |
| 1002 | C–C aromatic ring stretching phenylalanine (proteins) |
| 1031 | C–H bending mode in phenylalanine, C–N stretching of proteins |
| 1061 | C–C stretching in lipids, PO2− stretching (DNA/RNA) |
| 1125 | C–C, C–O stretching mode in carbohydrates, β-carotene |
| 1173 | Cytosine, guanine, tyrosine, phenylalanine (bending mode in plane of proteins) |
| 1206 | Amide III (proteins), C-C6H5 stretching mode in tyrosine, phenylalanine, and tryptophan |
| 1247 | Amide III (collagen assignment, β-sheet & random coils), guanine, cytosine (NH2) |
| 1313 | CH3CH2 twisting mode of lipids, amide III (α-helix) |
| 1334 | Guanine, CH3CH2 wagging of nucleic acid |
| 1447 | CH2 bending mode of proteins & lipids |
| 1524 | Carotenoids (β-carotene) |
| 1578 | Ring breathing modes in the DNA bases guanine, adenine |
| 1605 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C in-plane bending mode of phenylalanine & tyrosine, cytosine (NH2) C in-plane bending mode of phenylalanine & tyrosine, cytosine (NH2) |
| 1661 | Amide I vibration mode of structural proteins |
3.2 Characterization of the mean FTIR spectra of BxPC-3 and PANC-1 cells
FTIR imaging was carried out on aggregate fixed cells of each PDAC cell line, BxPC-3 and PANC-1, with typical spectral images and mean spectra depicted in Fig. 2. Visual images of PDAC cells are displayed in Fig. 2A on the top left for BxPC-3 and the bottom left for PANC-1, and their corresponding FTIR images are shown on the right. In Fig. 2B, the normalized mean spectrum is depicted for each cell line before and after PCF treatment. To enhance the spectral variations between the two cell lines, the second derivative spectra was computed and is depicted in Fig. 2C. Furthermore, the attributions of the main bands are displayed in Table 3.| Wavenumber (cm−1) | Peak assignments |
|---|---|
| 965 | C–C and C–O stretching mode in deoxyribose of DNA |
| 1058 | C–O stretching & C–O bending mode of carbohydrates |
| 1082 | Symmetric phosphate stretching (PO2−) (DNA/RNA) |
| 1121 | C–O stretching mode (proteins, carbohydrates) |
| 1171 | C–O stretching of proteins |
| 1234 | PO2− asymmetric stretching (DNA/RNA), amide III |
| 1310 | Amide III |
| 1390 | CH3 symmetric deformation (proteins) |
| 1452 | Asymmetric CH3 bending modes of proteins |
| 1550 | Amide II (proteins) |
| 1650 | Amide I (proteins) |
| 1742 | Fatty acid esters, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching mode of lipids O stretching mode of lipids |
| 2853 | CH2 symmetric stretching (lipids) |
| 2872 | CH3 symmetric stretching (lipids) |
| 2925 | CH2 asymmetric stretching (lipids) |
| 2958 | CH3 asymmetric stretching (lipids, proteins) |
| 3073 | Amide B |
| 3300 | Amide A (N–H stretching in proteins) |
As reported in Fig. 2B, the spectral profiles of the two cell lines, BxPC-3 and PANC-1, before and after exposure to PCF were nearly identical, with slight variations observed mainly in four areas over the 1800–900 cm−1 fingerprint region. The first region (area 1) between 930 cm−1 and 1100 cm−1 is related to vibrations of DNA/RNA and carbohydrates, with the bands at 965 cm−1 for the C–C and C–O stretching bands in deoxyribose of DNA, 1082 cm−1 for the symmetric phosphate stretching PO2− band of DNA and RNA, and the peak at 1058 cm−1 for the C–O stretching and bending of carbohydrates. The second region (area 2) between 1250 cm−1 and 1390 cm−1 is associated with signatures of protein with the bands at 1234 cm−1 and 1310 cm−1 associated with the amide III bond vibration. Moreover, the peak at 1390 cm−1 is associated with protein, specifically the CH3 symmetric deformation mode. The third region (area 3) between 1450 cm−1 and 1650 cm−1 is dominated by protein signatures from the amide I (1650 cm−1) and II bands (1550 cm−1); these bands are suggestive of changes in the secondary structure of proteins (α-helix, β-sheet). Lastly, the fourth (area 4) between 1700 and 1800 cm−1 is mainly associated with vibrations of lipids, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching and the bands associated with fatty acid esters.
O stretching and the bands associated with fatty acid esters.
3.3 BxPC-3 vs. PANC-1 prior to PCF treatment
PCA scores and loadings are displayed respectively in Fig. 3 and 4, with the first five component explaining 90% of the total variance present in Raman data (Fig. S3a†). PCA scores plot shows a strong phenotypic discrimination with minimal overlap between the two cell groups, principally along (PC1, PC3) and (PC3, PC5) planes. In contrast, no clear separation is observed in the (PC1, PC2) score plot as shown in Fig. S3b.† While PC1 and PC2 capture most of the spectral variance around 70% combined, in which the two cell lines do not show any phenotypic differences due to a possible shared biochemical characteristic. The 3rd component (PC3) on the other hand with 10.54% of the explained variance (low compared to PC1 and PC2) exhibits high phenotype discrimination, as a result, the features extracted by this component are likely responsible for this separation. This loading PC3 is predominated by numerous key Raman bands, including the C–C symmetric breathing of tryptophan at 747 cm−1, the C–C aromatic ring stretching phenylalanine at 1006 cm−1, the amide III and I vibrations at 1311 cm−1 and 1671 cm−1, and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C in-plane bending mode of phenylalanine, tyrosine and cytosine at 1632 cm−1. The band at 1585 cm−1 is attributed to the ring breathing modes in the DNA bases, and the band at 1441 cm−1 is linked to lipid bands (CH2 bending mode).
C in-plane bending mode of phenylalanine, tyrosine and cytosine at 1632 cm−1. The band at 1585 cm−1 is attributed to the ring breathing modes in the DNA bases, and the band at 1441 cm−1 is linked to lipid bands (CH2 bending mode).
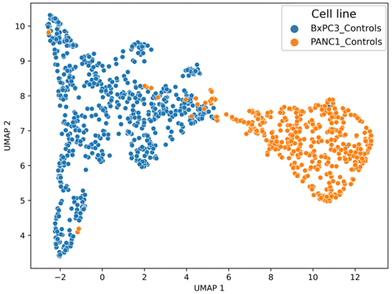
The 2D UMAP plot highlights clearly the differences between BxPC-3 and PANC-1 cell lines prior to PCF treatment using only two UMAP components (unlike PCA which required 5 PCs), with strong separation between the two groups, low intra-group variability and sufficient inter-group variability to allow group discrimination.
In contrast with the Raman spectral data, the variance explained by FTIR data over the first five PCs (almost 60%) is relatively low (Fig. S4a†), and as depicted in Fig. 6, there remains significant overlap between the two groups along the two PCA planes (PC1, PC5) and (PC2, PC5). Although, PC5 component only account for 4.16% of explained variance, it appears to be able to distinguish between the two groups of PDAC cells.
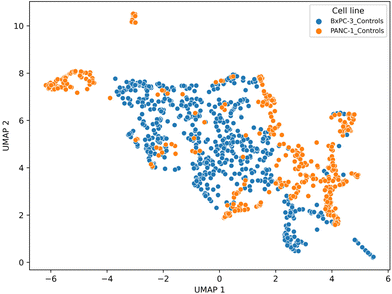
Again, UMAP analysis (using Euclidean distance as the distance metric, and with the number of neighbours set to 20) improves slightly the discrimination between the cell phenotype as shown in Fig. 7, though with the loss of interpretive information provided by PCA loadings.
3.4 BxPC-3, PANC-1 post exposure to PCF
The scores along PC3 (explaining 8.41% of the variance) for BxPC-3 cells are associated with loadings to nucleic acids bands at 748 cm−1 (which is attributed to the C–C symmetric breathing of tryptophan), and at 782 cm−1 related to the ring breathing modes of uracil, thymine, and cytosine in the DNA/RNA bases. Additionally, the spectral bands at 1120 cm−1 and 1200 cm−1 are respectively associated with C–C and C–O stretching mode in the carbohydrate and protein structural bands (amide III vibration), with a negative loading at 1670 cm−1 attributed to the amide I band. Lastly, lipid loadings in PC3 with the spectral features at 1377 cm−1 and 1484 cm−1 attributed to CH3CH2 twisting mode and CH2 bending mode of lipids signatures.
Contrastingly, the loadings to PC1 captured 72.3% of the variance, with discriminating bands mainly associated to carotenoids signatures with the three specific bands of β-carotenes at 1011 cm−1, 1158 cm−1, and 1524 cm−1 all with positive contribution to the formation of PC1.
As the two cell lines were exposed to the same PCF samples, the PCA scores plot of Raman data highlight different clustering across the two cell types and hence different loadings, this potentially suggests a cell type specific response to PCF treatment.
Again, for the FTIR, PCA was applied on the second derivative FTIR spectra over the 900–1800 cm−1 fingerprint region for both cell lines, with the scores plot depicted in Fig. 10 and their loadings displayed in Fig. S5.† Overall, poor discrimination of the cell line response to PCF was observed due to the high intra-group variability in both PDAC cell types leading to poor explained variance by the initial PCA components.
Raman-based UMAP analysis illustrates more accurately the impact of the PCF treatment on PDAC cells (BxPC-3 and PANC-1) than the PCA does. Indeed, UMAP scores plot highlighted spectral distinction within the different PCF-treated samples for both BxPC-3 and PANC-1, where some treated samples were clearly distinguished from the untreated ones, while for other treated cells no distinction with their counterpart was noticed. Moreover, PDAC cells appear to respond differently to PCF giving the results of UMAP clustering.
Unlike Raman, the impact of the PCF treatment on the cells is less noticeable in FTIR spectra, the discrimination of the cell line response to exposure to PCF appears to be more challenging. These results suggest a high intragroup variability within the treated groups for the two cell types with a low intergroup variability mainly due to baseline distortion.
3.5 BxPC-3 vs. PANC-1 pre- and post-exposure to PCF
 | ||
| Fig. 13 UMAP plot of Raman data for BxPC-3 and PANC-1 before and following PCF-treatment. (A) 2D-UMAP projection. (B) 3D-UMAP projection. | ||
According to PCA scores plot, the first two PCA components (PC1 = 60.57%, PC2 = 14.46%) capture almost 75% of the total variance within Raman data. BxPC-3_treated group appeared distinct and separated from BxPC-3_controls along PC2 axis in both (PC1, PC2) and (PC2, PC3) planes, indicating spectral alterations induced by the treatment. In contrast, both controls and PCF-treated PANC-1 spectra show minimal spectral differences and largely overlap, suggesting a less noticeable PCF-treatment effects in this cell line. On the other hand, Raman-based UMAP analysis highlight more effectively subtle spectral variations, that PCA unable to detect, particularly in PANC-1 groups. UMAP provides better class discrimination in complex datasets, reinforcing the capacity of Raman spectroscopy to detect delicate biochemical changes induced by PCF-exposure.
4 Discussion
As PC is usually discovered at a late stage it remains one of the most challenging malignancies for early detection, despite recent advancement in imaging technologies and rising clinical expertise. Patients with PCLs are considered at increased risk of developing PC, as some PCLs can be pre-cancerous and may progress to invasive malignancy, finding and monitoring these PCLs are essential for early PC detection,31.32 However, current diagnostics tools lack sufficient sensitivity and specificity to accurately distinguish between harmless and potentially pre-malignant PCLs.33,34Although, the use of PCF for malignancy risk stratification appears promising it remains at first stages. Prior research about the application of spectroscopy for pancreatic cancer diagnosis was primarily used for analysing pancreatic tissues,20–22 serum/plasma,35 or exosomes18 from healthy controls and PDAC patients or cell line. To our knowledge, no research has ever looked at the biological effects of PCF on the epithelial cells that surrounds PCLs. Thus, the role of PCF in the potential PCL to PC malignant transformation remains unclear and unexplored. Our approach addresses uniquely for the first time these aspects by applying FTIR and Raman spectroscopy combined with multivariate data analysis tools such PCA and UMAP techniques, to characterize these PCF-induced biomolecular changes in order to improve the risk stratification of PCL patient.
In the present work, BxPC-3 and PANC-1 PDAC cell lines were exposed ex vivo to PCF from patients with PCLs. The effect of exposure to PCF was elucidated spectrally using FTIR and Raman microspectroscopy together with dimensionality reduction methods, which demonstrated, on the one hand, the capacity of these techniques to discriminate PDAC cell phenotypes and, on the other hand, highlighted their response to PCF exposure. While our original intention was to profile the response across both normal and PDAC cell lines, we feel that the choice of PC cell lines as a reporter model here is better overall as this is a system that is primed to expand strongly to stimulants within the PCF. Importantly, Raman-based PCA and UMAP projections revealed a strong basal-level separation between BxPC-3 and PANC-1 prior to PCF-treatment. This separation is likely associated to their molecular, proteomic, and metabolic differences.36,37 For instance, BxPC-3 cells characterised by a wild-type KRAS and mutant SMAD4, whereas PANC-1 cell possess mutations in KRAS, CDKN2A, while retaining wild-type SMAD4. These genetic differences are also associated to a differential epithelial to mesenchymal transition (EMT) profiles, with BxPC-3 exhibit more epithelial features while PANC-1 harbour mesenchymal characteristics.24,36
In order to examine PCF-induced cellular responses, our work combined two complementary spectroscopic approaches (FTIR, Raman) with dimension reduction tools such as PCA and UMAP. While PCA, and UMAP were both effective in reducing the data dimension and identifying clustering patterns, there were some discernible differences between their findings. PCA as a linear technique allow the capture of linear variabilities inside the data linked the cell phenotype on the one hand and the cell response to PCF on the other hand, although the discrimination between the two cell lines post-exposure to PCF wasn't completely achieved especially for FTIR data with strong overlapping between the two groups. In contrast, UMAP projections was more efficient than PCA at detecting non-linear variabilities and smaller group separations particularly for Raman data as it preserve both the global and local structure of the data.27 This demonstrates the increasing utility of UMAP analysis over PCA in spectroscopy data exploration. Nevertheless, the superiority of Raman over FTIR was also noticed and elucidated in this study in capturing biomolecular variations between each cell line (BxPC-3 and PANC-1) before and following the PCF treatment. Similarly, between the two complementary techniques, Raman frequently outperformed FTIR in capturing PCF-induced alterations to the cell biology. Raman high spatial resolution and capability of detecting differences at the intracellular level might explain its superior performance over FTIR. While FTIR remains beneficial and helpful for global biomolecular profiling, it showed poor class discrimination compared to Raman.
While these preliminary findings appear promising and provide insights into how PCF alters the spectral profile of PDAC cells ex vivo, further investigations are still required to overcome the current limitations and allow advancement to clinical use. One of the primary limitations is the limited sample size as only 15 PCF patient samples were used to treat the cells. Expanding the sample size using larger cohort with diverse ethnicities and various PCLs subtypes is crucial to capture full PCF variability across a variety of PCLs and enhance the statistical validity. Additionally, the current methodology involved recording spectra from remaining cancer cells after 24 h of exposure to a low PCF concentration (5% v/v). Increased concentrations of PCF with a prolonged exposure time are still unexplored and may not yield to similar findings and remain to be elucidated. Interestingly, PCF can be extremely cytotoxic to the cells even at reduced concentrations leading to a significant cell death. As such, PCF samples included in this study should considered low cytotoxic and may not represent full spectrum of PCF effects on cancer cell biology.
Future work should investigate the biological effects of PCF on non-cancerous cell lines such as H6c7 and HPNE cell lines and its potential role during the PCL to PC malignant transformation. Multi-omics integration could greatly improve the biochemical interpretation of the detected spectral markers. Moreover, longitudinal investigations assessing the effects of PCF on normal cells may allow the examination of the ability of spectroscopy to monitor PCL progression overtime and assess its potential in the early detection of PC. By addressing the above aspects PCL risk classification and patient quality of life may improve tremendously.
5 Conclusion
Our approach demonstrates the capacity of vibrational spectroscopy particularly Raman technique in conjunction with unsupervised dimensionality reduction tools to discriminate PDAC cell phenotype and highlight PCF-induced spectral alterations on PDAC cell biology ex vivo. This approach, upon further validation, could have potential applications as a novel first-stage diagnostic screening method for PC and PCL risk stratification.Conflicts of interest
The authors declare no conflicts of interest.Data availability
Data for the control cell lines referred to in this article will be made available on Zenodo at the time of publication. Data collected from human participants, described throughout the publication are not available for confidentiality reasons.Acknowledgements
This work was conducted within the framework of the Graduate School NANO-PHOT (École Universitaire de Recherche, PIA3, contract ANR-18-EURE-0013), and the Taighde Eireann (Research Ireland) ADAPT Research Centre at TU Dublin. The ADAPT SFI Centre for Digital Media Technology is funded by Science Foundation Ireland through the Taighde Eireann (Research Ireland) Research Centers Programme and is co-funded under the European Regional Development Fund (ERDF) through Grant # 13/RC/2106_P2. The authors acknowledge that this work has been supported by doctoral scholarships from the “Grand-Est region” and the TU Dublin Fiosraigh Scholarship scheme. The authors also thank the Cellular and Tissular Imaging Platform (PICT) – URCATech, the Meath Foundation and Viatris Ireland for additional support.References
- H. Sung, et al., Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, CA-Cancer J. Clin., 2021, 71(3), 209–249, DOI:10.3322/caac.21660.
- R. L. Siegel, A. N. Giaquinto and A. Jemal, “Cancer statistics”, 2024. DOI:10.3322/caac.21820.
- L. Rahib, M. R. Wehner, L. M. Matrisian and K. T. Nead, Estimated Projection of US Cancer Incidence and Death to 2040, JAMA Network Open, 2021, 4(4), e214708, DOI:10.1001/jamanetworkopen.2021.4708.
- M. Hidalgo, et al., Addressing the challenges of pancreatic cancer: future directions for improving outcomes, Pancreatology, 2015, 15(1), 8–18, DOI:10.1016/j.pan.2014.10.001.
- P. Rawla, T. Sunkara and V. Gaduputi, Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors, World J. Oncol., 2019, 10(1), 10–27, DOI:10.14740/wjon1166.
- C. I. Puşcaşu, M. Rimbaş, R. B. Mateescu, A. Larghi and V. Cauni, Advances in the Diagnosis of Pancreatic Cystic Lesions, Diagnostics, 2022, 12(8), 8, DOI:10.3390/diagnostics12081779.
- N. C. M. van Huijgevoort, M. del Chiaro, C. L. Wolfgang, J. E. van Hooft and M. G. Besselink, Diagnosis and management of pancreatic cystic neoplasms: current evidence and guidelines, Nat. Rev. Gastroenterol. Hepatol., 2019, 16(11), 11, DOI:10.1038/s41575-019-0195-x.
- H. Aziz, A. W. Acher, S. G. Krishna, J. M. Cloyd and T. M. Pawlik, Comparison of Society Guidelines for the Management and Surveillance of Pancreatic Cysts: A Review, JAMA Surg., 2022, 157(8), 723–730, DOI:10.1001/jamasurg.2022.2232.
- D. R. Ardeshna, et al., Recent advances in the diagnostic evaluation of pancreatic cystic lesions, World J. Gastroenterol., 2022, 28(6), 624–634, DOI:10.3748/wjg.v28.i6.624.
- M. Aijazi and R. Das, Pancreatic Cyst Fluid Assessment: Updates in Genetic Analysis and Risk for Progression, Curr. Treat. Options Gastroenterol., 2024, 22(4), 287–301, DOI:10.1007/s11938-024-00446-4.
- X. Yang, et al., Diagnosis of Lung Cancer by FTIR Spectroscopy Combined With Raman Spectroscopy Based on Data Fusion and Wavelet Transform, Front. Chem., 2022, 10, 810837, DOI:10.3389/fchem.2022.810837.
- Who's Who? Discrimination of Human Breast Cancer Cell Lines by Raman and FTIR Microspectroscopy. Consulté le: 27 septembre 2024. [En ligne]. Disponible sur: https://www.mdpi.com/2072-6694/14/2/452.
- D. K. R. Medipally, et al., Vibrational spectroscopy of liquid biopsies for prostate cancer diagnosis, Ther. Adv. Med. Oncol., 2020, 12 DOI:10.1177/1758835920918499.
- C. A. Jenkins, et al., A high-throughput serum Raman spectroscopy platform and methodology for colorectal cancer diagnostics, Analyst, 2018, 143(24), 6014–6024, 10.1039/C8AN01323C.
- C. Kendall, et al., Vibrational spectroscopy : a clinical tool for cancer diagnostics, Analyst, 2009, 134(6), 1029–1045, 10.1039/B822130H.
- G. W. Auner, et al., Applications of Raman spectroscopy in cancer diagnosis, Cancer Metastasis Rev., 2018, 37(4), 691–717, DOI:10.1007/s10555-018-9770-9.
- C. T. Mandrell, et al., Machine Learning Approach to Raman Spectrum Analysis of MIA PaCa-2 Pancreatic Cancer Tumor Repopulating Cells for Classification and Feature Analysis, Life, 2020, 10(9), 181, DOI:10.3390/life10090181.
- J. Carmicheal, et al., Label-free characterization of exosome via surface enhanced Raman spectroscopy for the early detection of pancreatic cancer, Nanomedicine, 2019, 16, 88–96, DOI:10.1016/j.nano.2018.11.008.
- J. Li, et al., Abrogating cholesterol esterification suppresses growth and metastasis of pancreatic cancer, Oncogene, 2016, 35(50), 50, DOI:10.1038/onc.2016.168.
- K. Szymoński, et al., Variabilities in global DNA methylation and β-sheet richness establish spectroscopic landscapes among subtypes of pancreatic cancer, Eur. J. Nucl. Med. Mol. Imaging, 2023, 50(6), 1792–1810, DOI:10.1007/s00259-023-06121-7.
- K. Szymoński, et al., Combined analytical approach empowers precise spectroscopic interpretation of subcellular components of pancreatic cancer cells, Anal. Bioanal. Chem., 2023, 415(29), 7281–7295, DOI:10.1007/s00216-023-04997-w.
- V. Notarstefano, et al., Investigation of human pancreatic cancer tissues by Fourier Transform Infrared Hyperspectral Imaging, J. Biophotonics, 2020, 13(4), e201960071, DOI:10.1002/jbio.201960071.
- A. Sala, et al., Liquid Biopsy for Pancreatic Cancer Detection Using Infrared Spectroscopy, Cancers, 2022, 14(13), 13, DOI:10.3390/cancers14133048.
- E. L. Deer, et al., Phenotype and Genotype of Pancreatic Cancer Cell Lines, Pancreas, 2010, 39(4), 425–435, DOI:10.1097/MPA.0b013e3181c15963.
- C. Troein, S. Siregar, M. O. D. Beeck, C. Peterson, A. Tunlid and P. Persson, OCTAVVS: A Graphical Toolbox for High-Throughput Preprocessing and Analysis of Vibrational Spectroscopy Imaging Data, Methods Protoc., 2020, 3(2), 2, DOI:10.3390/mps3020034.
- D. Granato, J. S. Santos, G. B. Escher, B. L. Ferreira and R. M. Maggio, Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective, Trends Food Sci. Technol., 2018, 72, 83–90, DOI:10.1016/j.tifs.2017.12.006.
- L. McInnes, J. Healy and J. Melville, UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction, arXiv, 2020, preprint, arXiv:1802.03426. Consulté le: 15 février 2024. [En ligne]. Disponible sur: https://arxiv.org/abs/1802.03426.
- J. De Gelder, K. De Gussem, P. Vandenabeele and L. Moens, Reference database of Raman spectra of biological molecules, J. Raman Spectrosc., 2007, 38(9), 1133–1147, DOI:10.1002/jrs.1734.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Raman Spectroscopy of Biological Tissues, Appl. Spectrosc. Rev., 2007, 42(5), 493–541, DOI:10.1080/05704920701551530.
- Z. Movasaghi, S. Rehman and I. ur Rehman, Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues, Appl. Spectrosc. Rev., 2008, 43(2), 134–179, DOI:10.1080/05704920701829043.
- J. Rogowska, J. Semeradt, Ł. Durko and E. Małecka-Wojciesko, Diagnostics and Management of Pancreatic Cystic Lesions—New Techniques and Guidelines, J. Clin. Med., 2024, 13(16), 4644, DOI:10.3390/jcm13164644.
- H. Matthaei, R. D. Schulick, R. H. Hruban and A. Maitra, Cystic precursors to invasive pancreatic cancer, Nat. Rev. Gastroenterol. Hepatol., 2011, 8(3), 141–150, DOI:10.1038/nrgastro.2011.2.
- M. Distler, D. Aust, J. Weitz, C. Pilarsky and R. Grützmann, Precursor Lesions for Sporadic Pancreatic Cancer: PanIN, IPMN, and MCN, BioMed Res. Int., 2014, 2014(1), 474905, DOI:10.1155/2014/474905.
- G. H. Elta, B. K. Enestvedt, B. G. Sauer and A. M. Lennon, ACG Clinical Guideline: Diagnosis and Management of Pancreatic Cysts, Am. J. Gastroenterol., 2018, 113(4), 464–479, DOI:10.1038/ajg.2018.14.
- E. Duckworth, M. Mortimer, B. Al-Sarireh, V. Kanamarlapudi and D. Roy, Frontline clinical diagnosis—FTIR on pancreatic cancer, Cancer Med., 2023, 12(16), 17340–17345, DOI:10.1002/cam4.6346.
- Y. Kim, D. Han, H. Min, J. Jin, E. C. Yi and Y. Kim, Comparative Proteomic Profiling of Pancreatic Ductal Adenocarcinoma Cell Lines, Mol. Cells, 2014, 37(12), 888–898, DOI:10.14348/molcells.2014.0207.
- T. Tataranni, et al., Rewiring carbohydrate catabolism differentially affects survival of pancreatic cancer cell lines with diverse metabolic profiles, Oncotarget, 2017, 8(25), 41265–41281, DOI:10.18632/oncotarget.17172.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5an00230c |
| ‡ These authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2025 |