Fluorescent bionanoprobes based on quantum dot-chitosan–O-phospho-L-serine conjugates for labeling human bone marrow stromal cells
Christiane L. Salgadobc,
Alexandra A. P. Mansura,
Herman S. Mansur*a and
Fernando J. M. Monteirobc
aCenter of Nanoscience, Nanotechnology, and Innovation-CeNano2I, Department of Metallurgical and Materials Engineering, Federal University of Minas Gerais, Brazil. E-mail: hmansur@demet.ufmg.br
bINEB, Instituto Nacional de Engenharia Biomédica, Porto, Portugal
cFEUP, Faculdade de Engenharia, Departamento de Engenharia Metalúrgica e Materiais, Universidade do Porto, Porto, Portugal
First published on 30th September 2014
Abstract
Bone replacement materials might be promising alternatives to autogenous bone transplants during the repair and reconstruction of human bone tissues. However, bone healing is a very complex process, and the roles of phosphatidylserine (PS) and its moieties are not yet completely understood. In the present study, fluorescent quantum dots (QDs) functionalized with chitosan–O-phospho-L-serine (chi–OPS) conjugates have been synthesized and characterized while focusing on their potential applications as nanoprobes for labeling human bone marrow stromal cells (hBMSC). Essentially, chitosan was covalently linked to the peptide (O-phospho-L-serine, OPS) through the formation of amide bonds. In this sequence, these chi–OPS conjugates were utilized as direct capping ligands during CdS QDs (CdS/chi–OPS) biofunctionalization, which was achieved using a single-step process in an aqueous medium at room temperature. The core–shell nanostructures were characterized in detail by UV-visible spectroscopy (UV-Vis), photoluminescence spectroscopy (PL), atomic force microscopy (AFM), and transmission electron microscopy (TEM) with selected area electron diffraction (SEAD). The TEM images associated with the UV-vis optical absorption results indicated that ultra-small nanocrystals were formed with average diameters ranging from 2.2 to 2.8 nm. In addition, the PL results showed that the nanoconjugates exhibited “green” fluorescent activity under ultraviolet excitation. Cell viability was assessed in vitro via an MTT analysis, revealing that the bioconjugates were not cytotoxic after 3 days of incubation. Moreover, a quantitative flow cytometry (QFC) analysis and confocal fluorescence microscopy (CFA) were performed, verifying the fluorescence-labeling efficiency and the endocytosis of the bionanoprobes by hBMSC. In summary, innovative fluorescent conjugates were developed with properties for use as biomarkers when imaging and detecting bone tissue regeneration and metabolic events.
1. Introduction
Bone is a metabolically active tissue that undergoes continuous remodeling through two counteracting processes: bone formation and bone resorption. Under normal conditions, this continuous cycle of bone resorption and apposition (i.e., “bone turnover”) is balanced and regulated through the action of various systemic hormones and local mediators (e.g., cytokines, growth factors).1–3 In addition, human bone marrow stromal cells (hBMSC), which are also called mesenchymal stem cells (MSC), play a pivotal role during bone regeneration and repair in vivo.4,5 During the last 3–4 decades, biomaterials have been widely used in bone regenerative therapies, including in bone grafts and tissues engineering, with properties optimized for specific repair functions.6 Although most of these bone-related materials are biocompatible, they often require additional cells, growth factors, proteins (e.g. bone sialo proteins) and other agents to improve the cell response and subsequent mineralization, which usually renders this approach complicated and expensive.6–8 Bone sialo proteins (BSP) stimulate cell activities because they possess RGD motifs and phosphatidylserine (PS) residues influencing the tissue mineralization process, which mediates the specific interactions at the biomaterial-bone interfaces. Phosphatidylserine is known as an essential component of the cell membrane that plays a key role in cell signaling in the course of the tissue development, repair and function.9–11 Investigations involving cells cultured in vitro on PS-modified biomaterials suggested that PS is important for bone regeneration.11 Moreover, Reinstorf and collaborators12,13 have demonstrated that the levels of proliferation and differentiation in vivo increase when using rat calvaria osteoblasts on PS-modified cement composites. Thus, PS and its derivatives, such as the peptide O-phospho-L-serine (OPS, water-soluble PS motif), might be a more accessible, viable and inexpensive alternative to BSP for improving bone reconstruction processes.2 Nevertheless, despite the undeniable progress in biomaterials and bone tissue engineering, the mechanism and the intricate pathways involving PS and its derivatives during bone repair processes have not been fully elucidated, and sometimes the results are controversial.3,14,15 In this sense, nanomedicine (i.e., the application of science and nanotechnology to medicine and biology) can offer an entire set of very promising tools for a more in-depth understanding of the dynamic process of bone repair and the role of several signaling molecules involved in the events.16–23 The recent advances in nanotechnology have led to the development of innumerable multifunctional nanoparticles and nanoconjugates for biomedical applications16–22 including drug delivery, bioimaging, molecular imaging, cell tracking and cell labeling. In order to achieve the desired effect, the nanoparticles need to biocompatible and exert their actions directly on their target, i.e., cells, tissues and diseased-sites, without evoking side effects in other tissues. Thus, the interest in developing novel fluorescent nanoparticles, based on organic,16–19 inorganic and hybrid nanostructured systems24–28 for biomedicine and pharmaceutical applications has grown dramatically over the past two decades. Specifically, in the field of bone tissue engineering, several in vitro and in vivo studies highlight the great potential for utilizing nano-sized materials and nanostructures in bone research.24 In the vast realm of nanoparticles designed for biomedical applications, semiconductor nanocrystals, or quantum dots (QDs), have received an outstanding increase in interest from scientists and professionals because of their nanoscale dimensions and unique electronic, optical and biological properties.26 Compared to organics dyes, QDs offer considerably higher photochemical stability, brightness, and narrower emission spectra, improving the sensitivity and intervals of bioimaging at lower analyte concentrations. Moreover, the emission color of QDs can be tuned over a wide range of wavelengths, which range from ultraviolet to visible and infrared, by controlling the size and chemical composition of the fluorescent nanocrystals.26 These semiconductor nanoparticles are intrinsically hydrophobic, but they can be functionalized at their surfaces by applying water-soluble and biocompatible organic shells, such as peptides, polymers, carbohydrates, nucleotides sequences and other molecules, affixing new functionalities for nanomedicine.28 Therefore, peptides are an interesting choice when developing bioconjugates with QDs because they exhibit some important characteristics, such as biocompatibility, water solubility, chemical stability in physiological media, bio-affinity and the “smallest” possible molecular size among proteins and enzymes. Due to the complexity of synthesizing, stabilizing and bioconjugating QDs at the same time, only few studies have been reported using peptides, amino acids and polymer-based bioconjugates as direct capping agents with aqueous processing routes, which involve predominantly chelating metal ions from the inorganic core with the amine and carboxyl groups of ligands.28–30 Surprisingly, no studies were found in the consulted literature addressing the synthesis and characterization of CdS quantum dots that were simultaneously stabilized and biofunctionalized by OPS and chitosan–OPS conjugates, which were designed specifically for cell labeling and bioimaging bone-related biological events.In this study, it is presented for the first time the synthesis and characterization of fluorescent bio-nanoprobes based on CdS quantum dots functionalized with OPS and chitosan–OPS conjugates. Cell viability assays were used to ensure the cytocompatibility of the novel conjugates. In addition, quantitative flow cytometry and confocal fluorescence microscopy results have demonstrated the effectiveness of the QD/chi–OPS conjugates for targeting and endocytosis toward human bone marrow stromal cells. Consequently, these fluorescent nanostructures may be used as biomarkers when elucidating the role of PS during bone healing events and stages.
2. Experimental procedure
2.1. Materials
All of the reagents and precursors, including cadmium perchlorate hydrate (Aldrich, USA, Cd(ClO4)2·6H2O), sodium sulfide (Synth, Brazil, >98%, Na2S·9H2O), sodium hydroxide (Merck, USA, ≥99%, NaOH), acetic acid (Synth, Brazil, ≥99.7%, CH3COOH), 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (Sigma, USA, ≥98%, C8H17N3·HCl), and N-hydroxysulfosuccinimide sodium salt (Aldrich, USA, ≥98%, C4H4NNaO6S) were used as received. Chitosan powder (Aldrich Chemical, USA, catalog# 448869, low molecular weight, Mw = 50–190 kDa, lot supplied = 60–70 kDa; degree of deacetylation DD ≥ 75.0%, lot supplied = 96.1%; viscosity 20–300 cPoise, lot supplied = 35 cPoise, 1 wt% in 1% acetic acid) was used as a reference polysaccharide ligand. The peptide O-phospho-L-serine, OPS, (linear formula (OH)2P(O)OCH2CH(NH2)CO2H) was supplied by Sigma (USA). Unless specified otherwise, deionized water (DI water, Millipore Simplicity™) with a resistivity of 18 MΩ cm was used to prepare the solutions, and the procedures were performed at room temperature (23 ± 2 °C).2.2. Bioconjugation of the peptides (OPS) to chitosan (Chi)
O-Phospho-L-serine (OPS) was bioconjugated to chitosan (Chi) using 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC, Mw = 191.7 g mol−1) as a “zero-length” crosslinking agent in the presence of N-hydroxysulfosuccinimide sodium salt (sulfo-NHS, Mw = 217.1 g mol−1).31,32A chitosan solution (1%, w/v) was prepared by adding chitosan powder to aqueous acetic acid (2%, v/v). The solution was stirred at a constant, moderate rate at room temperature until solubilization was complete (pH ∼ 3.6). Subsequently, the pH of the chitosan acetate solution was adjusted to 6.0 ± 0.1 with NaOH (0.1 mol L−1), generating a sodium acetate buffer solution. EDC (1.0 wt%) and sulfo-NHS (2.0 wt%) were dissolved in phosphate saline buffer (pH 7.4). The OPS solution (20.0 mg mL−1) was prepared by dissolving OPS powder in DI water with moderate magnetic stirring for 10 min until complete solubilization had occurred (pH ∼ 1.7). The bioconjugation process proceeded as follows: the PS solution was added to a reaction flask containing a solution of EDC (0.5 mmol L−1) and sulfo-NHS (1.0 mmol L−1) with magnetic stirring for 10 min at 6 ± 2 °C. While stirring, the chitosan solution was added to the flask at a 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 molar ratio (chitosan monomer
1.5 molar ratio (chitosan monomer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide) to prepare the peptide-conjugated chitosan; the system was incubated at room temperature for 1.5 h. EDC converts the carboxyl group of OPS to amine-reactive sulfo-NHS esters in the presence of sulfo-NHS.33 These esters often react with the amine groups in chitosan, yielding a chitosan and peptide conjugate linked with stable covalent amide bonds (RC(O)NR′R′′) named chitosan–peptide (“Chi–OPS”). The schematic representation of the conjugation of chitosan with OPS peptides by amide bonds formation is depicted in Scheme 1.
peptide) to prepare the peptide-conjugated chitosan; the system was incubated at room temperature for 1.5 h. EDC converts the carboxyl group of OPS to amine-reactive sulfo-NHS esters in the presence of sulfo-NHS.33 These esters often react with the amine groups in chitosan, yielding a chitosan and peptide conjugate linked with stable covalent amide bonds (RC(O)NR′R′′) named chitosan–peptide (“Chi–OPS”). The schematic representation of the conjugation of chitosan with OPS peptides by amide bonds formation is depicted in Scheme 1.
2.3. Synthesis of CdS/polysaccharides conjugates
CdS nanoparticles stabilized by chitosan (Chi) and chitosan–peptide (Chi–OPS) were synthesized via an aqueous route in a reaction flask at room temperature as follows: 0.4 mg mL−1 of “chitosan–peptide solution” or “chitosan solution” (reference) in DI water were added to the flask (pH = 6.0 ± 0.1). With moderate magnetic stirring, the Cd2+ (Cd(ClO4)2·6H2O, 0.75 mmol L−1) and S2− precursor solutions (Na2S·9H2O, 0.37 mmol L−1) were added to the flask (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cd molar ratio was kept at 1
Cd molar ratio was kept at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and stirred for 3 min. The CdS QDs dispersions produced were clear, homogeneous and yellowish, which were named QD_Chi–OPS or QD_Chi (reference) based on the capping ligand used during the synthesis and biofunctionalization. The CdS QDs colloids were dialyzed for 24 h (water changes after 2 h and 4 h) against 3 L of DI water using a cellulose membrane with molecular weight cut-off (MWCO) of 14
2) and stirred for 3 min. The CdS QDs dispersions produced were clear, homogeneous and yellowish, which were named QD_Chi–OPS or QD_Chi (reference) based on the capping ligand used during the synthesis and biofunctionalization. The CdS QDs colloids were dialyzed for 24 h (water changes after 2 h and 4 h) against 3 L of DI water using a cellulose membrane with molecular weight cut-off (MWCO) of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da with moderate stirring at room temperature. After purification, the QD dispersions were stored at 6 ± 2 °C for further use.
000 Da with moderate stirring at room temperature. After purification, the QD dispersions were stored at 6 ± 2 °C for further use.
2.4. Synthesis of CdS–peptide conjugates (QD_OPS)
The CdS quantum dots colloidal dispersion directly stabilized by OPS, which is named QD_OPS, was produced via an aqueous route at room temperature. The OPS solution was added to a reaction flask, and the pH was adjusted to 11.5 ± 0.1 (NaOH, 0.1 mol L−1). With moderate magnetic stirring, Cd2+ (Cd(ClO4)2·6H2O, 0.75 mmol L−1) and S2− precursor solutions (CH3CSNH2, 0.37 mmol L−1) were added to the OPS solution (16.0 mmol L−1) and stirred for 3 min. The CdS QDs dispersion was purified and stored as described in Section 2.4.2.5. Physicochemical characterization of the CdS conjugates
The UV-visible spectra (UV-Vis) were measured using Perkin-Elmer equipment (Lambda EZ-210) in transmission mode using a quartz cuvette from 600 nm to 190 nm. The absorption spectra were used to monitor the formation of the CdS QDs during the reaction and their relative colloidal stability in the medium. The average nanoparticle size and their optical properties were estimated based on the “absorbance onset” of the curve. All of the experiments were conducted in triplicate (n = 3) unless specifically noted. A statistical analysis of the results was performed while assuming the mean and standard deviation when necessary.The photoluminescence (PL) characterization of the CdS QDs conjugates was performed based on spectra acquired at room temperature using a Nanodrop 3300 fluoro-spectrometer (Thermo Scientific, Blue LED with λexcitation = 470 ± 10 nm). The relative activity was calculated by subtracting the backgrounds of the samples without QDs. All of the tests were conducted using at least four replicates (n ≥ 4). Additionally, QD colloidal media were placed inside a “darkroom-chamber” where they were illuminated with a UV radiation emitting bulb (λexcitation = 254 nm and λexcitation = 365 nm, 6 W, Boitton Instruments). Digital color images were collected when the QDs fluoresced in the visible range.
The nanostructural characterizations of the QD based on the images and selected area electron diffraction patterns (SAED) were conducted using transmission electron microscopy (TEM) with a Tecnai G2-20-FEI microscope at an accelerating voltage of 200 kV. The energy-dispersive X-ray spectra (EDX) were collected for chemical analysis. The TEM samples were prepared by dropping the colloidal dispersion onto a holey carbon grid before the analysis. The QD sizes and distribution were obtained based on the TEM images by measuring at least 100 randomly selected nanoparticles using an image processing program (ImageJ, version 1.44, public domain, National Institutes of Health).
Atomic force microscopy (AFM) was conducted with an XE-70 (Parker) instrument operating in contact mode. The scanning rate was 1.0 Hz, and the images were acquired with a 512 × 512 pixel resolution. The samples were prepared by dropping the colloidal dispersion onto a mica plate after dilution in DI water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). The areas were randomly selected for statistical purposes.
5). The areas were randomly selected for statistical purposes.
Dynamic light scattering (DLS) analyses were performed using a Brookhaven ZetaPlus instrument with a laser light wavelength of 660 nm (35 mW red diode laser) and a thermostat with temperature stabilization. Standard square acrylic cells with a volume of 4.5 mL were used. For the DLS of the QDs, the colloidal solutions (2 mL) in the presence of 0.15 mol L−1 NaCl (1 mL) were filtered three times through a 0.45 μm aqueous syringe filter (Millex LCR 25 mm, Millipore) to remove any possible dust. Samples were measured at 25 ± 2 °C, and light scattering was detected at 90°. Each required approximately about 3 minutes, and 3 measurements were obtained for each system and averaged.
Zeta potential (ZP) measurements were performed on QD colloidal water media using a ZetaPlus instrument by applying the laser light diffusion method (Brookhaven Instruments). This instrument uses the laser Doppler electrophoresis technique (35 mW red diode laser at λ = 660 nm). All tests were performed using a minimum of three replicates (n = 3), and the values were averaged.
2.6. Biological characterization of CdS conjugates
The cytotoxicity of the samples was evaluated using a standard 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma-Aldrich). Briefly, osteoblast-like cells (MG63) were seeded into 96-well plates at 104 cells per well. After 24 h, various concentrations (0.5; 1.0; 2.5; 5.0; 10 and 50%) of the three QD-conjugate systems (QD_Chi, QD_OPS and QD_Chi–OPS) were added and incubated for 3 days. MTT (2.5 mg mL−1) was added to each well, and the plates were incubated for 4 h at 37 °C under 5% CO2. After the medium was removed, DMSO (100 μL, Merck) was added to each well. The optical density (OD) of the cell monolayer was measured at the wavelength = 550 nm on a microplate reader (Synergy Mx, BioTek). For the control, cells seeded in Tissue Culture Polystyrene (TCPS) wells with only complete medium were considered 100% viable after 3 days of culture.
3. Results and discussion
3.1. Physicochemical characterization of the CdS conjugates
The average CdS nanoparticle size was determined using Henglein's empirical model,34 which relates the diameter of the CdS nanoparticle (2R) to the exciton absorption transition onset (λexc) on the UV-Vis spectra according to eqn (1).
 | (1) |
The optical band gap (absorbance onset, EQD) and the “blue-shift” values were determined from the absorption coefficient data as a function of wavelength using the “Tauc relation”35 extracted from the UV-Vis spectra. The results of CdS diameter (2R) and EQD are summarized in Fig. 2 (Table 1) and support the formation of CdS quantum dots effectively capped by the ligands in the water media because the band gap energy of the semiconductor nanocrystals were significantly greater than the reference CdS bulk value (i.e., 2.42 eV).32
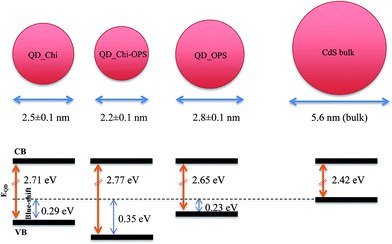 | ||
| Fig. 2 Schematically representation of band gap energy dependence on the surface capping ligands (not to scale); CB = conduction band; VB = valence band. | ||
| System | Parameters | Values after 5 days |
|---|---|---|
| QD_Chi | Band gap (eV) | 2.71 ± 0.02 |
| Blue shift (eV) | 0.29 ± 0.02 | |
| λexc (nm) | 398 ± 2 | |
| 2R (nm) | 2.5 ± 0.1 | |
| QD_Chi–OPS | Band gap (eV) | 2.77 ± 0.02 |
| Blue shift (eV) | 0.35 ± 0.02 | |
| λexc (nm) | 379 ± 2 | |
| 2R (nm) | 2.2 ± 0.1 | |
| QD_OPS | Band gap (eV) | 2.65 ± 0.02 |
| Blue shift (eV) | 0.23 ± 0.02 | |
| λexc (nm) | 417 ± 2 | |
| 2R (nm) | 2.8 ± 0.1 |
These results are considered very significant as far as the direct synthesis of semiconductor nanocrystals in aqueous media is concerned because CdS QDs were nucleated and stabilized at ultra-small sizes using water-soluble chitosan–peptide conjugates. In fact, to the best of our knowledge, there has been no reported literature where bioconjugates based on CdS QDs were directly produced and stabilized by chitosan conjugated with O-phospho-L-serine or directly with OPS as capping ligands that were synthesized at room temperature using strictly water colloidal chemistry.
 | ||
| Fig. 3 PL spectra of (a) QD_Chi, (b) QD_Chi–OPS, and (c) QD_OPS. Dark chamber images of QD_Chi–OPS fluorescing under 254 nm and 365 nm UV irradiation, as labeled. | ||
Based on the obtained spectra, a sharp green emission peak at approximately 509 nm could be observed for all samples and a broad orange-red emission band (600–750 nm) was verified only for QD_Chi and QD_Chi–OPS systems, in accordance with the darkroom fluorescence images. These PL profiles are compatible with the colloidal synthesis of quantum dots in aqueous medium and their ultra-small sizes. The green (509 nm) and orange emission centers are usually associated with metal atoms at interstitial positions (Cdi)36–38 and they were expected once in this study the synthesis was performed with an “excess” of cadmium ions compared to sulfides [S2−]/[Cd2+] = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. In addition, as the produced nanocrystals were very small, a large surface to volume ratio is observed and the number of defects causes the self-activated red fluorescence, which is characteristic of nanoparticles that contain a certain concentration of intrinsic defects of the VCd–VS (di-vacancies, i.e., cadmium: VCd and sulfur: VS)37 or VS type.38 Thus, the PL profiles and the quantum efficiency (QY ≈ 1.0%) obtained for the CdS QD nanoconjugates produced in this study are compatible with the colloidal synthesis of quantum dots in aqueous medium and their ultra-small sizes.26
2. In addition, as the produced nanocrystals were very small, a large surface to volume ratio is observed and the number of defects causes the self-activated red fluorescence, which is characteristic of nanoparticles that contain a certain concentration of intrinsic defects of the VCd–VS (di-vacancies, i.e., cadmium: VCd and sulfur: VS)37 or VS type.38 Thus, the PL profiles and the quantum efficiency (QY ≈ 1.0%) obtained for the CdS QD nanoconjugates produced in this study are compatible with the colloidal synthesis of quantum dots in aqueous medium and their ultra-small sizes.26
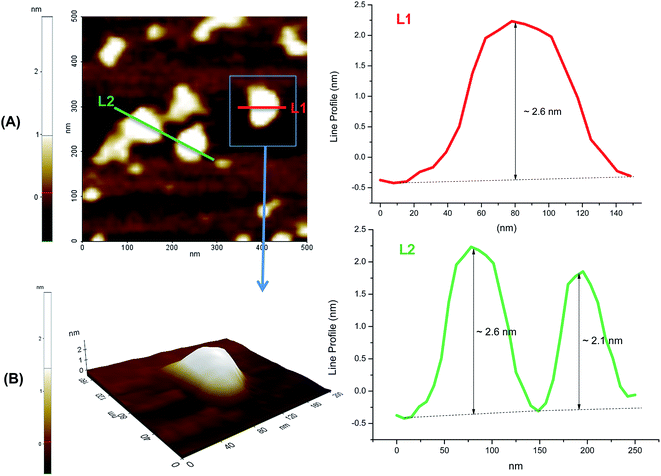
Moreover, morphological aspects of QDs were also investigated by examining three-dimensional AFM images of dispersions onto mica plates (Fig. 5). The QD_OPS bioconjugates were mostly isolated. The width measurement (x or y-axis) usually overestimates the actual size of the QDs due to the AFM tip convolution, however the height (z-axis) corresponds to a more accurate dimensional, with the diameter in the range of 2 to 3 nm, well-matched to TEM and UV-vis analyses.
The zeta potentials (ZP) measurements for the CdS_Chi and CdS_Chi–OPS samples (pH 6.0 ± 0.1) were approximately zero (0.0 ± 0.5 mV), indicating that the results were predominantly influenced by the surface charge of chitosan, which tends towards zero at pH values around the isoelectric point (pI) of the chitosan (pH range from 6.0 to 7.0, pKa ∼ 6.5). The ZP measured for CdS–OPS was −21 ± 0.4 mV (pH 7.3 ± 0.1) in agreement with the literature.39 These values indicated that the conjugates were not predominantly electrostatically stabilized but relied on the steric hindrance of the polymer or layered peptide chains adsorbed physically and chemically to prevent close contact between the fluorescent QDs.25
3.2. Cell toxicity assay
The cytotoxicity of the QD-conjugated nanoparticles was determined because QD nanocrystals are semiconductor materials (inorganic core) and the biological applicability of QDs relies primarily on how it affects biological functions and cell viability. The cytotoxicity experiment was performed using a standard methylthiazole tetrazolium (MTT) assay by incubating the osteoblast-like cells (MG63) with different concentrations of QD-conjugates for 3 days.40 The absorbance was measured at the wavelength = 550 nm, which is directly correlated with the cell quantity. The cell viability was calculated by assuming 100% viability of the control reference. The MTT results are summarized in Fig. 6, with no significant cellular toxicity observed at the different concentrations of QD nanoparticles conjugated with QD_Chi–OPS, QD_Chi or QD_OPS (after 3 days of culture). Despite some minor reduction on the cell viability at higher concentrations of nanoconjugates, they were not statistically different from the control sample (TCPS). These findings are very relevant as they confirmed the preliminary cytocompatibility of the novel nanoconjugates mostly due to the biocompatible shell capping the fluorescent CdS QDs. Moreover, the results are consistent with the work reported by Higuchi and collaborators41 using specific QD-dendrimers to label hMSC's cells after 1 and 2 days.The cytotoxicity of the QD-nanoconjugates was also evaluated by the resazurin assay, which offers a simple, rapid and sensitive measurement for the viability of mammalian cells. Besides the excellent correlation with MTT assay, resazurin assay indicates that living cells are metabolically active (i.e. mitochondrial metabolic activity) because they are able to reduce the non-fluorescent dye (resazurin) to the strongly-fluorescent dye (resorufin).42 Thus, the results in Fig. 7 indicated that all nanoconjugates produced in this study have presented cell viability statistically comparable to the control reference (100% viable cells, TCPS). These results endorsed the formazan-based assay (MTT) presented in the previous section, giving further evidence that the nanoconjugates made of CdS QDs were effectively functionalized by the biocompatible ligands (chitosan, chitosan–OPS and OPS). For that reason, the resazurin assay was always performed before the cells were evaluated by confocal microscopy, quantitative flow cytometry and TEM techniques to ensure that the cells remain viable and metabolically active after incubation with the different nanoconjugates.
3.3. Targeted uptake of QD-conjugated nanoparticles
In this research, the QD-based nanoconjugates were designed aiming at the potential use for cell-type targeting. For that reason, the peptide O-phospho-L-serine (OPS) was chosen as the specific bio-affinity molecule, which was conjugated with the chitosan polymer (Chi–OPS) and used as the surface-functionalization ligand of the CdS quantum dots (QD_Chi–OPS). The targeting behavior of Chi–OPS was compared to the system using chitosan but without covalently conjugation with OPS (QD_Chi). To test the cell targeting ability of the bio-nanoconjugates, human bone marrow stromal cells (hBMSC) were chosen because they possess the ability to differentiate into various tissues, including bone43 and they can expand in vitro easily. In addition, they have immunosuppressive and immunoevasive capabilities making them a very attractive candidate for bone regeneration in vivo and for clinical applications with a promising potential to be utilized for tissue engineering.44 However, the behavior regarding to the uptake of nanoparticles, intracellular trafficking and endosomal release mechanisms by these hBMSC cells is not properly understood yet.Thus, in this study, confocal fluorescence microscopy and quantitative flow cytometry characterization techniques were used to determine the uptake efficiency of the nanoconjugates (QD_Chi–OPS, QD_Chi and QD_OPS) by the culture of hBMSC cells. The cells were incubated with QD-conjugated nanoparticles for 1 h and, after washing, a microscopic inspection or flow cytometry was performed to examine the probe internalization. It can be observed in the images showed in Fig. 8 that a large amount of hBMSC cells were effectively labelled with the QD_Chi–OPS nanoprobes and fewer cells were probe-positive with the QD_Chi and QD_OPS conjugates. In addition, the fluorescent nanoprobes were accumulated within the cellular cytoplasm, giving strong indication that the QD_Chi–OPS were significantly internalized by the hBMSC cells through endocytic pathways and transported within the endosomal system. Subsequently, they might have been released into the cytoplasm.
These results were confirmed by following the percentage of cells that contained the nanoconjugates using quantitative flow cytometry. In Fig. 9, the systems were excited by a blue laser (λ = 405 nm) and evaluated in the Pacific blue and AmCyan-A channels. The hBMSC cells showed a fluorescent positive population with average intensities of approximately ∼20 times and ∼100 times higher for cells labelled with QD-Chi and QD_Chi–OPS, respectively, when compared to the control (untreated cells TCPS, unstained). Moreover, the percentage of cell positive population of approximately 19.5% for the QD_Chi–OPS conjugates was much higher than the value for the QD_Chi (10.3%), and almost no cells were positive for QD_OPS samples (0.5% – Fig. 9 and 10). These results provided supporting evidence that the nanoconjugates based on the combination of fluorescent QDs with surface-modified ligands of chitosan–OPS (QD_Chi–OPS) have presented the highest uptake efficiency by the hBMSC cells. Therefore, the two major goals of this study were achieved, i.e., to promote the cellular accumulation of QDs and to enhance the endosomal escape of the QDs, while preserving their fluorescent intensity as nanoprobes. Previously, other studies have modified QDs with other types of ligands, such as antibodies45 and sugars46 to enhance the cellular uptake through specific interactions with the target cells. However, these strategies are usually time-intensive and expensive. One of the main advantages of our work was that a biocompatible fluorescent QD was successfully produced for cell bioimaging using a facile process in aqueous medium and at relatively lower costs.
3.4. Visualization of the intracellular localization of the QD particles by TEM
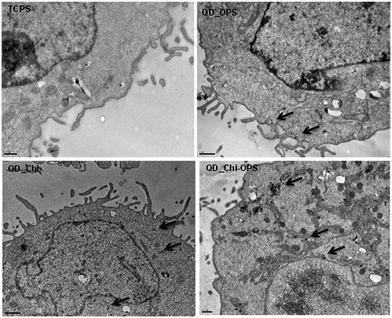
The intracellular localization of the QD-conjugated particles was investigated using transmission electron microscopy (TEM) after 1 h of incubation in the CdS-QDs aqueous colloidal solutions. As showed in Fig. 11, only a relative small portion of the QD nanoconjugates aggregated onto the surface of hBMSC cells, while the majority of the QD probes were transported into the cytoplasm. However, an endosomal structure was not observed inside the cells. The TEM images supported the confocal microscopy results presented in the previous section; only the QD_Chi–OPS conjugated particles were found inside a large number of cells and dispersed across the cytoplasm and membrane (small dark dots). At this point, it is important to highlight that cells are naturally resistant to the up-taking of foreign materials or they cannot completely permeate across cellular membranes.47 In this sense, it is essential to find selective binding systems for targeting the tissue of interest and simultaneously improving the fluorescent properties for cell bioimaging applications.48 Thus, the confocal microscopy and TEM results have clearly demonstrated the endosomal escape of QD_Chi–OPS and QD_Chi, despite the mechanism of the endosomal uptake and later release for these conjugates remains unclear requiring more future investigation.The entire envisioned application of the nanohybrid system developed in this study is schematically depicted in Fig. 12. The CdS/chitosan–OPS and CdS–OPS conjugates may be simultaneously serve as nanoprobes for the labeling and imaging of bone marrow stromal cells and potentially tracking the innumerous events and pathways involved in bone tissue repair and remodeling. Undoubtedly, further studies should be performed by scientists and other specialists to exploit the large number of in vitro and in vivo biomedical applications afforded by the novel fluorescent bioprobes developed in this work.
 | ||
| Fig. 12 Schematic representation of the hypothesized nanohybrid system (not to scale) based on fluorescent core and biofunctionalized surface with chitosan–O-phospho-L-serine peptide. | ||
4. Conclusions
In this study, the synthesis of CdS QDs directly capped with chitosan–O-phospho-L-serine peptide bioconjugates was demonstrated for the first time using “single-step” aqueous colloidal chemistry, and these conjugates exhibit functional properties as fluorophore probes whose in vitro biological affinity can be exploited for the labeling and imaging of bone marrow stromal cells (hBMSC). Moreover, biofunctionalizing QDs with Chi–OPS promoted the highest uptake of the conjugates into primary cultured hBMSCs. After the QDs conjugates were taken up by the cells (hBMSC), most likely via endocytosis pathways, the QDs were distributed throughout the cytoplasm. Larger amounts of labeled cells were observed after incubation with the QD_Chi–OPS solution. Moreover, this cytosolic delivery should extend the intracellular fluorescence intensity in primary cultured hBMSCs in vitro. This strategy may be potentially applied to avail of the numerous opportunities and address the challenges presented in the fields of bone tissue engineering and nanomedicine.Author contributions
The manuscript was written based on the contributions of every author. All of the authors have approved the final version of the manuscript.Conflict of interest
The authors declare that they have no competing or conflicting interests.Acknowledgements
The authors acknowledge the financial support from Brazilian agencies CAPES, FAPEMIG and CNPq. The authors are grateful to the staff of the Microscopy Centre/UFMG for the TEM analyses. This work was co-financed by FEDER funding through Programa Operacional Factores de Competitividade – COMPETE and by national (Portuguese) funds through FCT – Fundação para a Ciência e a Tecnologia within project PEst-C/SAU/LA0002/2013, and FCT again for the Post-Doc grant (SFRH/BPD/84443/2012). We also thank Rui Fernandes (IBMC) for the cellular TEM images and Catarina Leitão (AFCU/IBMC) for the quantitative flow cytometric analyses and the Bioimaging Center for Biomaterials and Regenerative Therapies (b.IMAGE) for help with confocal microscopy.References
- M. J. Seibel, Clin. Biochem. Rev., 2005, 26, 97 Search PubMed.
- A. Merolli and M. Santin, Molecules, 2009, 14, 5367 CrossRef CAS PubMed.
- L. Geris, J. Vander Sloten and H. Van Oosterwyck, Philos. Trans. R. Soc., A, 2009, 367, 2031 CrossRef CAS PubMed.
- C. Vater, A. Lode, A. Bernhardt, A. Reinstorf, C. Heinemann and M. Gelinsky, J. Biomed. Mater. Res., Part A, 2010, 92, 1452 Search PubMed.
- A. N. Tiaden, M. Breiden, A. Mirsaidi, F. A. Weber, G. Bahrenberg, S. Glanz, P. Cinelli, M. Ehrmann and P. J. Richards, Stem Cells, 2012, 30, 2271 CrossRef CAS PubMed.
- L. C. Palmer, C. J. Newcomb and S. R. Kaltz, Chem. Rev., 2008, 108, 4754 CrossRef CAS PubMed.
- E. S. Place, N. D. Evans and M. M. Stevens, Nat. Mater., 2009, 8, 457 CrossRef CAS PubMed.
- K. Aoki, N. Alles, N. Soysa and K. Ohya, Adv. Drug Delivery Rev., 2012, 64, 1220 CrossRef CAS PubMed.
- H. K. Varma, Y. Yokogawa, F. F. Espinosa, Y. Kawamoto, K. Nishizawa, F. Nagata and T. Kameyama, Biomaterials, 1999, 20, 879 CrossRef CAS.
- C. R. Yang, Y. J. Wang and X. F. Chen, Sci. China: Life Sci., 2012, 55, 236 CrossRef CAS PubMed.
- Z. Wu, H. M. Ma, T. Kukita, Y. Nakanishi and H. Nakanishi, J. Immunol., 2010, 184, 3191 CrossRef CAS PubMed.
- A. Reinstorf, M. Ruhnow, M. Gelinsky, W. Pompe, U. Hempel, K. W. Wenzel and P. Simon, J. Mater. Sci.: Mater. Med., 2004, 15, 451 CrossRef CAS.
- A. Reinstorf, U. Hempel, F. Olgemöller, H. Domaschke, W. Schneiders, R. Mai, B. Stadlinger, A. Rösen-Wolff, S. Rammelt, M. Gelinsky and W. Pompe, Materialwiss. Werkstofftech., 2006, 37, 491 CrossRef CAS.
- L. Offer, B. Veigel, T. Pavlidis, C. Heiss, M. Gelinsky, A. Reinstorf, S. Wenisch, K. S. Lips and R. Schnettler, J. Tissue Eng. Regener. Med., 2011, 5, 11 CrossRef CAS PubMed.
- R. Mai, R. Lux, P. Proff, G. Lauer, W. Pradel, H. Leonhardt, A. Reinstorf, M. Gelinsky, R. Jung, U. Eckelt, T. Gedrange and B. Stadlinger, Biomed. Tech., 2008, 53, 229 CrossRef CAS PubMed.
- X. Zhang, X. Zhang, L. Tao, Z. Chi, J. Xu and Y. Wei, J. Mater. Chem. B, 2014, 2, 4398 RSC.
- X. Zhang, X. Zhang, S. Wang, M. Liu, L. Tao and Y. Wei, Nanoscale, 2013, 5, 147 RSC.
- X. Zhang, M. Liu, B. Yang, X. Zhang, Z. Chi, S. Liu, J. Xu and Y. Wei, Polym. Chem., 2013, 4, 5060 RSC.
- X. Zhang, M. Liu, B. Yang, X. Zhang and Y. Wei, Colloids Surf., B, 2013, 112, 81 CrossRef CAS PubMed.
- X. Zhang, X. Zhang, B. Yang, J. Hui, M. Liu, Z. Chi, S. Liu, J. Xuc and Y. Wei, Polym. Chem., 2014, 5, 318 RSC.
- X. Zhang, X. Zhang, B. Yang, Y. Zhang and Y. Wei, ACS Appl. Mater. Interfaces, 2014, 6, 3600 CAS.
- X. Li, X. Zhang, Z. Chi, X. Chao, X. Zhou, Y. Zhang, S. Liu and J. Xu, Anal. Methods, 2012, 4, 3338 RSC.
- E.-S. Kim, E. H. Ahn, T. Dvir and D.-H. Kim, Int. J. Nanomed., 2014, 9, 1 CrossRef PubMed.
- A. Tautzenberger, A. Kovtun and A. Ignatius, Int. J. Nanomed., 2012, 7, 4545 CrossRef CAS PubMed.
- A. A. P. Mansur, J. B. Saliba and H. S. Mansur, Colloids Surf., B, 2013, 111, 60 CrossRef CAS PubMed.
- H. S. Mansur, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 113 CrossRef CAS PubMed.
- H. S. Mansur, A. A. P. Mansur, E. Curti and M. V. de Almeida, J. Mater. Chem. B, 2013, 1, 1696 RSC.
- H. S. Mansur, J. C. González and A. A. P. Mansur, Colloids Surf., B, 2011, 84, 360 CrossRef CAS PubMed.
- Z. Li, P. Huang, J. Lin, R. He, B. Liu, X. Zhang, S. Yang, P. Xi, X. Zhang, Q. Ren and D. Cui, J. Nanosci. Nanotechnol., 2010, 10, 4859 CrossRef CAS PubMed.
- Y. Guo, L. Wang, L. Yang, J. Zhang, L. Jiang and X. Ma, Mater. Lett., 2011, 65, 486 CrossRef CAS PubMed.
- C. K. Dixit, S. R. Vashist, B. D. MacCraith and R. O'Kennedy, Nat. Protoc., 2011, 6, 439 CrossRef CAS PubMed.
- H. S. Mansur, A. A. P. Mansur and J. C. González, Polymer, 2011, 52, 1045 CrossRef CAS PubMed.
- G. T. Hermanson, Bioconjugate Techniques, Elsevier Inc., Amsterdam, 2nd edn, 2008 Search PubMed.
- H. Weller, H. M. Schmidt, U. Koch, A. Fojtik, S. Baral, A. Henglein, W. Kunath, K. Weiss and E. Dieman, Chem. Phys. Lett., 1986, 124, 557 CrossRef CAS.
- J. Tauc and A. Menth, J. Non-Cryst. Solids, 1972, 8–10, 56 Search PubMed.
- V. M. Skobeeva, V. A. Smyntyna, O. I. Sviridova, D. A. Struts and A. V. Tyurin, J. Appl. Spectrosc., 2008, 75, 576 CrossRef CAS.
- V. Smyntyna, V. Skobeeva and N. Malushin, Radiat. Meas., 2007, 42, 693 CrossRef CAS PubMed.
- J. J. Ramsden and M. Gratzel, J. Chem. Soc., Faraday Trans., 1984, 80, 919 RSC.
- R. Khatik, P. Dwivedi, P. Khare, S. Kansal, A. Dube, P. R. Mishra and A. K. Dwivedi, Expert Opin. Drug Delivery, 2014, 11, 633 CrossRef CAS PubMed.
- H. S. Mansur, E. S. Costa Jr, A. A. P. Mansur and E. F. Barbosa-Stancioli, Mater. Sci. Eng., C, 2009, 29, 1574 CrossRef CAS PubMed.
- Y. Higuchi, C. Wu, K. L. Chang, K. Irie, S. Kawakami, F. Yamashita and M. Hashida, Biomaterials, 2011, 32, 6676 CrossRef CAS PubMed.
- S. Anoopkumar-Dukie, J. B. Carey, T. Conere, E. O'Sullivan, F. N. Van Pelt and A. Allshire, Br. J. Radiol., 2005, 78, 945 CrossRef CAS PubMed.
- W. E. Fibbe, Ann. Rheum. Dis., 2002, 61, ii29 CrossRef.
- A. H. Undale, J. J. Westendorf, M. J. Yaszemski and S. Khosla, Mayo Clin. Proc., 2009, 84, 893 CrossRef CAS.
- Y. Ohyabu, Z. Kaul, T. Yoshioka, K. Inoue, S. Sakai, H. Mishima, T. Uemura, S. C. Kaul and R. Wadhwa, Hum. Gene Ther., 2009, 20, 217 CrossRef CAS PubMed.
- Y. Higuchi, M. Oka, S. Kawakami and M. Hashida, J. Controlled Release, 2008, 125, 131 CrossRef CAS PubMed.
- R. L. Juliano, X. Ming and O. Nakagawa, Bioconjugate Chem., 2012, 23, 147 CrossRef CAS PubMed.
- Y. Yan, G. K. Such, A. P. R. Johnston, J. P. Best and F. Caruso, ACS Nano, 2012, 6, 3663 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |