Electrically regulated differentiation of skeletal muscle cells on ultrathin graphene-based films†
Samad Ahadian‡
a,
Javier Ramón-Azcón‡a,
Haixin Chang*a,
Xiaobin Lianga,
Hirokazu Kajib,
Hitoshi Shikuc,
Ken Nakajimaa,
Murugan Ramalingamadef,
Hongkai Wuf,
Tomokazu Matsueac and
Ali Khademhosseini*aghij
aWPI-Advanced Institute for Materials Research, Tohoku University, Sendai 980-8577, Japan. E-mail: hxchang@wpi-aimr.tohoku.ac.jp
bDepartment of Bioengineering and Robotics, Graduate School of Engineering, Tohoku University, Sendai 980-8579, Japan
cGraduate School of Environmental Studies, Tohoku University, Sendai 980-8579, Japan
dCentre for Stem Cell Research, A unit of the Institute for Stem Cell Biology and Regenerative Medicine, Christian Medical College Campus, Vellore 632002, India
eInstitut National de la Santé Et de la Recherche Médicale U977, Faculté de Chirurgie Dentaire, Université de Strasbourg, Strasbourg 67085, France
fDepartment of Chemistry, The Hong Kong University of Science and Technology, Clear water Bay, Hong Kong
gDepartment of Medicine, Center for Biomedical Engineering, Brigham and Women's Hospital, Harvard Medical School, Cambridge, Massachusetts 02139, USA
hHarvard–MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA
iWyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts 02115, USA
jDepartment of Maxillofacial Biomedical Engineering and Institute of Oral Biology, School of Dentistry, Kyung Hee University, Seoul 130-701, Republic of Korea. E-mail: alik@rics.bwh.harvard.edu
First published on 13th January 2014
Abstract
The electrical conductivity of graphene provides a unique opportunity to modify the behavior of electrically sensitive cells. Here, we demonstrate that C2C12 myoblasts that were cultured on ultrathin thermally reduced graphene (TR-Graphene) films had more favorable cell adhesion and spreading compared to those on graphene oxide (GO) and glass slide substrates, comparable with conventional Petri dish. More importantly, we demonstrate that electrical stimulation significantly enhanced myoblast cell differentiation on a TR-Graphene substrate compared to GO and glass slide surfaces as confirmed by the expression of myogenic genes and proteins. These results highlight the potential applications of graphene-based materials for cell-based studies, bioelectronics, and biorobotics.
1. Introduction
Graphene is a sheet of hexagonally bonded carbon atoms that forms a two-dimensional (2D) honeycomb lattice structure with a single-carbon-atom thickness. Since its discovery in 2004,1 graphene has found tremendous interest and applications in nanoscience and nanotechnology.2–5 Graphene and its derivatives have attracted much attention as functional materials in different biomedical applications, such as drug and gene delivery,6 photothermal tumor therapy,7 biosensing,8,9 cell imaging and monitoring,10,11 cell proliferation and differentiation,12–15 and regenerative medicine16 due to their unique electronic, mechanical, chemical, and optical properties. Graphene-based materials are highly stable in aqueous solutions17 and exhibit low inflammatory responses.18 In addition, physicochemical properties of these materials, such as surface chemistry, can be precisely controlled. Therefore, graphene-based materials can provide an attractive substrate for various cell studies. In a previous work, we demonstrated that skeletal muscle cells were able to proliferate and differentiate on both graphene oxide (GO) and thermally reduced graphene (TR-Graphene) substrates19 that is in agreement with the recent work by Ku and Park.20 Functional materials to regulate behaviors of skeletal muscle cells and other electrically active cells should be biocompatible, cell-adhesive, and conductive.21 This latter conductive property is often missing in the conventional materials for muscle cells.22 However, despite unique electrical properties of graphene,1 to our knowledge, graphene-based substrates have not been used to electrically alter skeletal muscle cell behaviors. In fact, it is still challenging to achieve electrical control of muscle cell behaviors on graphene-based materials.To address this challenge and assess the effect of electrical stimulation (ES) on the differentiation of muscle cells on graphene-based materials, we synthesized and characterized the ultrathin GO and TR-Graphene on glass slides and employed them as substrates to study the skeletal muscle cell adhesion, proliferation, and differentiation. Finally, a technique was developed to fabricate free-standing, mechanically robust, highly flexible, and contractile graphene cell sheets toward applications related to biosensing and bio-actuation.
