Recent advances and challenges in developing electrochemiluminescence biosensors for health analysis
Yuxi
Wei
,
Honglan
Qi
 and
Chengxiao
Zhang
and
Chengxiao
Zhang
 *
*
Key Laboratory of Analytical Chemistry for Life Science of Shaanxi Province, School of Chemistry and Chemical Engineering, Shaanxi Normal University, Xi’an, 710062, P. R. China. E-mail: cxzhang@snnu.edu.cn
First published on 13th February 2023
Abstract
This Feature Article simply introduces principles and mechanisms of electrochemiluminescence (ECL) biosensors for the determination of biomarkers and highlights recent advances of ECL biosensors on key aspects including new ECL reagents and materials, new biological recognition elements, and emerging construction biointerfacial strategies with illustrative examples and a critical eye on pitfalls and discusses challenges and perspectives of ECL biosensors for health analysis.
1. Introduction
World demography is changing (11% of the world's population is over 60 years of age by 2020; increased to 22% of the population by 2050).1 The expansion of the number of patients with chronic or acute diseases as well as cancer raises serious issues in the sustainability of health- and social-care systems.2 In 1998, the definition of a biomarker was: “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention”.3 Biomarkers are excreted from one or combination of body fluids (blood, urine, saliva, breath, sweat, seminal fluid, nipple aspirate fluid, tears, stool, interstitial fluid, and cerebrospinal fluid) and encompass a wide variety of molecules, including DNA, mRNA, enzymes, metabolites, transcription factors, and cell surface receptors (DNA, mRNA, enzymes, proteins, small molecules, cancer cells).4 For example, the goal of the cancer biomarker field is to develop reliable, cost-effective, powerful detection, and monitor strategies for cancer risk indication, early cancer detection, and tumor classification so that the patient can receive the most appropriate therapy and physicians can monitor the disease progression, regression, and recurrence. In the past few decades, significant and substantial progress has been made in this field. The early and accurate screening and detection of disease biomarkers is highly important for healthy life throughout lifestyle-related issues.From the view of analytical chemistry, the methods can be classified into two kinds including separation-based methods such as, high performance liquid chromatography, and electrophoresis, and molecular recognition-based methods, such as chemical sensors and biosensors. Biosensors are analytical devices incorporating a biorecognition material (e.g., nucleic acids, enzymes, antibodies, aptamers, and synthetic receptors) intimately associated with or integrated within a physicochemical transducer or transducing microsystem, which converts biological recognition events into a quantitative response, which may be optical, electrochemical, thermometric, piezoelectric, magnetic, or micromechanical. Biosensors offer new powerful analytical tools that are applicable for solving many challenging problems in health analysis. A level of sophistication and state-of-the art technology are commonly employed to produce easy-to-use, compact, and inexpensive devices.
Electrochemiluminescence (also electrogenerated chemiluminescence, ECL) is a kind of luminescence that occurs at/near the surface of electrode as a result of electrochemical reactions and chemiluminescence (CL) reactions. For example, ECL methods based on the Ru(bpy)32+-tripropylamine (TPA) system have extremely low detection limit at subpicomolar concentrations, extremely wide dynamic range, and easy application coupled with an automated ECL analyzer.5 ECL biosensors convert biological recognition events into a quantifiable ECL signal. From the view of the detection principle, ECL biosensors are different from electrochemical biosensors and fluorescent (FL) biosensor as well as chemiluminescence (CL) biosensors. In electrochemical biosensors, electrical signals directly come from the electrode, in which the conductivity of the electrode and electrode-modified materials as well as solution is required, while in FL and CL biosensors, the light transmissivity of the substrates and solution is required. In ECL biosensors, both conductivity and light transmissivity are required. In addition, ECL biosensors are scarcely affected by the background current and potential window of electrodes compared with electrochemical biosensors. Importantly, ECL biosensors exhibit much lower detection limits for signal reagents compared with fluorescent and electrochemical biosensors. In effect, in the design of the ECL biosensors, four issues must be clearly considered: (1) target analyte (biomarker), (2) biorecognition elements/material, (3) ECL signal regents to produce the detection signal (light), and (4) association or integration of the target, biorecognition elements, and ECL signal regents within an electrode transducing microsystem. In ECL measurement, the produced light as the detectable signal is recorded using light-electronics devices such as photomultiplier tube (PMT) and charge coupled device (CCD).
There are numerous reviews and features for ECL biosensors in recent years on ECL mechanisms,6,7 ECL nanomaterials,8–11 biosensing strategies,12–17 and ECL analytical applications.18,19 This Feature Article provides a didactic examination of the concepts and approaches related to new biological recognition elements, new ECL reagents and materials, and emerging construction of biointerfacial strategies in ECL biosensors for biomarkers, mainly contributed by our group and partly done by other groups recently, as shown in Fig. 1. The strengths and pitfalls of each approach are discussed with a critical eye. The challenges and perspectives of ECL biosensors for health analysis are discussed. It is hoped that this Feature Article will inspire broader interests across various disciplines and will stimulate more exciting developments in this still young yet very promising field for the benefit of human health analysis.
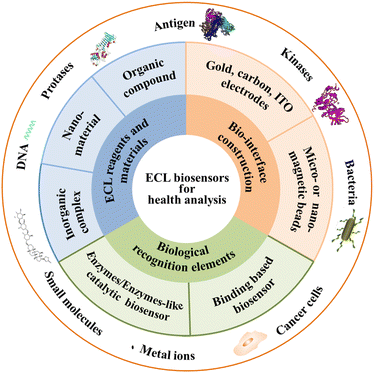 | ||
| Fig. 1 Scheme of the main structure and scope of ECL biosensors for health analysis in this article. | ||
2. New ECL reagents and nanomaterials
ECL reagents and materials (luminophores) play a significant role in fundamental research and applications of ECL biosensors since the sensitivity and detection limit of the ECL biosensors depended on the ECL properties of the ECL reagents and materials. From point of the ECL reaction mechanisms, there are ion annihilation process and co-reactant ECL processes.6,7 Ion annihilation ECL process involves the formation of an excited state as a result of an exergonic electron transfer between electrogenerated species, radical ions, at two electrodes, mainly involving organic compounds in organic solvents. The ion annihilation process is generally performed in organic solvents to obtain relatively stable radical cations and anions. The ECL in organic solvents is not capable of applications in ECL biosensors since biochemical interactions are mainly conducted in aqueous solution. In the ECL co-reactant process, ECL emissions are mainly generated with one directional potential scanning at an electrode in a solution containing luminophore and co-reactant, or at luminophore-modified electrode in co-reactant solution. Both the luminophore and the co-reactant species can be first oxidized or reduced, and then the intermediates produced from the oxidized or reduced co-reactant decompose to produce powerful reducing or oxidizing species, and finally a chemical reaction between the oxidized/reduced luminophore and the reducing/oxidizing species occurs to generate the excited state of the luminophore-related compound, which emits light. ECL biosensors for health analysis mainly conduct in this ECL co-reactant reaction pathway. In ECL biosensors, the luminophores can be mainly categorized into three types including small organic compounds, inorganic complexes, and nanomaterials, while the co-reactants mainly involve peroxydisulfate (S2O82−) for the cathodic co-reactant process and TPA and hydrogen peroxide (H2O2) for the anodic co-reactant process.6,7 These three types of the ECL reagents were widely employed in developing high performance ECL biosensors. On the basis of classic ECL reagents, here, new ECL reagents and nanomaterials contributed in ECL biosensors are highlighted.2.1 Small organic compounds
The ECL phenomenon of Grignard compounds was first reported in 1927. The ECL of luminol (5-amino-2,3-dihydro-1,4-phthalazinedione, MW = 177.16) was first observed by Harvey at 1929. Luminol is a classic CL reagent in aqueous alkaline solution containing H2O2 and a catalyst, such as Co2+, Cr3+, Fe2+, and heme. This point should be considered in design of ECL biosensors. In the absence of the catalyst, luminol is also a typical ECL reagent in aqueous neutral solution containing H2O2 or H2O2 which is generated by the reaction of the substrates with dissolved oxygen under the catalysis of the redox enzyme. The formats using luminol as ECL reagent can be classified into two categories in ECL biosensors, including conventional regent in solution for redox enzyme-labelled biorecognition elements and the labelling reagent for the binding biorecognition elements. In homogeneous ECL of the luminol–H2O2 system, the proposed ECL mechanism for luminol and H2O2 in alkaline solution is shown in Fig. 2A.6a Luminol can be electrochemically oxidized to generate the luminol radical while H2O2 can be electrochemically oxidized to generate HO2˙ radical, which deprotonates to produce O2˙ radical. The luminol radical reacts with HO2˙ and O2˙ to form excited 3-aminophthalic acid that is capable of emitting blue light (λmax = 425 nm). For luminol as a small labelling ECL reagent, the bio-affinity of luminol-labelled biorecognition elements (e.g., Ab and DNA) declines less; however, the ECL efficiency of luminol is greatly reduced and luminol is consumed.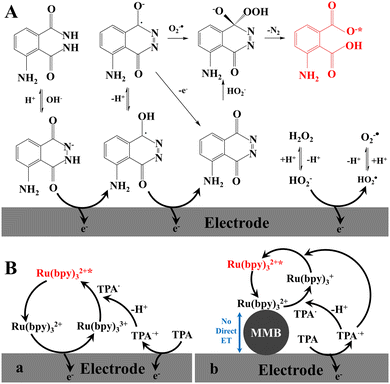 | ||
| Fig. 2 Schematic representation of ECL mechanisms. (A) Homogeneous, luminol–H2O2 system; (B) homogeneous (a), heterogeneous (b), Ru(bpy)32+–TPA system. | ||
On the basis of the properties of luminol as a labelling ECL reagent, we developed the homogeneous ECL immunoassay for the determination of digoxin antibody using luminol-labelled small digoxin hapten.20 To improve the sensitivity of the ECL biosensors, nanomaterials were applied as the carrier or modification of the electrodes. We further proposed using a nanocarrier loading several luminol and biorecognition elements for the determination of digoxin.21 Recently, Wang's group reported the boosted cathode luminol ECL via single-atom catalysts-tuned oxygen reduction reaction, exhibiting electron-transfer pathway-dependent response by adjusting the surrounding environment of the center metal atoms in a controlled way to selectively produce different active intermediates. Compared with the traditionally negligible cathodic ECL of luminol, about 70-fold enhancement was observed, attributed to the combination of electrochemical ORR catalyzed via SACs and chemical oxidation of luminol. This system was employed for the determination of the antioxidant capacities using ascorbic acid in the linear range of 0.10 nM to 100 µM with a detection limit of 0.10 nM.22
Luminol derivative N-(aminobutyl)-N-ethylisoluminol (ABEI, MW = 276.33) was employed as an ECL labelling reagent to obtain a high ECL efficiency of ABEI-labelled antibody since the linking amino group is far from the luminophore group. We developed homogenous ECL immunoassay for human IgG immunoglobulin G using ABEI as a label at gold nanoparticles-modified paraffin-impregnated graphite electrode.23 Based on this approach, Yuan group recently developed ECL biosensor using tetrahedron DNA dendrimer as a carrier and ABEI as ECL reporter for the ultrasensitive determination of lipopolysaccharides (LPS),24 and using metal–organic framework (MOF) loaded with ABEI for the sensitive detection of mucin1 on cancer cells.25
Recently, Mayer et al. synthesized a new ECL reagent, m-carboxy luminol, and demonstrated that m-carboxy luminol exhibited high solubility under physiological conditions and afforded a four-fold ECL signal increase compared with luminol.26 The entrapment of m-carboxy luminol in DNA-tagged liposomes enabled a DNA assay with a detection limit of 3.2 pM, which is 150-folds lower than the corresponding fluorescence approach. This remarkable sensitivity gain and the low excitation potential establish m-carboxy luminol as a superior ECL probe with direct relevance to CL and enzymatic bioanalytical approaches.26 This new ECL reagent will be widely employed as an ECL labelling reporter with a high ECL efficiency in ECL biosensors since the m-carboxy group in the m-carboxy luminol molecule far from the luminophore group can be covalently coupled with the amino group of the biorecognition elements.
Tu's group provided a review on the progress of the ECL biosensing strategy for clinical diagnosis with luminol as the ECL reporter.12 The luminol derivative-H2O2 systems are mainly used for the detection of the enzymes or the enzyme substrates on the basis of generation of hydrogen peroxide. These systems have intrinsic advantages, such as relatively low working potential and combination with the enzyme labels. However, the electrochemical oxidation of luminol moiety is irreversible and the luminol derivatives such as Ru(bpy)32+ cannot be regenerated; in addition, the ECL intensity strongly depends on the pH in the detection system.
2.2 Inorganic complexes
A classic ECL system of tris(2,2′-bipyridyl)-ruthenium(II) (Ru(bpy)32+,bpy = 2,2′-bipyridine)–TPA in aqueous solution was reported in 1990 due to the highest ECL efficiency of the Ru(bpy)32+–TPA system (λmax = 620 nm).27 For example, at the GC electrode, the initial ECL signal started at onset potentials where the direct oxidation of TPA occurs, and reached a first maximum at a potential of about 0.90 V vs. Ag/AgCl, about 50 mV less positive than the peak potential for TPA oxidation, and well before Ru(bpy)32+ oxidation. The second ECL signal has a peak potential value of 1.14 V vs. Ag/AgCl in the potential region of the direct oxidation of Ru(bpy)32+.28 The extensively studied ECL mechanisms of the Ru(bpy)32+–TPA system are presented in ref. 6 and 7. As showed in Fig. 2B, “homogeneous ECL mechanism” involves two ECL pathways. The direct oxidation of both TPA and Ru(bpy)32+ occurred at the surface of the electrode to generate TPA+˙ and Ru(bpy)33+, followed by the deprotonation reaction of the TPA+˙ to produce TPA˙. The generated TPA˙ and Ru(bpy)33+ react in the diffusion layer and generate the excited state Ru(bpy)32+*, which decays by emitting light. In most of the analytical methods, the Ru(bpy)32+ derivatives are constrained in proximity to the electrode since they are attached to a sensing element (i.e., antibody, bead, and DNA probe) and thus are not free to diffuse to the electrode. In these cases, “heterogenous ECL mechanism” concerns one pathway. ECL emission are therefore triggered exclusively by the TPA˙ radicals by diffusing from the electrode. TPA˙ was obtained in the same way described above.To label the Ru(bpy)32+ complex to the biorecognition elements, one of bpy ligand should contain one the connecting group with a suitable spacer, such as carboxyl group for amino group29 and amino group for carboxyl group,30 to form amido-bond. The N-hydroxysuccinimide ester (NHS)-contained Ru(bpy)32+ derivative is easily coupled covalently with the biorecognition elements bearing the contacted groups and a spacer (e.g., the propyl group, –(CH2)3–) can reduce the steric hindrance. The commercialized reagent bis(2,2-bipyridine)-4-methyl-4′-carboxybi-pyridine-ruthenium N-succinimidylester-bis(hexa-fluorophosphate [Ru(bpy)2(mcbpy-O-Su-ester)(PF6)2] (abbreviated as Ru1) was widely used as the labelling reagent. Ru(bpy)32+ applications will be presented in the following sections. Here, cyclometalated iridium(III) complexes as new ECL reagents are mainly presented.
Cyclometalated iridium(III) complexes as new ECL luminophors have received increasing attention because of their high ECL efficiencies, tuneable emission wavelengths, and working potentials through subtle changes in the structure of one or more ligands and easy synthesis.31 In the exploration of Ir(III) complexes, one aim is to improve their ECL efficiency, while another aim is to tune emission wavelengths and working potentials. In 2002, Bruce and Richter first reported the ECL emission of fac-tris(2-phenylpyridine)iridium(III) [Ir(ppy)3] with TPA as the co-reactant,32 opening up a new door of inorganic complexes. In 2005, Kim et al. synthesized two iridium complexes with high ECL efficiency, making a major breakthrough in cyclometalated Ir(III) complexes.33 Our group has done some efforts to synthesize and screen cyclometalated Ir(III) complexes to be utilized in ECL biosensors. One promising issue is to tune ECL emission wavelengths using different main ligand and ancillary ligands.34 We synthesized a new cyclometalated iridium(III) complex, [(bt)2Ir(dmphen)](PF6), using 2-phenylbenzothiazole (bt) as the C^N main ligand and 5,6-dimethyl-1,10-phenanthroline (dmphen) as the N^N ancillary ligand, which can be utilized as the ECL reagent inserted into the duplex DNA for label-free super-sandwich ECL biosensors for the detection of Micro-RNA.35 The other promising issue is that the cyclometalated iridium(III) complexes can be employed in potential-resolved strategy or wavelength-resolved strategy for multiple analytes since the ECL potentials or ECL wavelength of cyclometalated iridium(III) complexes bearing different ligands and Ru complex. We designed a potential-resolved ECL biosensor for the determination of matrix metalloproteinase (MMP)-2 and MMP-7 in a single run with detection limit as low as 5 ng mL−1 and 10 pg mL−1 using Ru1 (+0.9 V vs. SCE) and (dfppy)2Ir(dcbpy) (+1.4 V vs. SCE) as the ECL reporter.36 Guo et al., developed novel potential and spectrum dual-resolved ECL biosensors for the simultaneous determination of three antigens, namely, carcinoembryonic antigen (CEA), alpha-fetoprotein (AFP), and beta-human chorionic gonadotropin (β-HCG) using three new ruthenium(II) and iridium(III) complexes as the readout signal.37 Furthermore, we designed a chemical ECL sensor for hypochlorite using a new cyclometalated iridium complex, [(bt)2Ir(bpy-Fc)]PF6 (bpy-Fc = 4-ferrocenecarbonyl hydrazine-carbonyl-4′-methyl-2,2′-bipyridine), in which a hydrazine-carbonyl group was introduced into the ligand as a recognition group.38 Since Ma et al.39 in 2008 first reported iridium(III) solvent complexes as a highly selective luminescent switch-on probe for histidine/histidine-rich proteins, this type of cyclometalated iridium complex is getting increasing attention in analytical chemistry because of its flexible coordination with ligand or solvent.40,41 We synthesized carboxyl group bearing iridium(III) solvent complex [Ir(pbz)2(DMSO)Cl] (pbz = 3-(2-pyridyl)benzoic acid)] as PL and ECL for the detection of histidine.42 However, the ECL efficiency and working potentials of these cyclometalated iridium(III) complexes in competition to that of ruthenium(II) complexes[Ru(bpy)32+] are not obvious.
2.3 Nanomaterials
From the first report by Bard's group for the ECL behavior of nanomaterials, silicon nanocrystal quantum dots,43 CdSe NPs,44 and the first report on CdSe nanocrystal-based ECL biosensing for the detection of H2O2,45 a large number of nanomaterials have been reported as ECL signal materials so far. In the coreactant pathway, the coreactants in the ECL of quantum dots mainly involve S2O82−, TPA, H2O2, and C2O42−.46 In the quantum dots (QDs)–S2O82− systems, ECL is usually generated by negative potential scanning on the electrode, in which QDs and S2O82− are electrochemically reduced to generate the QDs˙− and SO4˙−, and then the excited state QDs* was generated by chemical reaction between QDs˙− and the strongly oxidizing intermediate SO4˙−.46We first investigated ECL of ZnS nanoparticles (NPs) in alkaline aqueous solution containing K2S2O8 and demonstrated that the surface passivation effect and the core/shell structure of ZnS/Zn(OH)2 played a significant role in the ECL process.47 The ZnS NPs was not applied in the ECL biosensors due to the very low ECL efficiency. In recent years, our focus shifts to aggregation-induced enhanced (AIE) ECL of organic nanoparticles. We found AIE ECL of organic nanoparticles (NPs, 5.82 nm) from donor–acceptor 6-[4-(N,N-diphenylamino)-phenyl]-3-ethoxy-carbonyl coumarin (DPA-CM) in organic solution.48 A strong and stable ECL emission was observed at the DPA-CM NPs modified glassy carbon electrode in the presence of TPA, and this ECL intensity was quenched by ascorbic acid, uric acid, and dopamine, respectively. The decreased ECL intensity was linear with concentration in the range of 0.05–50 µM with detection limit of 0.04, 0.2, and 0.4 µM for ascorbic acid, uric acid, and dopamine, respectively. This work shows an example of donor–acceptor-based organic NPs as ECL emitter and potential applications to monitor biomolecules.48 Furthermore, we reported the highly efficient AIE ECL of cyanophenyl-functionalized tetraphenylethene (tetra[4-(4-cyanophenyl)phenyl] ethene, TCPPE) and its application in biothiols analysis.49 TCPPE NPs with an average size of 15.84 nm showed high ECL efficiency (593%, CdS as standard) and stable ECL emission (over one month) in aqueous solution, ascribed to the efficient suppression of nonradiative transition, the decrease in the energy gap, and the increase in anionic radical stability. The “signal off” ECL sensors (fabricated by modifying TCPPE NPs on the surface of GCE) showed that the peak ECL intensity at −1.7 V (vs., Ag/AgCl) in 0.10 M phosphate buffered saline (PBS, pH = 7.40) containing 50 mM K2S2O8 was linear with the concentration in the range of 0.01–50 µM for cysteine or homocysteine, and 1–50 µM for glutathione with detection limits of 6, 7, and 300 nM for cysteine, homocysteine, and glutathione, respectively. We provided a mini-review on organic NPs for ECL assay.9 In addition, we reported a novel assembling method for the synthesis of sulphur quantum dots by simply treating sublimated sulphur powders with alkali using polyethylene glycol-400 as passivation agents. Significant ECL of the S dots is observed in an annihilation reaction.50 However, the ECL efficiency of organic NPs and the S dots mentioned above are too low to be employed as the ECL labelling reagents.
In recent years, ECL nanomaterials for ECL bioanalysis have received much attention. A number of mini-reviews on ECL nanomaterials for ECL bioanalysis are published in recent years, including quantum dots,51,52 metal nanoclusters,53 metal–organic frameworks (MOF),54 cabon-based nanomaterials (graphite carbon nitride, g-C3N4),55 and perovskite nanomaterials.56 The nanosheets (graphite carbon nitride, g-C3N4) and metal–organic frameworks are mainly employed as ECL reagents to be modified on the surface of electrode, performing in the quenching format, rather than as ECL labelling reagents. Zou's group reported a novel ECL wavelength-resolved ECL biosensor for the simultaneous determination of CEA, prostate specific antigen (PSA), and AFP using highly-passivated semiconductor nanocrystals, CdSe (λ 500–605, λmax = 550 nm), CdTe (λ 590–740, λmax = 650 nm), and CdTe (λd 680–900 nm, λmax = 776 nm) NCs as ECL tags.57 The attractive of semiconductor nanocrystals is the half bandwidth (<60 nm) of these semiconductor nanocrystals was much narrower than that (∼100 nm) of other ECL reagents including classic ECL reagents and g-C3N4. In addition, hybrid ECL NPs have also received much attention for the detection of multiple targets. We synthesized a hybrid ECL NPs by coupling bis(2,2′-bipyridine)-(5-aminophenanthroline)ruthenium (Ru-NH2) to the surface of CdS QDs.58 This hybrid ECL NPs showed one ECL peak with λmax = 420 nm at −1.6 V in K2S2O8 solution and another ECL peak CL peak with λmax = 620 nm at +1.3 V in TPA solution and applied to detect thrombin under different ECL measurement conditions with a low detection limit of 0.6 pM and 0.7 pM, respectively. Rusling's group reported a novel automated 3D-printed microfluidic array for the rapid detection of multiple proteins on the basis of nanomaterial enhancement using Ru(bpy)32+-silica nanoparticles (RuBPY–SiNP) with a diameter of 110 ± 13 nm for the detection of six proteins (IGF-1, PSA, PF-4, CD-14, VEGF-D, GOLM-1, PSMA, and IGFBP-3) with ultralow detection limits as low as 78–110 fg mL−1 in diluted serum.59 In addition, self-luminescent lanthanide metal–organic frameworks prepared from precursors containing Eu(III) ions and 5-boronoisophthalic acid (5-bop) were taken as ECL signal probes in ECL immunoassay. ECL emission mechanisms showed that 5-bop was excited with ultraviolet photons to generate a triplet-state, which then triggered Eu(III) ions toward red emission. The electron-deficient boric acid decreased the energy-transfer efficiency from the triplet-state of 5-bop to Eu(III) ions; consequently, both were excited with high efficiency at single excitation. This MOF displayed excellent performance characteristics in an ECL immunoassay with a detectable limit of 0.126 pg mL−1 cytokeratins21-1.60 Furthermore, arginine modification of black phosphorus quantum dots (R-BPQDs) was reported to enhance the ECL intensity 25-folds when R-BPQDs was modified on the GCE in 0.1 M PBS (pH = 7.4)–0.1 M K2S2O8 solution, attributed to the fact that arginine can passivate the surface oxidation defects of BPQD. The ECL mechanism of this system was proposed to be the same as the normal QDs–K2S2O8 system. Since integrin can inhibit the ECL of R-BPQDs, the application of R-BPQDs in ECL bioanalysis was demonstrated, the Arg-containing peptide RRGDS attached on BPQDs, where RGDS is a peptide specific to αV/β3 integrin on the A549 cell membrane, which were then coated on GCE to act as both the recognition unit and signal tag. Under optimal conditions, the IC50 of cyclo(RGDyK) for 1 × 106 A549 cells per mL was obtained to be 12.0 nM from the ECL response plot of cyclo(RGDyK)-treated A549 cells. This strategy could be extended for the evaluation of other inhibitors or the detection of cell surface groups by changing the Arg-containing peptide.61
A series of new ECL materials have been extensively investigated regarding the optical, electrochemical, and ECL properties, including 2D carbon nitride,62 covalent organic frameworks (COFs),62 and luciferase-free luciferin.62 However, these new ECL materials have not been applied in ECL biosensors for health analysis. Thus, they should be explored. However, the challenges of ECL nanomaterials in the design of ECL biosensors are their relatively low ECL efficiency, low bio-affinity of labelled probes, which is derived from large sizes, compared with molecular Ru(bpy)32+.
3. New biorecognition elements
Biorecognition elements play a significant role in ECL biosensors since they decide the selectivity, detection limit, and sensitivity of the ECL biosensors. The classical biological molecular recognition elements mainly include antibody, single-strand DNA (ss-DNA), enzyme, and biological receptor. According to the recognition principle, the ECL biosensors are mainly catalogued into binding (e.g. Ag/Ab), DNA hybridization, and enzyme catalytic ones.DNA hybridization ECL biosensors on the basis of Watson–Crick base-pairing rule have been employed in nucleic acid assays for the health diagnosis of a variety of hereditary and infective diseases. The target ss-DNA sequence (less than 50 bases) has been measured generally using a sandwich hybridization format, in which the target ss-DNA was hybridized with surface-tethered capture probes immobilized on the surface of electrode and then hybridized with ECL reagent-tagged complementary DNA sequence. Different hybridization strategies including structure switching, target-induced strand displacement, and super-hybridization have been developed. We first reported a hairpin DNA-based ECL hybridization ECL biosensors for the detection of the HIV gene with a detection limit of 9 × 10−11 M.29 The target HIV gene ss-DNA was hybridized with an ECL reagent-tagged hairpin DNA assembled on the surface of the gold electrode, which led to the conversion of the stem-loop of the ECL probe on the electrode into a linear double-helix configuration, and the tag was moved away from the electrode surface, which resulted in a decrease in the ECL signal.29 This approach can improve the discrimination of single-base mismatch. Furthermore, we also designed label-free super-sandwich hybridization ECL biosensors using the interacting ECL reagents into the double-stranded DNA to greatly enhance the ECL intensity for signal amplification, e.g., using Ru(phen)32+ for the detection of the HIV gene,63 and (bt)2Ir(dmphen) synthesized in our Lab for the detection of Micro-RNA.35 There are a large number of hybridization ECL biosensors published on the basis of DNA amplification strategies, mainly including a target DNA amplification strategy for specific genes on the basis of increasing amount of targets, reporter-DNA amplification strategy for specific genes, and micro-RNA and hybrid DNA amplification strategy for biomarker proteins on the basis of increasing the detected ECL signals. It is well known that polymerase chain reaction (PCR) is mostly used to amplify a single copy or a few copies of a piece of ss-DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence. This strategy using PCR is for increasing the amount of detected target ss-DNA in the detection system, resulting in increased ECL signal. Similarly, hybridization chain reaction (HCR) and rolling-circle amplification (RCA) can also be coupled with ECL biosensors. However, in view of the detection limit and real clinic applications, the methods with DNA hybridization ECL biosensors cannot compete with digital PCR fluorescence methods. Therefore, we pay little attention to the hybridization ECL biosensors with DNA amplification.
Antibody is mostly used in ECL immunosensors since it has high affinity for the vast majority of biomolecular binding pairs, such as most antibody–antigen pairs where the dissociation constants are of the order of 10−8–10−12 M.64 Thus, the detection limits of the methods using antibodies as biorecognition elements in the nanomolar to picomolar range are expected. However, it has some drawbacks including production from living animals and living cells, limited shelf life, temperature-sensitive denaturation, and cost. Therefore, extensive efforts have been devoted to developing new recognition elements by screening chemically synthetic compounds and natural compounds, such as aptamers, lectins, specific peptides, endopeptidases, DNAzymes, and nanozymes. Although a variety of ECL DNA hybridization biosensors and immunosensors have been reported for different biomarkers, herein, new biorecognition elements in both binding-based ECL biosensors and enzymes/enzymes-like catalytic ECL biosensors are presented.
3.1 Binding-based ECL biosensors
For the specific short peptides, the advantages are easily synthesized and designed to construct the ECL biosensors while the disadvantages are low bio-affinity and possible cleavage by some proteases in clinical samples.
3.2 Enzymes and enzymes-like catalytic ECL biosensors
Enzymes are proteins that catalyze biochemical reactions in living systems. Such catalysts are not only efficient but also extremely selective. Hence, enzymes combine the recognition with the amplification for many ECL biosensing applications. For examples, oxidases, a family of redox enzymes, can selectively catalyze the oxidation reaction of molecular oxygen to enzyme substrates (important clinic substances), such as glucose, cholesterol, and uric acid. Enzyme-based ECL biosensors (enzyme electrodes) are based on the coupling of a layer of an enzyme with an appropriate electrode. Such electrodes combine the specificity of the enzyme for its substrate with the ECL reagents such as luminol. As a result of such coupling, ECL enzyme-based ECL biosensors have shown to be useful for monitoring a wide variety of importance substrates in clinical samples. Several recent papers have addressed the application of nanomaterials in the typical luminol–H2O2 system, where luminol as ECL reagents was coupled with oxidases such as glucose oxidase (GOD) and cholesterol oxidase, and the determination of glucose and cholesterol. For examples, choline oxidase was employed for the ECL detection of choline77 and GOD-based ECL biosensor for the determination of glucose.78 However, the sensitivity and detection limit of this type ECL biosensors have no significant merits compared with that of the CL biosensors and electrochemical biosensors. In recent years, endopeptidase and protein kinases as well as enzymes-like materials including DNAenzyme and nanoenzyme have been employed as new catalytic recognition elements in ECL biosensors.![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) DDDDD) and a PKA-specific peptide (CLRRA
DDDDD) and a PKA-specific peptide (CLRRA![[S with combining low line]](https://www.rsc.org/images/entities/i_char_0053_0332.gif) LG) were employed as thio-phosphorylation sites of the substrates for CK2 and PKA, respectively. These two specific peptides were self-assembled onto the gold electrode via the Au–S bond to form a biosensor. In the presence of CK2 and PKA and co-substrate adenosine-5′-(γ-thio)-triphosphate (ATP-s), the two specific peptides on the biosensor were thio-phosphorylated, respectively, and then the ECL reagent Ru(bpy)32+-GNPs was assembled onto the thio-phosphorylated peptides via the Au–S bond. The Ru(bpy)32+-GNPs attached on the electrode surface produces a detectable ECL signal in the presence of TPA. This ECL assay was applied to the detection of CK2 in serum samples and the inhibition of CK2 and PKA. This strategy is promising for multiple protein kinase assay and kinase inhibitor profiling with high sensitivity, good selectivity, and versatility. Furthermore, to improve the sensitivity and selectivity, in the approach mentioned above, taking advantage of the ability of protein A binding with the Fc region of a variety of antibodies with high affinity, a ruthenium derivative-labelled protein A was utilized as a versatile ECL probe for the bioassay of these protein kinases (PKA and CK2). After the recognition of the phosphorylated peptide by monoclonal antiphosphoserine antibody, the versatile ECL probe was specifically bound to the antiphosphoserine antibody on the electrode surface. The ECL bioassay was developed successfully in the individual detection of PKA and CK2 with a detection limit of 0.005 U mL−1 and 0.004 U mL−1, respectively.86 In addition, we also combined this approach with bio-barcode probe to develop highly sensitive ECL biosensing method for monitoring CK2.87 A bio-barcode probe (h-DNA/AuNPs/p-DNA) was designed by conjugating phosphorylated DNA (p-DNA, 5′-SH-AAAAAAGTGT GTGTGT GTGTGTGTGTG-P-3′) and hairpin DNA (h-DNA, 5′-GCGCAACAAGA GTTCTTTTGAACTCTTGTTGCGCTTTTT-SH-3′) onto AuNPs for signal amplification using Ru(phen)32+ as an intercalator into h-DNA. This “signal on” ECL biosensor for the detection of CK2 showed a linear range of 0.005–0.2 U mL−1 with a detection limit of 0.001 U mL−1.
LG) were employed as thio-phosphorylation sites of the substrates for CK2 and PKA, respectively. These two specific peptides were self-assembled onto the gold electrode via the Au–S bond to form a biosensor. In the presence of CK2 and PKA and co-substrate adenosine-5′-(γ-thio)-triphosphate (ATP-s), the two specific peptides on the biosensor were thio-phosphorylated, respectively, and then the ECL reagent Ru(bpy)32+-GNPs was assembled onto the thio-phosphorylated peptides via the Au–S bond. The Ru(bpy)32+-GNPs attached on the electrode surface produces a detectable ECL signal in the presence of TPA. This ECL assay was applied to the detection of CK2 in serum samples and the inhibition of CK2 and PKA. This strategy is promising for multiple protein kinase assay and kinase inhibitor profiling with high sensitivity, good selectivity, and versatility. Furthermore, to improve the sensitivity and selectivity, in the approach mentioned above, taking advantage of the ability of protein A binding with the Fc region of a variety of antibodies with high affinity, a ruthenium derivative-labelled protein A was utilized as a versatile ECL probe for the bioassay of these protein kinases (PKA and CK2). After the recognition of the phosphorylated peptide by monoclonal antiphosphoserine antibody, the versatile ECL probe was specifically bound to the antiphosphoserine antibody on the electrode surface. The ECL bioassay was developed successfully in the individual detection of PKA and CK2 with a detection limit of 0.005 U mL−1 and 0.004 U mL−1, respectively.86 In addition, we also combined this approach with bio-barcode probe to develop highly sensitive ECL biosensing method for monitoring CK2.87 A bio-barcode probe (h-DNA/AuNPs/p-DNA) was designed by conjugating phosphorylated DNA (p-DNA, 5′-SH-AAAAAAGTGT GTGTGT GTGTGTGTGTG-P-3′) and hairpin DNA (h-DNA, 5′-GCGCAACAAGA GTTCTTTTGAACTCTTGTTGCGCTTTTT-SH-3′) onto AuNPs for signal amplification using Ru(phen)32+ as an intercalator into h-DNA. This “signal on” ECL biosensor for the detection of CK2 showed a linear range of 0.005–0.2 U mL−1 with a detection limit of 0.001 U mL−1.
New recognition elements described above have been demonstrated to have broad prospects. However, compared with the reported antibodies, the number of new recognition elements are limited and their specific bio-affinity is relatively low.97,98 Therefore, the synthesis and screening of new recognition elements should be continued and the specific bio-affinity of new recognition elements should be further improved.
4. Emerging constructing biointerfacial strategies
After presenting typical and new ECL reagents/materials and biorecognition elements as two basic components, we step forward to the construction of biointerfacial strategies for the design of ECL biosensors. This part was taken as one section in this article due to the following reasons: (1) in Sections 2 and 3, new ECL reagents/materials and biorecognition elements are only presented, and constructing biointerfacial strategies was not addressed. (2) Constructing biointerfacial strategies are significant for designing ECL biosensors. In this section, important and representative emerging construction biointerfacial strategies are presented and overlap with the sections above was avoided.Designing biointerfaces of ECL biosensors for the determination of given targets is a very important issue because it decides the accuracy, sensitivity, and applications of ECL biosensors. To put it simply, the problem is how to together organize target analyte, biorecognition element, ECL reagents/materials (mainly ECL probes), and electrode/magnetic beads (MBs) for forming ECL biosensors. The immobilization of biorecognition elements onto the solid surfaces involves two kinds of the molecular recognition elements including easily oriented ones (ss-DNA, aptamers, specific peptides, etc.) and randomly oriented ones (antibodies and enzymes) on two kinds of solid surfaces including electrodes and MBs. In effect, the conventional strategies for the construction of biointerfaces in electrochemical and optical biosensors can all be utilized in the construction of biointerfaces for ECL biosensors, such as physical adsorption and entrapment, self-assembly, and covalent coupling methods. Physical adsorption and entrapment strategies are rarely reported in the construction of biointerfaces for ECL biosensors due to the leakage and fall off of the capture probes. More targeted, the construction of biointerfacial strategies for ECL biosensors will be presented on different solid phases including electrodes and MBs.
4.1 Electrode surfaces
From the viewpoint of clinical diagnosis, a group of biomarkers is better than one biomarker. Therefore, the strategies for the simultaneous detection of multiple biomarkers matrix in “one shot” are expected. A series of ECL resolution strategies including space resolution, potential resolution, and wavelength resolution ones have been developed for ECL biosensor array in our Lab. In space resolution, an ECL peptide-based biosensor array containing four gold electrodes was designed for the bioassay of two protein kinases (PKA, CK2). The four gold electrodes were divided into two groups, in which two gold electrodes were self-assembled with PKA specific peptide while other electrodes were done with the CK2 specific peptide. After the specific peptide phosphorylations under the action of the corresponding kinase, the universal ECL probe (antiphosphoserine antibody and Ru(bpy)32+ derivative-labelled protein A) was added to form four sandwich bioconjugates. An ECL imaging bioassay on the ECL peptide-based biosensor array using CCD as the detector was developed for the simultaneous detection of PKA and CK2 in the linear range from 0.01 U mL−1 to 0.4 U mL−1 at one time using the versatile ECL probe.86 Furthermore, the ECL aptamer-based biosensor array for the detection of multiple acute myocardial infarction (AMI) biomarkers including such as Myo,cTnI and cTnT was designed by self-assembling three kinds of specific aptamers for a corresponding biomarker on four gold electrodes (diameter = 1 mm), in which one kind of specific aptamer was immobilized on the two gold electrodes to check the reliability of the fabricated biosensor and the ECL measurements. To obtain a high bio-affinity of the target, three kinds of corresponding biotinylated antibodies and one kind of versatile ECL probe, Ru(bpy)32+ derivative-labelled streptavidin (Ru1-SA) was utilized. Therefore, three kinds of AMI biomarkers can be quantified in “one shot” by ECL imaging using CCD.102 Another approach based on the potential resolution on one Au electrode for the detection of two kinds of matrix metalloproteinases was developed using two kinds of ECL tagging reagents including conventional ruthenium complex Ru1 and cyclometalated iridium complex (dfppy)2Ir(dcbpy) (Ir1). In 0.1 M PBS containing 50 mM TPA, the ECL peak of Ru1 and Ir1 at Au electrode appeared at 1.1 V and 1.4 V (vs., Ag/AgCl), respectively, suggesting that this system can be possibly employed in potential resolution, while the ECL wavelength of Ru1 and Ir1 was 621 nm and 558 nm, respectively, which is possible for wavelength resolution. Considering that wavelength resolution needs a monochromator, potential resolution was designed. Ru1-tagged MMP-2 peptide and Ir1-tagged MMP-7 peptide were self-assembled on the surface of one Au electrode. When the sample solution containing MMP-2 and MMP-7 was introduced, the corresponding peptide on the electrode was cleaved and the corresponding ECL reagent-tagged was removed from the electrode, resulted in a decrease in the ECL intensity.36
Nanomaterials-modified GCE electrode can be employed in ECL biosensors for enhancing ECL signals. For example, based on electrostatic interaction and Au–thiol-self-assembling, a “signal off” ECL biosensor for the determination of PSA was fabricated by casting the mixture of Nafion and GNPs onto the surface of glassy carbon electrode to form GNPs/Nafion film, and then, Ru(bpy)32+ was electrostatically adsorbed into the GNPs/Nafion film; finally, the specific peptide (CHSSKLQK)-tagged with ferrocene carboxylic acid (Fc-peptide) was self-assembled onto the surface of the AuNPs. An extremely low detection limit of 8 × 10−13 g mL−1 was achieved because of the signal amplification through GNPs and the ECL background suppression through Fc as the ECL quencher.84
Glassy carbon electrode can be employed in the cell culture. We developed a negative ECL imaging method for the morphological characterization of living cells cultured on the surface of GCE in the Ru(bpy)3Cl2–TPA solution. We found that the lower the ECL intensity, the higher the degree of inhibition of electron transfer of TPA and Ru(bpy)32+ in cells cultured on GCE. The ECL images reveal the morphological characteristics of living cells, including cell shape, cell area, average cell boundary, and the junction distance between two adjacent cells under H2O2 stimulation and electrical stimulation. In addition, quantitative analysis was performed with an extremely limit of detection of 29 MCF-7 cells per mL. Compared with the ECL imaging methods reported that employ an ECL reagent that is attached to the cell, the proposed negative ECL imaging method does not require cell labelling and can be used to obtain in situ morphological characteristics information. Compared with common fluorescence imaging methods, the proposed method does not require cell staining with fluorescent dyes.105
Importantly, ITO electrodes as basic electrodes have been employed for the ECL imaging of cells and biomarkers. ITO electrodes were also employed in the cell culture for ECL imaging. Jiang's group reported an ECL-based capacitance microscopy with a square wave voltage for label-free imaging of CEA antigen on single MCF-7 cells at ITO electrode. After the exposure of the MCF-7cells that are cultured at an ITO electrode in PBS solution containing (8-amino-5-chloro-7-phenylpyrido[3,4-d]pyridazine-1,4(2H,3H)-dione (L012) taken as ECL reagent, the label-free ECL images were collected under applied square wave voltage. The developed method can visualize CEA at amounts as low as 1 pg.109 Jiang's group also developed a single biomolecule imaging method on ITO electrode using Ru(bpy)32+-doped silica/Au nanoparticles (RuDSNs/AuNPs) as the ECL nanoemitters. Using MCF-7 cells as a model and CK19 as the target analyte, after labelling with RuDSN/AuNPs/Ab2 nanoemitters, ECL light was emitted from the labeled cells when a high potential was applied to the ITO electrode in a TPA solution. The average S/N ratio of ECL at single cell shows a ∼6.5-fold increase in comparison to that of PL, exhibiting an improved contrast to visualize the protein at the cells using the proposed ECL imaging methodology.110 Feng's group combined wide field optical imaging and electrochemical recording system for the monitoring of single-molecule ruthenium complex-based ECL reactions in aqueous solution at ITO electrode to allow for synchronized electrochemical measurement and optical imaging via an inverted microscope. They explored single-molecule ECL for imaging live human embryonic kidney 293 (HEK293) cell, revealing specific information in contrast to the bright-field image and the diffraction-limited ECL image.111
For ITO electrodes, the advantages are high optical transparency and strong adhesion compared with gold electrodes and GCE while the disadvantage is a corrosive effect when the applied potentials is higher than 1 V vs. Ag/AgCl.6b
The strategies for the construction of a biointerface on the surface of the working electrode described above played an important role in the fundamental research on ECL biosensors. However, the ECL biosensors based on these strategies have not been commercially used in real clinical health analysis. More efforts are needed to push forward applications.
4.2 Interfaces of magnetic nanoparticles and microbeads
Magnetic nanoparticles (MNPs) and magnetic microbeads (MMBs) act as solid supports for the immobilization of biomarkers to facilitate the isolation and detection of proteins, enzymes, and DNA, which has led to the development of sensitive and efficient automated immunoassays.112,113 Gooding's group demonstrated that MNPs acted as “dispersible electrode”.114 The MNPs that are modified with biorecognition molecules are dispersed in the sample matrix, where they capture the analyte. Application of a magnetic field rapidly bring the MNPs back to a macroscale electrode, where the quantitation of captured analytes can be realized electrochemically. From 1991, Blackburn et al., reported ECL detection for the development of immunoassays and DNA probe assays for clinical diagnostics using functional MMBs with a ruthenium complex (Ru(bpy)32+) as the reporter.5 The commercial automated ECL immunoassay analyzers such as Roche and Elecsys have been widely applied in clinical diagnosis for the determination of a variety of biomarkers since the excess ECL probes can be easily washed away in the flow system and batch synthesized functional MBs can be used in multiple tests. However, in these commercial ECL immunoassay analyzers, a pump is required and Au working electrode was employed since it is easily pretreated by electrochemical oxidation. The sensitivity of the ECL method based on the Ru(bpy)32+–TPA ECL system at the Au working electrode (Origen I analyzer) is lower than that at GCE.103 In addition, the price of the automated ECL immunoassay analyzers is much expensive than that of a simple ECL analyzer equipped with an electrochemical workstation and a PMT. To develop highly sensitive dispersive ECL biosensors using MBs for developing countries in the world and to establish a bridge between fundamental research and real applications in clinical diagnosis, we designed a series of dispersive ECL biosensors for the determination of multiple protein biomarkers.Feng's group developed a single-molecule ECL imaging bioassay at ITO electrode using single-molecule ECL microscopy. Imaging single biomolecules was first realized by localizing the ECL events of the Ru1-labeled MB@SA (φ = 2.8 µm). They further quantified biomolecules by spatiotemporally merging the repeated reactions at one molecule site and then counting the clustered molecules. The proposed single-molecule ECL bioassay was used to detect CEA via sandwich format with a calibration curve of CEA quantification from 0.5 to 50 ng mL−1, showing a limit of detection of 67 attomole concentration, which is 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times better than conventional ECL bioassays. This spatial resolution and sensitivity make single-molecule ECL bioassay a new toolbox for both specific bioimaging and ultrasensitive quantitative analysis.116
000 times better than conventional ECL bioassays. This spatial resolution and sensitivity make single-molecule ECL bioassay a new toolbox for both specific bioimaging and ultrasensitive quantitative analysis.116
Furthermore, we developed a gold nanoelectrode ensembles (Au-NEE) platform taken as a disposable ECL platform with immunomagnetic microbeads for the determination of carbohydrate antigen 19-9 (CA 19-9).117 Au NEE is a collection of individual gold nanoelements randomly spaced and unevenly embedded in an insulating substrate using electroless deposition. The average diameter of the prepared Au NEE was estimated to be 120 ± 4 nm and the average density of the individual gold nanoelectrodes was estimated to be (3.38 ± 0.09) × 108 pore per cm2. The prepared Au NEE showed peak-shaped voltammogram rather than an S-shaped one, attributed to the total diffusional overlap. It was found that the ECL intensity at Au-NEE was 12.9-fold in the Ru(bpy)32+–TPA ECL system and 19.6-fold in the luminol–H2O2 system, compared with that at the Au macroelectrode using the normalized active area of the electrodes, mainly attributed to the diffusion overlap of the Au-NEE and the edge effect of the individual gold nano-disks of the Au-NEE. The magnetic capture probe (MBs@SA/biotin-Ab1) was synthesized by binding the biotinylated CA 19-9 antibody with streptavidin (SA)-coated magnetic micro-beads (MBs@SA, 2.8 µm), while Ru1-labelled CA 19-9 antibody (Ru1-Ab2) was used as the ECL probe. “Sandwich” bioconjugates (Ru1-Ab2· CA 19-9·Ab1-biotin) on MMBs were transferred onto the ECL platform, and then the ECL measurements were performed in TPA solution. The developed method showed that the ECL peak intensity was directly in proportion to the concentration of CA 19-9 in the range from 0.5 to 20 U mL−1 with a limit of detection of 0.4 U mL−1. This work demonstrates that Au-NEE can be employed as a useful disposable ECL platform with the merits of cheapness, low nonspecific adsorption, and practical application. The proposed approach will open a new avenue in the point-of-care test for the determination of protein biomarkers.
Furthermore, we developed highly efficient ECL quenching on lipid-coated multifunctional MNPs for the determination of protease PSA incorporating membrane-confined quenching with a specific cleavage reaction.119 A new two hydrophobic long alkyl chains-containing ruthenium complex [Ru(bpy)2(ddcbpy)](PF6)2 (bpy = 2,2′-bipyridine, ddcbpy = 4,4′-didodecyl-carbonyl-2,2′-bipyridine) was synthesized as a signal probe, while [cholesterol-(CH2)6-HSSKLQK(peptide)-ferrocene (quencher)] was designed as a specific peptide-quencher probe. The MNPs were prepared by inserting both the signal probe and the peptide quencher probe into the cholesterol–phospholipid-coated Fe3O4 MNPs (diameter, ∼200 nm). When the analyte PSA was introduced into the suspension of MNPs, PSA cleaved the amide bond of SK in cholesterol-(CH2)6-HSSKLQK-Fc, and then the cleaved peptide motif-Fc-quencher was deviated from the MNPs, resulting in an increase in the ECL intensity on bare GCE. It was found that the ECL quenching constant (Fc-COOH to [Ru(bpy)2(ddcbpy)]) on the prepared MMNP (2.68 × 107 M−1) was 137-fold higher than that on the lipid coated electrode (1.95 × 105 M−1) and 391-folds higher than that in the solution (6.86 × 104 M−1). Based on the highly efficient ECL quenching of the ruthenium complex by ferrocene on the MNPs, a new “signal on” ECL method was developed for free PSA with a linear range from 0.01 to 1.0 ng mL−1 and a limit of detection of 3 pg mL−1. This work demonstrates that the approach of ECL quenching by ferrocene on lipid-coated MNPs is promising and could be easily extended to determine other proteases.
5. Challenges and perspectives
ECL biosensors, specifically, MBs-based ECL biosensors have been applied in real clinic analysis. As described above, although great achievements have been made in the three respects on ECL reagents and materials, biorecognition elements, and constructing biointerfacial strategies, the challenges in developing ECL biosensors for health analysis remain, such as other biosensors in sensitivity (detection limit), selectivity, speediness,64 and applications.In the design of the ECL biosensors, we should do what we need to do. To design ECL biosensors with good performance, two issues regarding the basic concepts should be kept in mind. The first issue is the biorecognition reaction of the analytes with the biorecognition elements. The molecular structure, size (molecular weight), and shapes of the biorecognition elements must be considered. Affinity ECL biosensors are based on the selective binding of certain biorecognition elements (e.g., antibodies, receptors, or oligonucleotides) toward specific target analytes for triggering useful ECL signals. The biorecognition process is controlled primarily by the shape and size of the receptor pocket and the analyte ligand of interest. Such a binding process is governed by thermodynamics (in contrast to the kinetic control exhibited by biocatalytic systems).120 The high specificity and affinity of biochemical binding reactions (such as DNA hybridization and antibody–antigen complexation) lead to highly selective and sensitive biosensors. The second issue is electrode reaction and CL reactions. As described above, three ECL co-reactant systems are employed in ECL biosensors so far. The electrochemical behaviors of the ECL reagent precursors and co-reactants are much different on different bare electrodes and modified electrodes while the rate constants of the CL reactions are generally independent of the electrode reactions. For Ru(bpy)32+–TPA systems, Ru(bpy)32+ can be oxidized on Au, Pt, and carbon electrodes with reversible electrochemical behaviors, while TPA can also be oxidized on Au, Pt, and carbon electrodes with irreversible electrochemical behaviors, and the electrochemical oxidation behaviors of TPA are much different at different electrodes103 and at different electrode surface hydrophobicities.121 As shown in Fig. 2, the Ru(bpy)32+ derivative attached on the surface of the electrodes and MMBs cannot be directly oxidized on the electrodes.6c This basic knowledge seems to be ignored in many published papers. Regarding the development of ECL biosensors for health analysis, without doubt, developing high ECL efficiency of the ECL systems including the synthesis of new signal reagents and finding new efficient co-reactants and new biorecognition elements are eternal themes.
Here, the pitfalls and challenges of developing the ECL sandwich immunosensors taken as a model are mainly discussed. The ECL sandwich immunosensors are based on immunological reactions involving the shape recognition of the antigen (Ag) by the antibody (Ab) binding site (epitope) to form the antibody·antigen (Ab·Ag) complex and then binding by the ECL signal probes (e.g., Ru1-Ab2). The remarkable selectivity of antibodies is based on the stereospecificity of the binding site for the antigen and is reflected by large binding constants. Antibody (5–6 nm) (monoclonal or polyclonal) bears the three-dimensional structure illustrated by models as Y-shaped molecules. Antibody (e.g., IgG, 5–6 nm) interacts with the antigen through the antigen-binding epitope (0.2–0.4 nm) located at the tip of the fragment of antigen binding (Fab) “arms” while fragment of crystallization (Fc) has no binding function.120 Therefore, three issues should be considered in the sandwich ECL Immunosensors.
5.1 Orient immobilization of capture antibody
A main challenge for using antibody employed as a capture probe on a surface of the electrode and MBs for the target of interest is to develop the oriented immobilization methods for the antibody fragment of crystallization (Fc) to the surface (in contrast to ss-DNA and peptide). Protein A or protein G is covalently immobilized on the surface of the solid phase and then the Fc of the antibody is orientally bound with the immobilized protein A or protein G.122 In addition, heptapeptide HWRGWVC can also act as a mediated substance for the oriented immobilization of the antibody.123 To improve the sensitivity of the biosensor, HWRGWVC heptapeptide (HWR) was utilized to capture the antibody Fc portion via specific interaction to realize site-oriented immobilization. After connecting with ABEI-Ft@Au via Au–S bonding, HWR improved the incubation efficiency of the antibody with a better maintained biological activity.123 However, the binding constants of the mediated substance with the antibody are lower than that of the target with the antibody. The more stable and oriented immobilization of the antibody should be explored. Besides, the surface density of the capture probes on the electrodes and MBs should be controlled and measured since it decides the sensitivity.5.2 High bio-affinity and high ECL efficiency of the ECL signal probes
Here, the ECL signal probes, which are ECL reagents labelled biorecognition elements, are mainly discussed. The ECL signal probes are generally synthesized by covalently coupled (labelling) Ru(bpy)32+ derivative of the antibody. High labelling ratio (e.g., >6) of the Ru(bpy)32+ derivative to the antibody is in favor of high ECL efficiency, rather than high bio-affinity. Especially, Ru(bpy)32+ derivative (MW, about 1000) is tagged to specific peptides (<30 amino acids) or ss-DNA (<30 bases), which possibly changes the bio-affinity. It should be argued that in many published papers, nanosheets (e.g., C3N4) were taken as ECL nanomaterials for labeling antibody, DNA, and peptide used as ECL probes, and carbon nanotubes and metal organic frameworks (MOFs) were taken as a carrier for Ru(bpy)32+ derivative and bio-recognition elements since these nanomaterials obstruct the binding reactions due to the steric hindrance effect. Without doubt, labelling methods must remain the binding sites of the antibody for the target. Besides, for MNPs and MMBs, challenges are raised from the nonspecific adsorption of the target protein biomarkers and the ECL signal probes (protein), which gives rise to a high background signal. The anti-nonspecific adsorption strategies should be further developed.5.3 Signal amplification and applications
For developing high performance ECL biosensors, when the capture probe on the electrode/MMB and the ECL signal probes are ready, the main work is to develop efficient formats. A large number of research papers have been published on ECL biosensors with signal amplification using multilayers. These ECL biosensors may seem to have wide linear ranges and low detection limits. However, the sensitivity (slope of the calibration curve) was much low since the change in the ECL is only several times when the change in the analyte concentration is several orders. This is attributed to the fact that multilayers strongly reduce the conductivity, electron transfer, luminescence efficiency, and transmission. Multilayers for the construction of biointerfaces should be avoided since they lead to large accumulation errors. In addition, different ECL detectors (PMT and CCD) can get different ECL signal for one ECL biosensor. To prepare the ECL biosensor reported in the work with that in other works published, ECL detectors should be standardized. The calibration curve of relative ECL intensity (I/I0 − 1, in signal-on) vs. the concentration of the analyte for the ECL biosensor should be recommended. Furthermore, most of the ECL biosensors in published papers, which obtained good results, were only applied in excessively diluted blood/serum samples rather than real samples. One challenge of the ECL biosensors is real applications for health analysis. Commercial biosensors and ECL kits for real applications for health analysis should be developed. Emerging ECL biosensors fields including ECL bioimaging and ECL biosensing devices with smartphone for point-of-care testing (POCT) can be seen in our previous Feature Article,7 if readers are interested.This Feature Article mainly highlights our contributions to ECL biosensors for health analysis, including new ECL reagents and materials, new biological recognition elements and emerging construction biointerfacial strategies for biomarkers, while it presents a balanced discussion of related work to set our contributions within a wider context. The pitfalls, challenges, and perspectives of developing ECL biosensors are discussed for health analysis. We hope that this Article will attract more attention to design innovative ECL biosensors for health analysis. We believe that ECL biosensors as powerful devices for the highly sensitive and selective quantification of important biomarkers will be widely applied in health analysis in the future.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 21974081, 22274093, 22074087) and the Fundamental Research Funds for the Central Universities (No. GK202202002).Notes and references
- Y. Y. Broza, X. Zhou, M. M. Yuan, D. Y. Qu, Y. B. Zheng, R. Vishinkin, M. Khatib, W. W. Wu and H. Haick, Chem. Rev., 2019, 119, 11761–11817 CrossRef CAS PubMed.
- G. A. Kwong, S. Ghosh, L. Gamboa, C. Patriotis, S. Srivastava and S. N. Bhatia, Nat. Rev. Cancer, 2021, 21, 655–668 CrossRef CAS PubMed.
- Biomarkers Definitions Working Group, Clin. Pharmacol. Ther., 2001, 69, 89–95 CrossRef PubMed.
- C. L. Sawyers, Nature, 2008, 452, 548–552 CrossRef CAS PubMed.
- G. F. Blackburn, H. P. Shah, J. H. Kenten, J. Leland, R. A. Kamin, J. Link, J. Peterman, M. J. Powell, A. Shah, D. B. TaHey, S. K. Tyagi, E. Wilkins, T. G. Wu and R. J. Massey, Clin. Chem., 1991, 37, 1534–1539 CrossRef CAS.
- (a) M. M. Richter, Chem. Rev., 2004, 104, 3003–3036 CrossRef CAS PubMed; (b) W. J. Miao, Chem. Rev., 2008, 108, 2506–2553 CrossRef CAS PubMed; (c) S. Rebeccani, A. Zanut, C. I. Santo, G. Valenti and F. Paolucci, Anal. Chem., 2022, 94(1), 336–348 CrossRef CAS PubMed.
- H. L. Qi and C. X. Zhang, Anal. Chem., 2020, 92, 524–534 CrossRef CAS PubMed.
- H. L. Qi, Y. G. Peng, Q. Gao and C. X. Zhang, Sensors, 2009, 9, 674–695 CrossRef CAS PubMed.
- H. L. Qi and C. X. Zhang, Curr. Opin. Electrochem., 2022, 34, 101023 CrossRef CAS.
- Y. Cao, J.-L. Zhou, Y. W. Ma, Y. Zhou and J. J. Zhu, Dalton Trans., 2022, 51, 8927–8937 RSC.
- L. Y. Hu, Y. Wu, M. Xu, W. L. Gu and C. Z. Zhu, Chem. Commun., 2020, 56, 10989–10999 RSC.
- C. Fang, H. L. Li, J. L. Yan, H. M. Guo and Y. F. Tu, ChemElectroChem, 2017, 4, 1587–1593 CrossRef CAS.
- R. A. Husain, S. R. Barman, S. Chatterjee, I. Khan and Z.-H. Lin, J. Mater. Chem. B, 2020, 8, 3192 RSC.
- H. Nasrollahpour, B. Khalilzadeh, A. Naseri, M. Sillanpaa, C. H. Chia, R. A. Husain, S. R. Barman, S. Chatterjee, I. Khanc and Z.-H. Lin, J. Mater. Chem. B, 2020, 8, 3192–3212 RSC.
- H. Nasrollahpour, B. Khalilzadeh, A. Naseri, M. Sillanpää and C. H. Chia, Anal. Chem., 2022, 94, 349–365 CrossRef CAS PubMed , formats.
- X.-L. Huo, H.-J. Lu, J.-J. Xu, H. Zhou and H.-Y. Chen, J. Mater. Chem. B, 2019, 7, 6469–6475 RSC.
- L. L. Li, Y. Chen and J.-J. Zhu, Anal. Chem., 2017, 89(1), 358–371 CrossRef CAS PubMed.
- Y. Zhang, R. Zhang, X. L. Yang, H. L. Qi and C. X. Zhang, J. Pharm. Anal., 2019, 9, 9–19 CrossRef PubMed.
- C. Ma, Y. Cao, X. D. Gou and J.-J. Zhu, Anal. Chem., 2020, 92, 431–454 CrossRef CAS PubMed.
- C. X. Zhang, H. H. Zhang and M. L. Feng, Anal. Lett., 2003, 36, 1103–1114 CrossRef CAS.
- H. L. Qi and C. X. Zhang, Anal. Chim. Acta, 2004, 501, 31–35 CrossRef CAS.
- H. Y. Xia, X. L. Zheng, J. Li, L. G. Wang, Y. Xue, C. Peng, Y. C. Han, Y. Wang, S. J. Guo, J. Wang and E. K. Wang, J. Am. Chem. Soc., 2022, 144, 7741–7749 CrossRef CAS PubMed.
- H. L. Qi, Y. Zhang, Y. G. Peng and C. X. Zhang, Talanta, 2008, 75, 684–690 CrossRef CAS PubMed.
- S. B. Xie, Y. W. Dong, Y. L. Yuan, Y. Q. Chai and R. Yuan, Anal. Chem., 2016, 88, 5218–5224 CrossRef CAS PubMed.
- X. Y. Jiang, Z. L. Wang, H. J. Wang, Y. Zhuo, R. Yuan and Y. Q. Chai, Chem. Commun., 2017, 53, 9705–9708 RSC.
- M. Mayer, S. Takegami, M. Neumeier, S. Rink, A. Jacobivon, S. Schulte, M. Vollmer, A. G. Griesbeck, A. Duerkop and A. J. Baeumner, Angew. Chem., Int. Ed., 2018, 57, 408–411 CrossRef CAS PubMed.
- J. K. Leland and M. J. Powell, J. Electrochem. Soc., 1990, 137, 3127–3131 CrossRef CAS.
- W. Miao, J. P. Choi and A. J. Bard, J. Am. Chem. Soc., 2002, 124, 14478–14485 CrossRef CAS PubMed.
- J. Zhang, H. L. Qi, Y. Li, J. Yang, Q. Gao and C. X. Zhang, Anal. Chem., 2008, 80, 2888–2894 CrossRef CAS PubMed.
- B. Sun, H. L. Qi, F. Ma, Q. Gao, Ch. X. Zhang and W. J. Miao, Anal. Chem., 2010, 82, 5046–5052 CrossRef CAS PubMed.
- M. A. Haghighatbin, S. E. Laird and C. F. Hogan, Curr. Opin. Electrochem., 2018, 7, 216–223 CrossRef CAS.
- D. Bruce and M. M. Richter, Anal. Chem., 2002, 74, 1340–1342 CrossRef CAS PubMed.
- J. I. Kim, I. S. Shin, H. Kim and J. K. Lee, J. Am. Chem. Soc., 2005, 127, 1614–1615 CrossRef CAS PubMed.
- Y. Y. Zhou, H. F. Gao, X. M. Wang and H. L. Qi, Inorg. Chem., 2015, 54, 1446–1453 CrossRef CAS PubMed.
- Y. Zhao, X. L. Yang, D. J. Han, H. L. Qi, Q. Gao and C. X. Zhang, ChemElectroChem, 2017, 4, 1775–1782 CrossRef CAS.
- H. F. Gao, Q. Dang, S. Q. Xia, Y. Zhao, H. L. Qi, Q. Gao and C. X. Zhang, Sens. Actuators, B, 2017, 253, 69–76 CrossRef CAS.
- W. Guo, H. Ding, C. Gu, Y. Liu, X. Jiang, B. Su and Y. Shao, J. Am. Chem. Soc., 2018, 140, 15904–15915 CrossRef CAS PubMed.
- D. J. Han, M. P. Qian, H. F. Gao, B. Wang, H. L. Qi and C. X. Zhang, Anal. Chim. Acta, 2019, 1074, 98–107 CrossRef CAS PubMed.
- D. L. Ma, W. L. Wong, W. H. Chung, F. Y. Chan, P. K. So, T. S. Lai, Z. Y. Zhou, Y. C. Leung and K. Y. Wong, Angew. Chem., Int. Ed., 2008, 47, 3735–3739 CrossRef CAS PubMed.
- C. Li, M. Yu, Y. Sun, Y. Wu, C. Huang and F. Li, J. Am. Chem. Soc., 2011, 133, 11231–11239 CrossRef CAS PubMed.
- Y. Zhou, Y. Ding, Y. Huang, L. Cai, J. Xu and X. Ma, ACS Omega, 2020, 5, 3638–3645 CrossRef CAS PubMed.
- M. Y. Zhang, H. Huang, M. P. Qian, Q. Gao, C. X. Zhang and H. L. Qi, J. Electroanal. Chem., 2022, 920, 116578 CrossRef CAS.
- Z. F. Ding, B. M. Quinn, S. K. Haram, L. E. Pell, B. A. Korgel and A. J. Bard, Science, 2002, 296, 1293–1297 CrossRef CAS PubMed.
- N. Myung, Z. F. Ding and A. J. Bard, Nano Lett., 2002, 2, 1315 CrossRef CAS.
- G. Z. Zou and H. X. Ju, Anal. Chem., 2004, 76, 6871–6876 CrossRef CAS PubMed.
- J. P. Lei and H. X. Ju, Trends Anal. Chem., 2011, 30, 1351–1359 CrossRef CAS.
- L. H. Shen, X. X. Cui, H. L. Qi and C. X. Zhang, J. Phys. Chem. C, 2007, 111, 8172–8175 CrossRef CAS.
- H. W. Liu, L. F. Wang, H. F. Gao, H. L. Qi, Q. Gao and C. X. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 44324–44331 CrossRef CAS PubMed.
- X. F. Wang, H. W. Liu, J. X. Jiang, M. P. Qian, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Chem., 2022, 94, 5441–5449 CrossRef CAS PubMed.
- L. H. Shen, H. N. Wang, S. N. Liu, Z. W. Bai, S. C. Zhang, X. R. Zhang and C. X. Zhang, J. Am. Chem. Soc., 2018, 140, 7878–7884 CrossRef CAS PubMed.
- E. Yang, Y. Zhang and Y. Shen, Anal. Chim. Acta, 2022, 1209, 339140 CrossRef CAS PubMed.
- S. O'Connor, L. Dennany and E. O'Reilly, Bioelectrochemistry, 2023, 149, 108286 CrossRef PubMed.
- Y. Cao, J. L. Zhou, Y. Ma, Y. Zhou and J. J. Zhu, Dalton Trans., 2022, 51, 8927–8937 RSC.
- C. Wang, S. Liu and H. Ju, Bioelectrochemistry, 2023, 149, 108281 CrossRef CAS PubMed.
- J. Jiang, X. Lin, D. Ding and G. Diao, RSC Adv., 2018, 8, 19369–19380 RSC.
- Y. Cao and J. J. Zhu, Front. Chem., 2021, 9, 629830 CrossRef CAS PubMed.
- J. Zhou, L. Nie, B. Zhang and G. Z. Zou, Anal. Chem., 2018, 90, 12361–12365 CrossRef CAS PubMed.
- X. F. Wang, H. W. Liu, H. L. Qi, Q. Gao and C. X. Zhang, J. Mater. Chem. B, 2020, 8, 3598–3605 RSC.
- K. Kadimisetty, S. Malla, K. S. Bhalerao, I. M. Mosa, S. Bhakta, N. H. Lee and J. F. Rusling, Anal. Chem., 2018, 90, 7569–7577 CrossRef CAS PubMed.
- Y. G. Wang, G. H. Zhao, H. Chi, S. H. Yang, Q. F. Niu, D. Wu, W. Cao, T. D. Li, H. M. Ma and Q. Wei, J. Am. Chem. Soc., 2021, 143, 504–512 CrossRef CAS PubMed.
- S. Q. Yu, Y. Du, X. H. Niu, G. M. Li, D. Zhu, Q. Yu, G. Z. Zou and H. X. Ju, Nat. Commun., 2022, 13, 7302 CrossRef CAS PubMed.
- (a) J. J. Ji, J. Wen, Y. F. Shen, Y. Q. Lv, Y. L. Chen, S. Q. Liu, H. B. Ma and Y. J. Zhang, J. Am. Chem. Soc., 2017, 139, 11698–11701 CrossRef CAS PubMed; (b) Y. J. Li, W. R. Cui, Q. Q. Jiang, Q. Wu, R. P. Liang, Q. X. Luo and J. D. Qiu, Nat. Commun., 2021, 12, 4735 CrossRef CAS PubMed; R. A. Luo, H. F. Lv, Q. B. Liao, N. N. Wang, J. R. Yang, Y. Li, K. Xi, X. J. Wu, H. X. Ju and J. P. Lei, Nat. Commun., 2021, 12, 6808 Search PubMed; (c) M. Belotti, M. M. T. El-Tahawy, L. J. Yu, I. C. Russell, N. Darwish, M. L. Coote, M. Garavelli and S. Ciampi, Angew. Chem., Int. Ed., 2022, 61, e202209670 CAS.
- S. P. Ruan, Z. J. Li, H. L. Qi, Q. Gao and C. X. Zhang, Microchim. Acta, 2014, 181, 1293–1300 CrossRef CAS.
- Y. F. Wu, R. D. Tilley and J. J. Gooding, J. Am. Chem. Soc., 2019, 141, 1162–1170 CrossRef CAS PubMed.
- L. L. Wu, Y. D. Wang, X. Xu, Y. L. Liu, B. Q. Lin, M. X. Zhang, J. L. Zhang, S. Wan, C. Y. Yang and W. H. Tan, Chem. Rev., 2021, 121, 12035–12105 CrossRef CAS PubMed.
- J. G. Bruno and J. L. Kiel, Biosens. Bioelectron., 1999, 14, 457–464 CrossRef CAS PubMed.
- Y. Li, H. Qi, Y. Peng, J. Yang and C. X. Zhang, Electrochem. Commun., 2007, 9, 2571–2575 CrossRef CAS.
- Y. Li, H. L. Qi, Q. Gao and C. X. Zhang, Biosens. Bioelectron., 2011, 26, 2733–2736 CrossRef CAS PubMed.
- F. Ma, Y. Zhang, H. L. Qi, Q. Gao, C. X. Zhang and W. J. Miao, Biosens. Bioelectron., 2012, 32, 37–42 CrossRef CAS PubMed.
- C. L. Nilsson, Anal. Chem., 2003, 75, 348A–353A CrossRef PubMed.
- H. Y. Yang, Y. Q. Wang, H. L. Qi, Q. Gao and C. X. Zhang, Biosens. Bioelectron., 2012, 35, 376–381 CrossRef CAS PubMed.
- H. Y. Yang, Z. J. Li, M. Shan, C. C. Li, H. L. Qi, Q. Gao, J. Y. Wang and C. X. Zhang, Anal. Chim. Acta, 2015, 863, 1–8 CrossRef CAS PubMed.
- E. Han, L. Ding, H. Z. Lian and H. X. Ju, Chem. Commun., 2010, 46, 5446–5448 RSC.
- H. L. Qi, X. Y. Qiu, D. P. Xie, C. Ling, Q. Gao and C. X. Zhang, Anal. Chem., 2013, 85, 3886–3894 CrossRef CAS PubMed.
- Z. J. Li, H. Y. Yang, L. J. Sun, H. L. Qi, Q. Gao and C. X. Zhang, Sens. Actuators, B, 2015, 210, 468–474 CrossRef CAS.
- H. L. Qi, Q. Dang, M. M. Dong, H. F. Gao and M. Li, Rev. Anal. Chem., 2014, 33, 255–263 CrossRef CAS.
- B. Leca and L. J. Blum, Analyst, 2000, 125, 789–791 RSC.
- X. Tian, S. Lian, L. Zhao, X. Chen, Z. Huang and X. Chen, J. Solid State Electrochem., 2014, 18, 2375–2382 CrossRef CAS.
- S. R. Denmeade, W. Lou, J. Lovgren, J. Maim, H. Lilja and J. T. Isaacs, Cancer Res., 1997, 57, 4924–4930 CAS.
- H. L. Qi, C. Wang, X. Y. Qiu, Q. Gao and C. X. Zhang, Talanta, 2012, 100, 162–167 CrossRef CAS PubMed.
- H. L. Qi, M. Li, M. M. Dong, S. P. Ruan, Q. Gao and C. X. Zhang, Anal. Chem., 2014, 86, 1372–1379 CrossRef CAS PubMed.
- X. L. Yang, Y. X. Wei, Z. M. Wang, J. X. Wang, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Chem., 2022, 94, 2305–2312 CrossRef CAS PubMed.
- Q. Dang, H. F. Gao, Z. J. Li, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Bioanal. Chem., 2016, 408, 7067–7075 CrossRef CAS PubMed.
- M. Y. Zhang, L. Shi, X. R. Liu, M. Qian and H. L. Qi, Electroanalysis, 2022, 34, 281–285 CrossRef CAS.
- M. M. Dong, X. Liu, Q. Dang, H. L. Qi, Y. Huang, Q. Gao and C. X. Zhang, Anal. Chim. Acta, 2016, 906, 72–79 CrossRef CAS PubMed.
- X. Liu, M. M. Dong, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Chem., 2016, 88, 8720–8727 CrossRef CAS PubMed.
- L. F. Wang, J. J. Song, X. F. Wang, H. L. Qi, Q. Gao and C. X. Zhang, Chin. Chem. Lett., 2020, 31, 2520–2524 CrossRef CAS.
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1, 223–229 CrossRef CAS PubMed.
- E. M. McConnell, I. Cozma, Q. B. Mou, J. D. Brennan, Y. Lu and Y. F. Li, Chem. Soc. Rev., 2021, 50, 8954–8994 RSC.
- F. Ma, B. Sun, H. L. Qi, H. G. Zhang, Q. Gao and C. X. Zhang, Anal. Chim. Acta, 2011, 683, 234–241 CrossRef CAS PubMed.
- F. Manea, F. B. Houillon, L. Pasquato and P. Scrimin, Angew. Chem., Int. Ed., 2004, 43, 6165–6169 CrossRef CAS PubMed.
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. L. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- Q. Wang, H. Wei, Z. Zhang, E. Wang and S. Dong, Trends Anal. Chem., 2018, 105, 218–224 CrossRef CAS.
- D. Jiang, D. Ni, Z. T. Rosenkrans, P. Huang, X. Yan and W. Cai, Chem. Soc. Rev., 2019, 48, 3683–3704 RSC.
- H. Y. Yan, Y. Wu, W. L. Gu, C. Z. Zhu, D. Du and Y. H. Lin, Angew. Chem., Int. Ed., 2020, 59, 2565–2576 CrossRef PubMed.
- Y. X. Song, S. Y. Lu, J. Hai, K. Liang, S. H. Sun, G. P. Meng and B. D. Wang, Anal. Chem., 2021, 93, 11470–11478 CrossRef CAS PubMed.
- V. Crivianu-Gaita and M. Thompson, Biosens. Bioelectron., 2016, 85, 32–45 CrossRef CAS PubMed.
- I. Bazin, S. A. Tria, A. Hayat and J.-L. Marty, Biosens. Bioelectron., 2017, 87, 285–298 CrossRef CAS PubMed.
- Y. Li, H. L. Qi, Y. G. Peng, Q. Gao and C. X. Zhang, Electrochem. Commun., 2008, 10, 1322–1325 CrossRef CAS.
- Y. Li, H. L. Qi, Q. Gao, J. Yang and C. X. Zhang, Biosens. Bioelectron., 2010, 26, 754–759 CrossRef CAS PubMed.
- M. Shan, M. Li, X. Y. Qiu, H. L. Qi, Q. Gao and C. X. Zhang, Gold Bull., 2014, 47, 57–64 CrossRef CAS.
- X. L. Yang, Y. Zhao, L. J. Sun, H. L. Qi, Q. Gao and C. X. Zhang, Sens. Actuators, B, 2018, 257, 60–67 CrossRef CAS.
- Y. B. Zu and A. J. Bard, Anal. Chem., 2000, 72, 3223–3232 CrossRef CAS PubMed.
- B. Wang, S. W. Shi, X. L. Yang, Y. Wang, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Chem., 2020, 92, 884–891 CrossRef CAS PubMed.
- H. F. Gao, W. J. Han, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Chem., 2020, 92, 8278–8284 CrossRef CAS PubMed.
- X. N. Mi, H. Li, R. Tan and Y. F. Tu, Anal. Chem., 2020, 92, 14640–14647 CrossRef CAS PubMed.
- D. Li, Y. Li, F. Luo, B. Qiu and Z. Y. Lin, Anal. Chem., 2020, 92, 12686–12692 CrossRef CAS PubMed.
- J. W. Gong, T. T. Zhang, T. Luo, X. Luo, F. Yan, W. Z. Tang and J. Y. Liu, Biosens. Bioelectron., 2022, 215, 114563 CrossRef CAS PubMed.
- J. J. Zhang, R. Jin, D. C. Jiang and H. Y. Chen, J. Am. Chem. Soc., 2019, 141, 10294–10299 CrossRef CAS PubMed.
- Y. J. Liu, H. D. Zhang, B. X. Li, J. W. Liu, D. C. Jiang, B. H. Liu and N. Sojic, J. Am. Chem. Soc., 2021, 143, 17910–17914 CrossRef CAS PubMed.
- J. Dong, Y. Lu, Y. Xu, F. Chen, J. Yang, Y. Chen and J. Feng, Nature, 2021, 596, 244–249 CrossRef CAS PubMed.
- M. A. M. Gijs, F. Lacharme and U. Lehmann, Chem. Rev., 2010, 110, 1518–1563 CrossRef CAS PubMed.
- L. H. Reddy, J. Arias, J. Nicolas and P. Couvreur, Chem. Rev., 2012, 112, 5818–5878 CrossRef CAS PubMed.
- I. Y. Goon, L. M. H. Lai, M. Lim, R. Amal and J. J. Gooding, Chem. Commun., 2009, 8821–8823 Search PubMed.
- C. Wang, H. L. Qi, X. Y. Qiu, Q. Gao and C. X. Zhang, Anal. Methods, 2012, 4, 2469–2474 RSC.
- W. X. Zhu, J. R. Dong, G. X. Ruan, Y. Zhou and J. D. Feng, Angew. Chem., Int. Ed., 2023, e202214419 CAS.
- X. L. Yang, Y. X. Wei, Y. J. Du, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Chem., 2020, 92, 15837–15844 CrossRef CAS PubMed.
- J. Zhang, H. L. Qi, Z. J. Li, N. Zhang, Q. Gao and C. X. Zhang, Anal. Chem., 2015, 87, 6510–6515 CrossRef CAS PubMed.
- X. L. Yang, Y. X. Wei, Z. M. Wang, J. X. Wang, H. L. Qi, Q. Gao and C. X. Zhang, Anal. Chem., 2022, 94, 2305–2312 CrossRef CAS PubMed.
- N. Trier, P. Hansen and G. Houen, Int. J. Mol. Sci., 2019, 20, 6289 CrossRef CAS PubMed.
- Y. B. Zu and A. J. Bard, Anal. Chem., 2001, 73, 3960–3964 CrossRef CAS PubMed.
- H. L. Qi, C. Wang and N. Cheng, Microchim. Acta, 2010, 170, 33–38 CrossRef CAS.
- L. Yang, D. W. Fan, Y. Zhang, C. F. Ding, D. Wu, Q. Wei and H. X. Ju, Anal. Chem., 2019, 91, 7145–7152 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |



