Computational insight into conformational states of glucagon-like peptide-1 receptor (GLP-1R) and its binding mode with GLP-1
Abstract
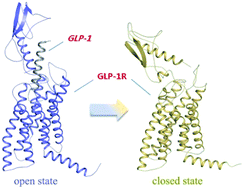
The glucagon-like peptide-1 receptor (GLP-1R) has captivated researchers because of its tremendous therapeutic effects for the treatment of type 2 diabetes mellitus (T2DM). However, since the full-length crystal structure of GLP-1R has not been revealed yet, the molecular binding mode and the activation mechanism remain unclear, which will be the obstacle for the discovery of novel potent GLP-1R agonists. In the present study, we constructed the model of GLP-1R in its full length and explored the binding modes between GLP-1 and GLP-1R by means of a bunch of computational methods including homology modeling, protein–protein docking, and molecular dynamics simulations. Our model is in agreement with previous experiment and the results from our MD simulations that verified the binding modes between GLP-1 and GLP-1R are reasonable. What's more, we found the absence or presence of GLP-1 significantly affected the conformation of extracellular domain (ECD) of GLP-1R. The GLP-1R in the apo form stabilized in a ‘closed’ state which is unfavorable to the binding of GLP-1, resembling as the GCGR. By contrast, in the GLP-1/GLP-1R complex, GLP-1R maintained an ‘open’ state.

 Please wait while we load your content...
Please wait while we load your content...