2. Results and discussion
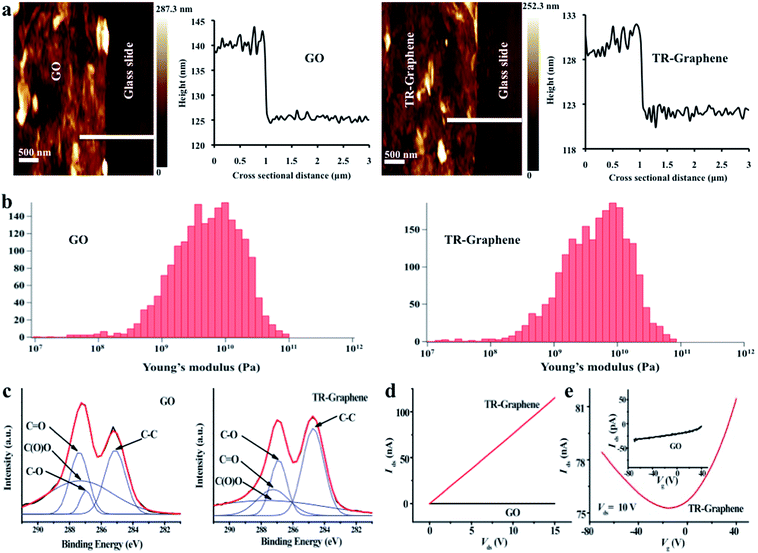
Ultrathin graphene-based films were fabricated by solution casting of GO on the glass substrates and then thermally reducing them at low temperature. The thermal reduction of GO to TR-Graphene was confirmed by Raman spectroscopy as reported in our previous work.2 The intensity ratio of D-band to G-band in Raman spectra decreased from 1.26 for GO to 1.16 for TR-Graphene due to the removal of oxygenous groups after the reduction process. Morphology, thickness, and mechanical properties of ultrathin GO and TR-Graphene films on the glass slide were determined by atomic force microscopy (AFM). Fig. 1a and b show the thickness and mechanical properties of GO and TR-Graphene films. The GO thickness was 13.8 ± 3.3 nm, while the TR-Graphene had a thickness of 8.0 ± 4.3 nm indicating a few-layer stacking of both materials on the glass slide. In addition, AFM studies showed that the morphology of GO and TR-Graphene films on the glass slide had a roughness of 6.3 ± 2.1 and 6.0 ± 3.6 nm for the GO and TR-Graphene substrates, respectively (ESI, Fig. S1†). The thickness and roughness values reported above were the average of 15 independent measurements of 5 different AFM images. The lateral dimension of both materials also varied from a few hundred nanometers to a few micrometers as measured by the AFM images (Fig. S1†). Both GO and TR-Graphene substrates exhibited high and almost similar stiffness. The average Young's modulus for GO and TR-Graphene were determined to be ∼4.9 and 4.2 GPa, respectively (Fig. 1b). The Young's modulus for the bare glass slide has been reported to be ∼70 GPa.23 Therefore, the graphene substrates had a lower stiffness compared to glass slide. The average Young's modulus for GO and TR-Graphene reported here are in agreement with previously reported measurements.24 It is known that substrate stiffness is a key factor for regulating cell behaviors, such as cell adhesion and differentiation.25We modified the reduction states of the graphene films by thermal reduction, which removed some of the oxygenous groups on the GO surfaces as confirmed by X-ray photoelectron spectroscopy (XPS) measurements (Fig. 1c). This is in accordance with our previous results in which water contact angle measurements on the GO and TR-Graphene substrates (26.5 ± 7.3° and 60.4 ± 3.3°, respectively)19 indicated that TR-Graphene substrates were more hydrophobic compared to the GO substrates. It is important to note that our TR-Graphene substrates were only partially reduced and contained some remaining oxygen-containing side groups on their surface.
The electrical conductivity of the ultrathin GO and TR-Graphene films was also analyzed (Fig. 1d and e). The TR-Graphene substrate showed over 3 orders of magnitude higher conductivity than the GO substrate. Field effect measurements of GO and TR-Graphene substrates were performed by using gold as source and drain electrodes, and highly doped silicon as the gate. Interestingly, the TR-Graphene substrate here showed a bipolar field effect behavior, similar with the single-layer graphene,26 while no such field effect was found in GO substrate. These measurements indicated that TR-Graphene substrates were more conductive compared to GO substrates.
We assessed the biocompatibility of ultrathin GO and TR-Graphene films for their use as substrates for C2C12 myoblast cell culture. In particular, we analyzed cell viability on various substrates to determine the cytotoxicity of these materials. The results showed that the GO and TR-Graphene substrates were non-toxic to C2C12 myoblasts as well as resulted in cell proliferation during the five days of culture (ESI, Fig. S2†). Note that the calcein AM/ethidium homodimer live/dead assay was used for quantifying the muscle cell proliferation as a standard method to measure the cell proliferation on graphene substrates.27,28 Interestingly, there was no statistical difference between the C2C12 cell growth on the GO and TR-Graphene substrates and that of cells on the glass slide. These results are in agreement with those shown for other cell types, such as human neural29 and mesenchymal stem cells.13
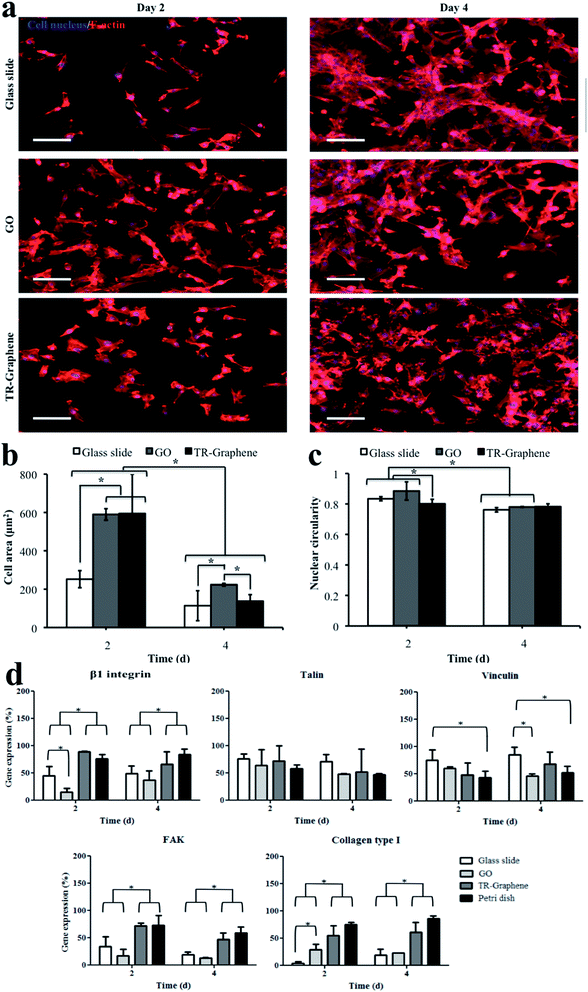
We also analyzed the effects of various substrates on C2C12 morphology and adhesion on days 2 and 4 of culture by visualizing cytoskeletal structures (Fig. 2). C2C12 myoblasts that were cultured on the glass slides showed a spindle-like shape, while those on the GO and TR-Graphene substrates were more spread (Fig. 2a). This was probably due to higher surface roughness and consequently increased number of anchoring sites of the GO and TR-Graphene substrates compared to the glass slide substrate that promoted the cell adhesion and extension.30,31 The cell area quantification on days 2 and 4 of culture confirmed these observed morphologies as to have smaller cell area on the glass slide compared to those on the GO and TR-Graphene substrates (Fig. 2b). The cell area values at day 4 of culture were smaller than those at day 2 because the cells were more confluent, which limited the spreading of individual cells. In the previous work,19 we showed that TR-Graphene substrates were able to adsorb a higher amount of proteins (i.e., fibronectin, fetal bovine serum, and fibroblast growth factor proteins) compared with GO and glass slide substrates. Cell attachment, spreading, and proliferation phenomena on a substrate have a direct relationship with the amount of proteins adsorbed on the substrate because cell–substrate interactions are the result of complex biological phenomena of protein adsorption, receptor–ligand binding, and signal transduction.32 Adsorbed proteins on the substrate increase the cell adhesion and regulate consequent cellular behaviors.33 Since TR-Graphene substrates were able to adsorb more proteins compared with GO and glass slide substrates, they could adsorb more proteins from the culture medium and these proteins promoted the muscle cell attachment and spreading on the TR-Graphene compared with GO and glass slide. It was also possible that the adsorbed proteins changed the surface energy of TR-Graphene substrates because it has been shown that the amount of proteins adsorbed on the materials could change their surface energy.33 The surface energy of a substrate has a substantial effect on regulating cell attachment, spreading, and proliferation.34 Therefore, it seems that changing the surface energy of TR-Graphene due to the adsorbed proteins from the culture medium favored higher muscle cell attachment and spreading compared with that of GO and glass slide. Interestingly, the nuclear shape index (circularity) at day 2 of culture for the myoblasts on the TR-Graphene substrates was significantly lower than those for the GO and glass slide substrates (Fig. 2c). Nuclear circularity has been demonstrated to have correlations with different cell behaviors, such as adhesion, spreading, and proliferation.35 In particular, it has been shown that the C2C12 cell elongation occurs during the early stages of muscle cell differentiation.36,37 Therefore, it was expected that the cells on the TR-Graphene substrate would have higher differentiation rate compared to those on the GO and glass slide substrates. The nuclear circularity values at day 4 of culture for the cells on the GO and glass slide substrates (0.781 ± 0.003 and 0.761 ± 0.014, respectively) significantly decreased compared to those at day 2 (0.885 ± 0.059 and 0.834 ± 0.013, respectively; p < 0.01 for both cases). However, there was no statistical difference between the nuclear circularities at day 4 for all substrates, indicating the C2C12 myoblasts reached the minimum nuclear circularity for growth and differentiation.
Fig. 2d shows the gene expression profiles for adhesion- and growth-related genes (i.e., β1 integrin, collagen type I, and focal adhesion kinase (FAK)) and focal adhesion components, including talin and vinculin for the C2C12 myoblasts on the glass slide, GO, and TR-Graphene substrates and conventional Petri dish as revealed by the conventional RT-PCR technique. Integrin is a receptor that causes the attachment between a cell and its surrounding matrix and it plays a role in migration and invasion.38 Collagen type I provides mechanical strength and adhesive domains in the extracellular matrix.28 FAK plays an important role in regulating downstream signaling of integrin, integrin-dependent signaling related to cell survival, and growth factor receptors.39 Talin and vinculin are required for focal adhesion assembly to increase the coupling strength between cell cytoskeleton and integrin and help in formation of initial nanocomplex clusters. Both vinculin and talin are responsible for the cell adhesion response. Most importantly, with regards to the gene analysis results, the TR-Graphene substrates were a suitable substrate for the C2C12 muscle cell adhesion and proliferation and its performance was comparable with that of conventional Petri dish substrate, particularly in the case of β1 integrin, FAK, and collagen type I genes. Both GO and glass slide substrates exhibited less gene expression of β1 integrin, FAK, and collagen type I compared to the TR-Graphene substrate. The observed differences in the muscle cell adhesion and spreading on the TR-Graphene and GO substrates likely stem from the combined discrepancies between the morphological, mechanical, and surface chemical properties of the two substrates.
We further studied the performance of GO and TR-Graphene substrates to differentiate C2C12 myoblasts into myotubes (Fig. 3). In these experiments, after 1 day of culture, the growth medium for the cells was changed to the differentiation medium and the cells were induced to undergo differentiation process for 9 days of culture. During the differentiation period, the cells expressed myosin heavy chain protein and fused together to form C2C12 myotubes.40 The myotube analysis was performed for two groups of myotubes at day 10 of culture; the first group was subjected to ES at day 8 of culture for two continuous days (voltage 8 V, frequency 1 Hz, and duration 10 ms) whereas no ES was applied to the second group as shown in Fig. 3a. The presence of an electric field within the Petri dish due to the ES was verified by the simulation of the potential and current density (Fig. S3†). Enhanced differentiation of C2C12 myoblasts on the TR-Graphene substrate compared to the GO and glass slide substrates was observed even without applying the electric field. The length for the C2C12 myotubes on the GO and TR-Graphene substrates was significantly longer than those on the glass slide, while there was more positive regions of the myosin heavy chain on the TR-Graphene substrate compared to the GO and glass slide substrates. Differences between the morphological, mechanical, chemical, and electrical properties of two substrates could cause this observed difference in the muscle cell differentiation on the GO and TR-Graphene substrates. However, it seems that the morphological changes of graphene substrates had minor effect on this phenomenon because a recent study demonstrated that C2C12 myoblasts differentiated more on the GO substrate compared to the reduced GO substrates despite a similar morphology.20 In agreement with our results, Jun et al.41 demonstrated that more C2C12 myoblasts were able to differentiate on the conductive fibers composed of poly (L-lactide-co-ε-caprolactone) (PLCL) and polyaniline compared to non-conductive PLCL fibers even without the application of electrical stimulus, indicating the important role of the substrate conductivity on myogenic differentiation. High performance of graphene substrates for the differentiation of other electrophysiologically active cells has been also shown,42 where the growth and neurite sprouting of mouse hippocampal cells was promoted on the graphene compared to those on the conventional tissue culture plates, while there was no obvious disparity between the morphology of two substrates.
Chemical properties of GO and TR-Graphene substrates also appear to play a crucial role in C2C12 myoblast differentiation. As shown previously,19 TR-Graphene substrates were able to physisorb higher amounts of fibronectin, fetal bovine serum, and fibroblast growth factor compared to GO and glass slide substrates. This result is in agreement with previous studies indicating that graphene-based materials were able to bind to serum proteins.14 Note that precise tuning of oxygen-contained functional groups on the GO substrate is needed to have the optimum cellular adhesion, proliferation, and phenotype. In this regard, we previously found that moderately reduced GO substrate (90 min treatment), similar to the TR-Graphene substrate in this study, demonstrated an improved performance relative to GO and highly reduced GO (260 min treatment) for inducing C2C12 myoblast differentiation.19 This is also consistent with a recent report in which highly reduced graphene, as generated by a chemical method, induced less myogenic differentiation relative to GO.20
Importantly, TR-Graphene substrates appeared to accelerate the differentiation of C2C12 myoblasts upon the application of an ES compared to the GO and glass slide substrates. Both myotube length and coverage area values for the C2C12 myotubes electrically stimulated on the TR-Graphene substrate (272 ± 128 μm and 29 ± 2%, respectively) were significantly increased compared to those for the non-stimulated myotubes (179 ± 77 μm and 18 ± 3%, respectively). Although the same trend was observed when comparing the electrically stimulated C2C12 myotubes to non-stimulated ones cultured on the GO and glass slide substrates, the coverage area changes for TR-Graphene due to the ES is about 2 folds of that for GO and glass slide.
Genes related to the differentiated myotubes and contraction ability (i.e., muscle regulatory factor 4 (MRF4), sarcomeric actin, α-actinin, and myosin heavy chain isofactor IId/x (MHC-IId/x)) were significantly up-regulated for the C2C12 myotubes obtained on the TR-Graphene substrate compared to those on the GO and glass slide substrates with and without applying the ES (Fig. 4). A significant effect of ES for the muscle tissue maturation and contraction was observed for all underlying cases. However, this effect was more profound for the TR-Graphene substrate compared to the GO and glass slide substrates. For instance, the expression level of MHC-IId/x for electrically stimulated C2C12 myotubes on TR-Graphene substrates increased over 10-fold compared to that for non-stimulated myotubes, while the expression levels of MHC-IId/x did not significantly change for C2C12 myotubes on GO and glass slide before and after ES (Fig. 4d). The contractility of C2C12 myotubes cultured on the TR-graphene substrate was also observed and recorded (see ESI, Movie S1†).
Recently, there has been great interest to increase the conductivity of currently used scaffolds for the electrically active cells using nanomaterials.43 For instance, gold nanostructures were impregnated into degradable biomaterials to increase the electrical conductivity and cellular excitability of both cardiomyocyte and neural cells.21,44 Also, poly(lactic-co-glycolic-acid) (PLGA)-carbon nanofiber composite materials were shown to have higher conductivity compared to the pure PLGA fibers.45 These hybrid materials also showed higher adhesion and proliferation of both cardiomyocytes and neural cells. Although graphene possesses high electrical conductivity, to our knowledge, this feature has not been exploited to enhance muscle cell differentiation. Park et al. used a graphene substrate as the stimulating electrode for neural stem cells.29 They showed that these cells were more electrophysiologically active upon applying the electrical field through the graphene electrodes compared to non-stimulated samples. Here, we used the graphene for an efficient cell differentiation and consequently myofiber fabrication particularly upon applying the ES that has not been previously reported. Indeed, various cell behaviors, such as cell migration, proliferation, differentiation, and apoptosis can be regulated by applying the electrical fields.46 Therefore, many research opportunities exist to increase the yield of cell and tissue responses to direct and alternative electric fields using the graphene substrates. Other advantages of these materials include their exceptional optical transparency and biocompatibility. In addition, the electronic properties of graphene-based materials can be precisely controlled.47 Therefore, they can be adjusted for various cellular interfacing, stimulation regimes, and monitoring applications.48
As a potential application of our system we generated contractile, free-standing graphene/muscle myofiber structures. To do so, poly(N-isopropylacrylamide) (PNIPAm) was deposited on a glass slide and TR-Graphene was then fabricated on the PNIPAm-coated glass slides. We cultured C2C12 myoblasts on these substrates and induced the differentiation process at day 1 of culture. After 10 days of culture, the muscle myofibers were formed on the TR-Graphene substrates according to the same ES protocol to fabricate C2C12 myotubes on TR-Graphene substrates as detailed above and in experimental section (ESI, Fig. S4†). The resulted cell sheet-graphene was stable at 37 °C on the glass slide and was then released from the glass slide at room temperature to form free-standing graphene–cell sheets by dissolving PNIPAm in culture medium at room temperature. The free-standing muscle myofibers on TR-Graphene exhibited contractility upon applying the electric field (ESI, Movie S2†). Note that here the electric field was employed just to demonstrate the contractility of prefabricated muscle myofibers on free-standing graphene. A rough calculation showed that the myotube displacement on the free-standing TR-Graphene was ∼10 times more than that on TR-Graphene substrate. This is most likely due to a more deformable substrate in the free-standing sheets compared to the rigid substrate, as shown in the ESI, Movie S3.† Taken together, this strategy suggests fabricating free-standing, mechanically robust, highly flexible, and contractile graphene cell sheets toward applications in bio-actuators.
3. Conclusions
In summary, we showed electrical stimulation as a powerful tool to enhance the differentiation of skeletal muscle cells on ultrathin graphene-based films compared to those on the counterpart GO and glass slide substrates. In addition, C2C12 myoblasts on TR-Graphene showed similar adhesion and proliferation behaviors to those on the conventional Petri dish. A strategy was also proposed to fabricate free-standing, mechanically robust, highly flexible, and contractile graphene cell sheets toward applications in bio-actuators. These results open a new path to control the cell behavior on graphene-based scaffolds for biological and bio-electronic applications.Acknowledgements
Samad Ahadian acknowledges Dr Toshinori Fujie, Tohoku University for the help with the PNIPAm. This work was supported by the World Premier International Research Center Initiative (WPI), MEXT, Japan.Notes and references
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang and S. V. Dubonos, et al., Science, 2004, 306, 666 CrossRef CAS PubMed.
- H. Chang, Z. Sun, Q. Yuan, F. Ding, X. Tao and F. Yan, et al., Adv. Mater., 2010, 22, 4872 CrossRef CAS PubMed.
- H. Chang, G. Wang, A. Yang, X. Tao, X. Liu and Y. Shen, et al., Adv. Funct. Mater., 2010, 20, 2893 CrossRef CAS.
- K. S. Novoselov, V. I. Fal'ko, L. Colombo, P. R. Gellert, M. G. Schwab and K. Kim, Nature, 2012, 490, 192 CrossRef PubMed.
- H. Chang and H. Wu, Adv. Funct. Mater., 2013, 23, 1984 CrossRef.
- L. Feng, S. Zhang and Z. Liu, Nanoscale, 2011, 3, 1252 RSC.
- K. Yang, S. Zhang, G. Zhang, X. Sun, S.-T. Lee and Z. Liu, Nano Lett., 2010, 10, 3318 CrossRef CAS PubMed.
- H. Chang, L. Tang, Y. Wang, J. Jiang and J. Li, Anal. Chem., 2010, 82, 2341 CrossRef PubMed.
- S. He, B. Song, D. Li, C. Zhu, W. Qi and Y. Wen, et al., Adv. Funct. Mater., 2010, 20, 453 CrossRef.
- A. Kolmakov, D. A. Dikin, L. J. Cote, J. Huang, M. K. Abyaneh and M. Amati, et al., Nat. Nanotechnol., 2011, 6, 651 CrossRef CAS PubMed.
- T. Cohen-Karni, Q. Qing, Q. Li, Y. Fang and C. M. Lieber, Nano Lett., 2010, 10, 1098 CrossRef CAS PubMed.
- H. Chen, M. B. Müller, K. J. Gilmore, G. G. Wallace and D. Li, Adv. Mater., 2008, 20, 3557 CrossRef CAS.
- T. R. Nayak, H. Andersen, V. S. Makam, C. Khaw, S. Bae and X. Xu, et al., ACS Nano, 2011, 5, 4670 CrossRef CAS PubMed.
- W. C. Lee, C. H. Y. X. Lim, H. Shi, L. A. L. Tang, Y. Wang and C. T. Lim, et al., ACS Nano, 2011, 5, 7334 CrossRef CAS PubMed.
- S. Park, N. Mohanty, J. W. Suk, A. Nagaraja, J. An and R. D. Piner, et al., Adv. Mater., 2010, 22, 1736 CrossRef CAS PubMed.
- Y. Zhang, T. R. Nayak, H. Hong and W. Cai, Nanoscale, 2012, 4, 3833 RSC.
- D. Li, M. B. Müller, S. Gilje, R. B. Kaner and G. G. Wallace, Nat. Nanotechnol., 2008, 3, 101 CrossRef CAS PubMed.
- V. C. Sanchez, A. Jachak, R. H. Hurt and A. B. Kane, Chem. Res. Toxicol., 2012, 25, 15 CrossRef CAS PubMed.
- X. Shi, H. Chang, S. Chen, C. Lai, A. Khademhosseini and H. Wu, Adv. Funct. Mater., 2012, 22, 751 CrossRef CAS.
- S. H. Ku and C. B. Park, Biomaterials, 2013, 34, 2017 CrossRef CAS PubMed.
- S. Ahadian, S. Ostrovidov, V. Hosseini, H. Kaji, M. Ramalingam and H. Bae, et al., Organogenesis, 2013, 9, 87 CrossRef PubMed.
- J. Ramón-Azcón, S. Ahadian, M. Estili, X. Liang, S. Ostrovidov and H. Kaji, et al., Adv. Mater., 2013, 25, 4028 CrossRef PubMed.
- A. Schneider, G. Francius, R. Obeid, P. Schwinté, J. Hemmerlé and B. Frisch, et al., Langmuir, 2006, 22, 1193 CrossRef CAS PubMed.
- O. C. Compton, D. A. Dikin, K. W. Putz, L. C. Brinson and S. B. T. Nguyen, Adv. Mater., 2010, 22, 892 Search PubMed.
- D. E. Discher, P. Janmey and Y. Wang, Science, 2005, 310, 1139 CrossRef CAS PubMed.
- C. Gómez-Navarro, R. T. Weitz, A. M. Bittner, M. Scolari, A. Mews, M. Burghard and K. Kern, Nano Lett., 2007, 7, 3499 CrossRef PubMed.
- C. Cha, S. R. Shin, X. Gao, N. Annabi, M. R. Dokmeci and X. Tang, et al., Small DOI:10.1002/smll.201302182 , in press.
- S.-R. Ryoo, Y.-K. Kim, M.-H. Kim and D.-H. Min, ACS Nano, 2010, 4, 6587 CrossRef CAS PubMed.
- S. Y. Park, J. Park, S. H. Sim, M. G. Sung, K. S. Kim and B. H. Hong, et al., Adv. Mater., 2011, 23, H263 CrossRef CAS PubMed.
- H. N. Kim, A. Jiao, N. S. Hwang, M. S. Kim, D. H. Kang and D.-H. Kim, et al., Adv. Drug Delivery Rev., 2013, 65, 536 CrossRef CAS PubMed.
- T. Fujie, S. Ahadian, H. Liu, H. Chang, S. Ostrovidov and H. Wu, et al., Nano Lett., 2013, 13, 3185 CrossRef CAS PubMed.
- X. Li, H. Liu, X. Niu, B. Yu, Y. Fan and Q. Feng, et al., Biomaterials, 2012, 33, 4818 CrossRef CAS PubMed.
- M. Rouahi, O. Gallet, E. Champion, J. Dentzer, P. Hardouin and K. Anselme, J. Biomed. Mater. Res., Part A, 2006, 78A, 222 CrossRef CAS PubMed.
- S. A. Redey, M. Nardin, D. Bernache-Assolant, C. Rey, P. Delannoy and L. Sedel, et al., J. Biomed. Mater. Res., Part A, 2000, 50, 353 CrossRef CAS.
- J. Ramón-Azcón, S. Ahadian, R. Obregón, G. Camci-Unal, S. Ostrovidov and V. Hosseini, et al., Lab Chip, 2012, 12, 2959 RSC.
- N. T. Swailes, M. Colegrave, P. J. Knight and M. Peckham, J. Cell Sci., 2006, 119, 3561 CrossRef CAS PubMed.
- H. Musa, C. Orton, E. E. Morrison and M. Peckham, J. Muscle Res. Cell Motil., 2003, 24, 301 CrossRef CAS.
- J. D. Hood and D. A. Cheresh, Nat. Rev. Cancer, 2002, 2, 91 CrossRef PubMed.
- S. K. Mitra, D. A. Hanson and D. D. Schlaepfer, Nat. Rev. Mol. Cell Biol., 2005, 6, 56 CrossRef CAS PubMed.
- S. Ahadian, J. Ramón-Azcón, S. Ostrovidov, G. Camci-Unal, V. Hosseini and H. Kaji, et al., Lab Chip, 2012, 12, 3491 RSC.
- I. Jun, S. Jeong and H. Shin, Biomaterials, 2009, 30, 2038 CrossRef CAS PubMed.
- N. Li, X. Zhang, Q. Song, R. Su, Q. Zhang and T. Kong, et al., Biomaterials, 2011, 32, 9374 CrossRef CAS PubMed.
- T. Dvir, B. P. Timko, D. S. Kohane and R. Langer, Nat. Nanotechnol., 2011, 6, 13 CrossRef CAS PubMed.
- J.-O. You, M. Rafat, G. J. C. Ye and D. T. Auguste, Nano Lett., 2011, 11, 3643 CrossRef CAS PubMed.
- D. A. Stout, B. Basu and T. J. Webster, Acta Biomater., 2011, 7, 3101 CrossRef CAS PubMed.
- M. Hronik-Tupaj and D. L. Kaplan, Tissue Eng., Part B: Rev., 2012, 18, 167 CrossRef CAS PubMed.
- N. A. Kotov, J. O. Winter, I. P. Clements, E. Jan, B. P. Timko and S. Campidelli, et al., Adv. Mater., 2009, 21, 3970 CrossRef CAS.
- R. Obregón, S. Ahadian, J. Ramón-Azcón, L. Chen, T. Fujita and H. Shiku, et al., Biosens. Bioelectron., 2013, 50, 194 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S4, Table S1, Experimental Section, and Movies S1–S3. See DOI: 10.1039/c3ra46218h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |