Sensitive determination of formamidopyrimidine DNA glucosylase based on phosphate group-modulated multi-enzyme catalysis and fluorescent copper nanoclusters†
Junyao
Li
a,
Mengyang
Zhang
a,
Huaisheng
Wang
 b,
Jie
Wu
b,
Jie
Wu
 a,
Ruixue
Zheng
a,
Jiahui
Zhang
a,
Yan
Li
a,
Zhaoyin
Wang
a,
Ruixue
Zheng
a,
Jiahui
Zhang
a,
Yan
Li
a,
Zhaoyin
Wang
 *a and
Zhihui
Dai
*a and
Zhihui
Dai
 *a
*a
aJiangsu Collaborative Innovation Center of Biomedical Functional Materials and Jiangsu Key Laboratory of Biofunctional Materials, School of Chemistry and Materials Science, Nanjing Normal University, Nanjing, 210023, P. R. China. E-mail: daizhihuii@njnu.edu.cn; inzo@163.com
bDepartment Of Chemistry, Liaocheng University, Liaocheng, 252059, P. R. China
First published on 17th June 2020
Abstract
In this work, a method for quantifying the activity of formamidopyrimidine DNA glucosylase (Fpg) was designed based on phosphate group (P)-modulated multi-enzyme catalysis and fluorescent copper nanoclusters (CuNCs). By eliminating 8-oxoguanine from double-stranded DNA, Fpg generates a nick with P at both 3′ and 5′ termini. Subsequently, part of the DNA is digested by 5′P-activated lambda exonuclease (λ Exo), and the generated 3′P disables exonuclease I (Exo I), resulting in the generation of single-stranded DNA containing poly(thymine) (poly(T)). Using poly(T) as templates, CuNCs were prepared to emit intense fluorescence as the readout of this method. However, in the absence of Fpg, the originally modified 5′P triggers the digestion of λ Exo. In this case, fluorescence emission is not obtained because CuNCs cannot be formed without DNA templates. Therefore, the catalysis of λ Exo and Exo I can be tuned by 5′P and 3′P, which can be further used to determine the activity of Fpg. The fluorescent Fpg biosensor works in a “signal-on” manner with the feature of “zero” background noise, and thus shows desirable analytical features and good performance. Besides, Fpg in serum samples and cell lysate could be accurately detected with the biosensor, indicating the great value of the proposed system in practical and clinical analysis.
Introduction
DNA is the carrier of genetic information,1–3 and thus it is crucial to maintain the stability of DNA.4,5 However, cellular DNA is constantly attacked by endogenous and exogenous factors, which may destroy the structural integrity of bases and lead to the dysfunction of cells.6 Because it possesses the lowest redox potential in four bases of DNA, guanine (G) is susceptible to oxidative damage.7 Typically, cellular reactive oxygen species oxidize the G group in the C8 position, producing 8-oxoguanine (8-oxoG).8,9 Due to the slight change in the C8 position, 8-oxoG may hybridize with adenine (A) rather than cytosine (C) during DNA replication, thereby resulting in the mutation of DNA.10–12 Accordingly, an abnormal level of 8-oxoG may lead to various neurodegenerative diseases, such as multiple sclerosis, Parkinson's disease, or Alzheimer's disease.13,14 Fortunately, damaged DNA can be recovered by enzyme-enabled DNA repair pathways in most cases.15–19 For instance, certain DNA N-glycosylases are able to recognize 8-oxoG in double-stranded DNA (dsDNA), and then cleave the N-glycosidic bond, removing 8-oxoG from damaged DNA.20 Depending on the base excision repair (BER) pathway, 8-oxoG can be eliminated, which avoids the accumulation of base mutation.21 Because formamidopyrimidine DNA glucosylase (Fpg), one of the DNA N-glycosylases, is essential to BER, the dysregulation of Fpg is considered an indicator of apoptosis- or mutations-related diseases.22 Therefore, the quantification of Fpg is helpful to give more insights into the BER pathway and to monitor DNA oxidative damage.To the best of our knowledge, strategies for Fpg quantification can be classified into three categories: (1) direct recognition with an antibody; (2) discrimination of the produced apurinic–apyrimidinic site; (3) differentiation of the generated DNA in terms of structural features, sequences, and the terminal phosphate group (P). Among these strategies, P-based methods are relevant to enzyme catalysis, and thus reveal superiority in the analytical performances. According to the principle of Fpg catalysis, nicks with P at both the 3′ and 5′ termini are generated after eliminating 8-oxoG from dsDNA.23 Since 5′P is able to activate some enzymes containing lambda exonuclease (λ Exo) and T4 DNA ligase, some analytical approaches have been proposed based on DNA with 5′P.24,25 However, owing to the lack of understanding of 3′P, DNA with 3′P is useless in Fpg biosensors. Recently, we demonstrated that 3′P is capable of preventing DNA digestion by exonuclease I (Exo I),26 rendering ignored DNA with 3′P valuable. On the basis of our work, novel biosensors may be designed by integrating 3′P-induced inhibition and 5′P-induced activation.
From a technology perspective, Fpg activity has been mainly detected up to now with standard biological methods, including gel electrophoresis, western blot, and comet assay.20 Apart from these means, fluorescent methods have attracted increasing attention in recent years. By fixing an analog of cytosine (pyrrolo-dC) opposite 8-oxoG, a fluorescence switch was designed to determine the activity of Fpg.23 However, one base-induced fluorescence variation is not so sensitive, which could affect the reliability and detection limit of this method. Another fluorescent Fpg biosensor was fabricated using rolling circle amplification and magnetic nanoprobes.22 In this method, multiplex DNA hybridization was employed to obtain fluorescent signals. Nevertheless, the DNA hybridization efficiency is difficult to be quantitatively controlled, and so is prone to inducing false-negative results. To be noted, fluorescent dyes are toxic and costly. In addition, the excitation and emission spectra of fluorescent dyes normally overlap, limiting their applications. In contrast, fluorescent metallic nanomaterials are safe, cost-effective, and involve a facile preparation.27 Meanwhile, due to the large Stokes shift, fluorescent metallic nanomaterials may display intact excitation and emission spectra, thus providing more information than fluorescent dyes. More importantly, some fluorescent metallic nanomaterials are synthesized with DNA as templates, greatly facilitating the construction of DNA-related biosensors.28 To address the aforementioned issues in the current fluorescent Fpg biosensors, the determination of Fpg with DNA-templated metallic nanomaterials is urgently needed.
Herein, resulting from the distinct effects of P on λ Exo and Exo I, multi-enzyme catalysis was tuned and used to quantify the activity of Fpg together with fluorescent copper nanoclusters (CuNCs). In the absence of Fpg, λ Exo and Exo I triggered a thorough digestion of the DNA used in this method, and as the CuNCs cannot be prepared without DNA templates, “zero” background noise was achieved. However, in the presence of Fpg, Fpg produces both 3′P and 5′P; where 5′P facilitates the digestion of λ Exo, while 3′P induces direct resistance to Exo I. As a result, DNA templates are generated to form fluorescent copper nanoclusters (CuNCs). Relying on the intense fluorescence of CuNCs, a platform for “signal-on” Fpg detection was established. Benefitting from the multi-enzyme catalysis and fluorescent metallic nanomaterials, the biosensor was capable of determining Fpg with a low detection limit, and could sensitively quantify Fpg in real human serum.
Experimental
Chemicals and materials
Exonuclease I (Exo I), exonuclease III (Exo III), T4 DNA ligase, and terminal deoxynucleotidyl transferase (TdT) were obtained from Takara Biotechnology Co., Ltd (Dalian, China). S1 nuclease was provided by Promega Biological Engineering Technology & Services Co., Ltd (Beijing, China). Formamidopyrimidine DNA glucosylase (Fpg), lambda exonuclease (λ Exo), and uracil–DNA–glycosylase (UDG) were acquired from New England Biolabs Ltd (Beijing, China). Glucose oxidase and GelRed were purchased from Shanghai Sangon Biological Engineering Technology & Services Co., Ltd (Shanghai, China). Dulbecco's modified eagle's medium (DMEM), penicillin–streptomycin, fetal bovine serum (FBS), trypsin–EDTA, and phosphate-buffered saline (PBS) were bought from Thermo Fisher Scientific Inc. (Shanghai, China). Cell lysis solution (P0013) and phenylmethanesulfonyl fluoride were purchased from Beyotime Institute of Biotechnology (Shanghai, China). All other chemicals were provided by Sigma-Aldrich Co., Ltd (Shanghai, China).All the oligonucleotides were provided by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd (Shanghai, China). The sequences of DNA are listed as follows. Single-stranded DNA-1 (ssDNA-1): (5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGT/i8oxodG/CGATCAGTGCTGA-3′), and single-stranded DNA-2 (ssDNA-2): (5′-P-CACTGATCGCAC-3′).
Instruments
Fluorescence assays were recorded on a Fluoromax-4 spectrometer (Horiba, France). The morphology characterizations of CuNCs were collected using a JEOL JEM-2100F apparatus with a 200 kV accelerating voltage. Circular dichroism (CD) analysis was achieved by using a Chirascan circular dichroism spectrometer (Applied Photophysics, Britain). Polyacrylamide gel electrophoresis (PAGE) analysis was performed on a Gel Doc XR system (Bio-Rad, USA).Construction of a fluorescence Fpg biosensor
To fabricate the fluorescence Fpg biosensor, 20 μL of 5 μM ssDNA (ssDNA-1 and ssDNA-2) and 10 μL hybridization buffer (10 mM Tris-HCl, 500 mM NaCl, 100 mM MgCl2, pH 7.6) were heated to 90 °C and kept for 5 min, and then slowly cooled to room temperature. Afterwards, 5 μL Fpg at varying concentrations, 0.5 μL of 10 mg mL−1 bovine serum albumin (BSA), 5 μL 10× PBS buffer (38 mM KH2PO4, 62 mM K2HPO4, 100 mM MgCl2, pH 7.0), and an extra 9.5 μL ultrapure water were mixed and kept at 37 °C for 1 h. Then, 5 U λ Exo and 6 μL 10× PBS buffer were incubated with the mixture at 37 °C for 30 min. After that, the digestion of Exo I was realized by adding 6 U Exo I and 2 μL 10× PBS buffer at 37 °C for another 30 min. Copper nanoclusters (CuNCs) were synthesized by orderly mixing the foregoing solution with 15 μL of 20 mM ascorbic acid (AA), 10 μL of 1 mM CuCl2, and adequate 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (10 mM MOPS, pH 7.5) to a volume of 300 μL, and then the mixture was retained for 15 min without light to synthesize fluorescent CuNCs. Fluorescence emission spectra were immediately obtained with the excitation wavelength of 340 nm and by using a 400 nm optical filter.Optimization of detection conditions
To obtain better detection results, the amounts of λ Exo and Exo I used in this biosensor were optimized. The activity of λ Exo was surveyed in the absence of Fpg. First, 20 μL of 5 μM (ssDNA-1 and ssDNA-2) and 10 μL hybridization buffer were treated according to the typical procedure of DNA hybridization. Second, 0.5 μL of 10 mg mL−1 BSA, 5 μL 10× PBS buffer, and 14.5 μL ultrapure water were added and kept at 37 °C for 1 h. Third, λ Exo in the range from 0 to 10 U and 6 μL 10× PBS buffer were mixed with the above solution at 37 °C for 30 min. After that, 6 U Exo I and 2 μL 10× PBS buffer were added into the mixture at 37 °C for another 30 min. Finally, CuNCs were prepared and applied for the fluorescence measurements. The optimization of Exo I was accomplished by adjusting the activity of Exo I in the range from 0 to 10 U, while fixing λ Exo as 5 U. The other treatment was unchanged.Investigation of selectivity
In order to verify the specificity of the biosensor, TdT, Exo III, glucose oxidase, pepsin, T4 DNA ligase, S1 nuclease, and UDG were used as the substitutes of Fpg. Briefly, 20 μL of 5 μM ssDNA (ssDNA-1 and ssDNA-2) and 10 μL hybridization buffer were incubated at 90 °C for 5 min, and then cooled gradually to room temperature. Afterwards, 10 U different enzymes, 0.5 μL of 10 mg mL−1 BSA, 5 μL 10× PBS buffer, and 9.5 μL ultrapure water were mixed with DNA solution at 37 °C for 1 h. The subsequent procedure was consistent with the fabrication of the biosensor.CD assays and PEGE analysis of DNA treated by multi-enzyme catalysis enzymatic reaction products
CD assays and PAGE analysis were carried out to confirm the effect of multi-enzyme catalysis on DNA. Samples for the CD assays were prepared in accordance with those for the fluorescence measurements, but a higher concentration of DNA was required. The final concentration of DNA was 2.8 μM. Measurement of the CD spectra was performed with the bandwidth of 1 nm, step size of 1 nm, and the scan range from 200 to 300 nm.Samples for PAGE analysis were similar to those for the CD tests. The obtained samples were resolved in 10% polyacrylamide gel. Electrophoresis was accomplished at 160 V for about 3 h using 1× TBE buffer (9 mM Tris-HCl, 9 mM H3BO3, 0.2 mM EDTA, pH 8.0) as a running buffer. After staining with GelRed, the gel was finally imaged.
Analysis of Fpg in human serum
To evaluate the anti-interference performance and clinical applicability of the biosensor, human serum samples were acquired from Jiangsu Cancer Hospital. The human serum samples were diluted to 10% and mixed with Fpg at various concentrations. After that, the prepared samples were detected using the proposed biosensor.Culture, lysis of cells and analysis of Fpg in the cell lysate
MCF-7 cells, F98 cells, U87 cells and A549 cells were obtained from Cell Bank, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM with 10% (v/v) FBS and 1% penicillin–streptomycin at 37 °C in humidified air containing 5% CO2. The cells in the logarithmic phase of growth were washed three times with sterile PBS and collected for the following experiments. Then, the cells were lysed with ice-cold cell lysis solution for the florescence assays. Finally, the cell lysates spiked with different concentrations Fpg were diluted to 20% and detected referring to the detection in the human serum samples.Results and discussion
Mechanism of Fpg sensing based on P-modulated multi-enzyme catalysis and CuNCs
As is well known, 5′P in dsDNA is the optimum substrate of λ Exo. Our study verified that 3′P in single-stranded DNA (ssDNA) can effectively resist the digestion by Exo I. Taking these two aspects into account, we consider that a nick with 3′P and 5′P may enable λ Exo, but disable Exo I. This principle was employed to design an Fpg biosensor composed of two ssDNA (ssDNDA-1 and ssDNA-2) and two enzymes (λ Exo and Exo I) (Fig. 1). The ssDNA-1 possesses the sequence of poly(T), which can be used as a template to form fluorescent CuNCs, and an 8-oxoG site, which can be cleaved by Fpg. SsDNA-2 contains P at the 5′ terminus, and thus will be digested by λ Exo after the formation of dsDNA (dsDNA-1) with ssDNA-1. In the presence of Fpg, a portion of ssDNA-1 is separated from dsDNA-1 at the 8-oxoG site, generating 3′P-poly(T) ssDNA (ssDNA-3) and 5′P-dsDNA (dsDNA-2). The generated dsDNA-2 is throughout digested by λ Exo, while ssDNA-3 is reserved due to the impediment of Exo I. As a result, ssDNA-3 serves as a template for the formation of CuNCs, and the intense fluorescence of CuNCs is obtained as the readout of the Fpg biosensor. However, in the absence of Fpg, ssDNA-2 and ssDNA-1 are sequentially digested by λ Exo and Exo I. Consequently, CuNCs cannot be prepared without templates, rendering the background noise as zero. Therefore, modulation of multi-enzyme catalysis is achieved based on the opposite effects of P on λ Exo and Exo I. In combination with CuNCs, a fluorescent biosensor was established to quantify the activity of Fpg sensitively. Actually, endonuclease VIII (Endo VIII) is also capable of generating a nick with P at both the 3′ and 5′ termini. However, different from Fpg, Endo VIII releases damaged pyrimidine from dsDNA. Because 8-oxoG can only be recognized by Fpg, the detection of Fpg will not be affected by other enzymes, like Endo VIII, which ensures the selectivity of this method.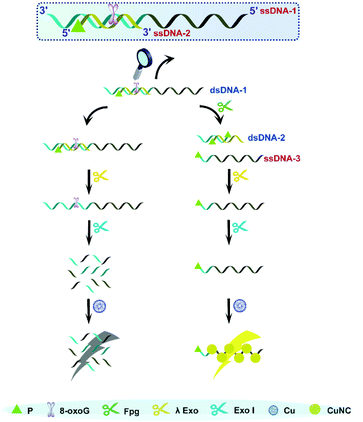 | ||
| Fig. 1 Illustration of Fpg detection based on the distinct effects of P on λ Exo and Exo I along with CuNCs. | ||
Manipulation of DNA with multi-enzyme catalysis
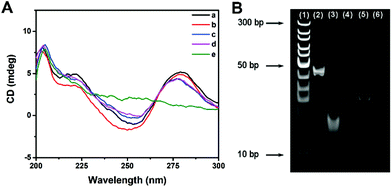
The effect of multi-enzyme catalysis on dsDNA-1 was studied with CD spectrum and PAGE image. In CD spectrum, the positive peak at 280 nm and the negative peak at 253 nm manifested the existence of dsDNA as B-form helices. Compared with those of dsDNA, the peak intensities of ssDNA were evidently decreased. Curve a in Fig. 2A suggests that dsDNA-1 is the hybridization of long ssDNA-1 and short ssDNA-2. After the addition of Fpg, dsDNA-2 and ssDNA-3 are generated by the cleavage of dsDNA-1. In this case, only a slight change could be observed in the CD spectrum due to the mixture of dsDNA and ssDNA (curve b). When λ Exo was further added, dsDNA-2 was completely digested. As a result, the CD peaks displayed a trend in the change from dsDNA to ssDNA (curve c). Since ssDNA-3 is protected by 3′P against Exo I, the CD peaks of ssDNA were not affected by Exo I (curve d). In contrast, dsDNA-1 was digested by λ Exo and Exo I in the absence of Fpg, resulting in the disappearance of both ssDNA and dsDNA peaks (curve e). Similar results could be obtained from the PEGE image (Fig. 2B). The intense band in lane 2 originated from dsDNA-1. In lane 3, the band representing dsDNA-1 was replaced with two new bands, implying that dsDNA-1 was cleaved by Fpg into dsDNA-2 and ssDNA-3. Furthermore, only the band corresponding to ssDNA-3 was reserved in the presence of Fpg and λ Exo, certifying the digestion of λ Exo to dsDNA-2 (lane 4). In lane 5, a band at the same position of lane 4 indicates that the terminal protection of 3′P works well, resulting in the persistence of ssDNA-3. However, no band could be observed in the absence of Fpg, because ssDNA-2 and ssDNA-1 in dsDNA-1 were digested by λ Exo and Exo I, respectively. The CD spectra and PAGE image both prove that terminal P possessed distinct effects on λ Exo and Exo I, which could be utilized in the design of a novel Fpg biosensor.Fabrication of a fluorescent Fpg biosensor
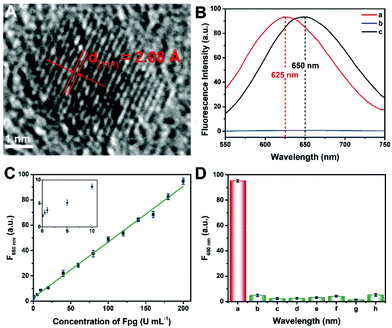
Since the fluorescence of CuNCs served as the readout of this Fpg biosensor, the morphological and optical properties of the CuNCs were further investigated. As shown in Fig. S1 and S2,† ssDNA-3-templated CuNCs were monodisperse with a mean diameter of 3.89 nm. According to the high-resolution transmission electron microscopy (HRTEM) image (Fig. 3A), the lattice spacing was 2.08 Å, which is consistent with the (111) plane of the face-centred-cubic phase of copper. According to our previous results, both dsDNA and ssDNA can be employed as templates for the synthesis of CuNCs, but the maximum emission wavelengths of the two kinds of CuNCs are different.29 Without any enzymes, CuNCs were formed taking dsDNA-1 as templates, and the wavelength of the emission peak was 625 nm (curve a in Fig. 3B). In the presence of λ Exo and Exo I, dsDNA-1 was digested throughout. Accordingly, fluorescence could not be obtained due to the lack of DNA templates (curve b). However, with the participation of Fpg, ssDNA-3 was retained to replace dsDNA-1 as the templates. As a result, ssDNA-templated CuNCs with intense fluorescence were prepared, and the maximum emission wavelength was changed to 650 nm (curve a). Therefore, a “signal-on” Fpg biosensor with “zero” background noise was constructed based on the variation of fluorescence induced by the effect of P on multi-enzyme catalysis. The dosages of λ Exo and Exo I in this biosensor are essential to the determination of Fpg. These detection conditions were optimized, and the relevant data are shown in Fig. S3 and S4.† With the increase in Fpg concentration from 0 to 200 U mL−1, more ssDNA-3 was generated, causing a gradual enhancement of the fluorescence (Fig. S5†). In light of the data shown in Fig. 3C, this biosensor displayed a linear relationship between the concentration of Fpg ([Fpg]) and the fluorescence intensity at 650 nm (F650 nm) in the wide range from 0.1 to 200 U mL−1. The calibration equation is F650 nm = 0.439 × [Fpg] + 2.79, with a correlation coefficient of R = 0.998. The detection limit can be as low as 0.01 U mL−1 (S/N = 3). The selectivity of this Fpg biosensor was further evaluated with several other enzymes instead of Fpg. As shown in Fig. 3D, besides Fpg, the other enzymes did not lead to evident fluorescence at 650 nm, indicating the high specificity of this biosensor. In comparison with the reported approaches for analyzing enzymes eliminating 8-oxoG, the biosensor revealed a lower detection limit and a wider linear range (Table S1†), which were ascribed to the following reasons. First, the distinct effects of P on λ Exo and Exo I enable the quantification of Fpg in a “signal-on” manner, providing the basis for high-performance detection. Second, multi-enzyme catalysis is integrated into the analytical strategy, which ensures the high efficiency and selectivity of this biosensor. Third, nanomaterials provide enhanced fluorescent signals, rendering the readout of this biosensor more sensitive. Forth, the analytical method possesses the feature of “zero” background noise, which further improves the analytical performances.Analysis of Fpg in real samples
To investigate its practicality, the biosensor was first used to analyze Fpg in real human serum. Fpg at different concentrations was mixed with human serum samples diluted 10 times, and were then detected by the proposed method. The results of the spike and recovery tests are shown in Table 1. It was found that the relative standard deviations (RSDs) at any concentration of Fpg were all below 2%, and the recovery rates were between 98% and 102%, suggesting the reliability of this approach. In order to further investigate its clinical applicability, the biosensor was challenged with cell lysate. Cell lysate was obtained from four kinds of tumour cells (MCF-7 cells, F98 cells, U87 cells and A549 cells), and different concentrations of Fpg were added. As shown in Fig. 4, even if mixed with cell lysate, exogenous Fpg could still be detected with our method. Although the compositions of the serum samples and cell lysate were complex, Fpg in these samples could be measured accurately, indicating the good potential of the biosensor in clinical applications.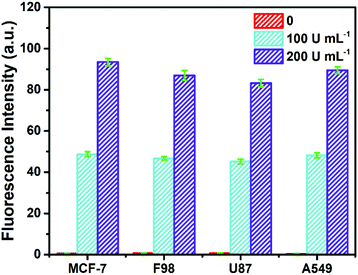 | ||
| Fig. 4 Determination of Fpg activity in diluted cell lysate with exogenous Fpg at different concentrations. | ||
| Added Fpg (U mL−1) | Measure Fpg (U mL−1) | RSD (%, n = 3) | Recovery (%) |
|---|---|---|---|
| 1 | 1.02 | 1.31 | 102 |
| 10 | 9.8 | 0.60 | 98 |
| 50 | 49.7 | 1.78 | 99.4 |
| 100 | 99.8 | 0.84 | 99.8 |
| 200 | 200.4 | 1.97 | 100.2 |
Conclusions
In this work, a novel biosensor was developed to analyze the activity of Fpg relying on multi-enzyme catalysis and fluorescent metallic nanoparticles. Since P at different termini exhibits opposite effects on λ Exo and Exo I, the biosensor works in a “signal-on” mode. In the absence of Fpg, DNA is totally digested by λ Exo and Exo I, and thus CuNCs cannot be prepared, resulting in “zero” background noise. In the presence of Fpg, ssDNA is generated and used to form CuNCs, which emit intense fluorescence as the readout of this approach. Benefitting from the high efficiency of enzyme catalysis and the intense fluorescence of CuNCs, the biosensor revealed a capability to quantify Fpg in a wide linear range from 0.1 to 200 U mL−1 with a low detection limit of 0.01 U mL−1. In addition, Fpg in serum samples and cell lysate could also be quantified accurately, manifesting the value of this method in clinical diagnosis. Overall, distinct effects of the same group on different enzymes were employed to construct a biosensor, which provides an alternative perspective for designing analytical methods using multi-enzyme catalysis and fluorescent nanomaterials.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China for the projects (No. 21625502 and 21974070), Natural Science Foundation of Jiangsu Province of China (No. BK20192008 and BK20191367) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_1168). All experiments involving human serum were performed in accordance with the guidelines of Safety and Welfare of Human Research of Nanjing Normal University and approved by the Ethics Committee of Nanjing Normal University. Informed consent was obtained from human participants of this study.Notes and references
- G. Padroni, J. M. Withers, A. Taladriz-Sender, L. F. Reichenbach, J. A. Parkinson and G. A. Burley, J. Am. Chem. Soc., 2019, 141, 9555–9563 CrossRef CAS PubMed.
- J. Buck, P. Grossen, P. R. Cullis, J. Huwyler and D. Witzigmann, ACS Nano, 2019, 13, 3754–3782 CrossRef CAS PubMed.
- Y. Wang, S. Wei, K. Wang, R. Xu and H. Zhao, Acta Chim. Sin., 2020, 78, 271–278 CrossRef.
- L. J. Wang, F. Ma, B. Tang and C. Y. Zhang, Anal. Chem., 2016, 88, 7523–7529 CrossRef CAS PubMed.
- Z. Wu, Z. K. Wu, H. Tang, L. J. Tang and J. H. Jiang, Anal. Chem., 2013, 85, 4376–4383 CrossRef CAS PubMed.
- M. Ren, J. Bai, Z. Xi and C. Zhou, Sci. China: Chem., 2019, 62, 561–570 CrossRef CAS.
- E. N. Thomas, C. L. Simms, H. E. Keedy and H. S. Zaher, Nucleic Acids Res., 2019, 47, 9857–9870 CrossRef CAS PubMed.
- J. Shang, Z. Li, L. Liu, D. Xi and H. Wang, ACS Sens., 2018, 3, 512–518 CrossRef CAS PubMed.
- S.-C. Liu, H.-W. Wu, J.-H. Jiang, G.-L. Shen and R.-Q. Yu, Anal. Methods, 2013, 5, 164–168 RSC.
- L. J. Wang, H. X. Wang, L. Jiang and C. Y. Zhang, ACS Sens., 2018, 3, 2675–2683 CrossRef CAS PubMed.
- X. Wang, T. Hou, T. Lu and F. Li, Anal. Chem., 2014, 86, 9626–9631 CrossRef CAS PubMed.
- F. Liu, T. Gao, Z. Ye, D. Yang, Z. Wang and G. Li, Electrochem. Commun., 2015, 50, 51–54 CrossRef CAS.
- A. Kreppel, I. D. Blank and C. Ochsenfeld, J. Am. Chem. Soc., 2018, 140, 4522–4526 CrossRef CAS PubMed.
- J. Fukae, M. Takanashi, S. Kubo, K. Nishioka, Y. Nakabeppu, H. Mori, Y. Mizuno and N. Hattori, Acta Neuropathol., 2005, 109, 256–262 CrossRef CAS PubMed.
- A. C. Drohat and C. T. Coey, Chem. Rev., 2016, 116, 12711–12729 CrossRef CAS PubMed.
- Q. Xi, J. J. Li, W. F. Du, R. Q. Yu and J. H. Jiang, Analyst, 2016, 141, 96–99 RSC.
- M. Flaender, G. Costa, G. Nonglaton, C. Saint-Pierre and D. Gasparutto, Analyst, 2016, 141, 6208–6216 RSC.
- J. Wang, Y. Wang, S. Liu, H. Wang, X. Zhang, X. Song and J. Huang, Analyst, 2018, 143, 3951–3958 RSC.
- A. Esadze and J. T. Stivers, Chem. Rev., 2018, 118, 11298–11323 CrossRef CAS PubMed.
- X. J. Kong, S. Wu, Y. Cen, R. Q. Yu and X. Chu, Biosens. Bioelectron., 2016, 79, 679–684 CrossRef CAS PubMed.
- H. Zhang, L. Wang, Y. Xie, X. Zuo, H. Chen and X. Chen, Analyst, 2019, 144, 3064–3071 RSC.
- J. Song, F. Yin, X. Li, N. Dong, Y. Zhu, Y. Shao, B. Chen, W. Jiang and C. Z. Li, Analyst, 2018, 143, 1593–1598 RSC.
- C. Y. Lee, K. S. Park and H. G. Park, Biosens. Bioelectron., 2017, 98, 210–214 CrossRef CAS PubMed.
- L. J. Wang, Q. Zhang, B. Tang and C. Y. Zhang, Anal. Chem., 2017, 89, 7255–7261 CrossRef CAS PubMed.
- X. Zhang, Y. Wang and X. Zhou, Anal. Chem., 2019, 91, 1665–1670 CrossRef CAS PubMed.
- J. Li, W. Fu, Z. Wang and Z. Dai, Chem. Sci., 2019, 10, 5616–5623 RSC.
- X. R. Song, N. Goswami, H. H. Yang and J. Xie, Analyst, 2016, 141, 3126–3140 RSC.
- M. Madsen and K. V. Gothelf, Chem. Rev., 2019, 119, 6384–6458 CAS.
- F. Xu, H. Shi, X. He, K. Wang, D. He, Q. Guo, Z. Qing, L. Yan, X. Ye, D. Li and J. Tang, Anal. Chem., 2014, 86, 6976–6982 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Morphology of ssDNA-3-templated CuNCs, diameter analysis of ssDNA-3-templated CuNCs, optimization of the dosage of λ Exo, optimization of the dosage of Exo I, fluorescence spectra of CuNCs with different concentrations of Fpg, comparison of analytical performances of different relevant biosensors and related references. See DOI: 10.1039/D0AN00928H |
| This journal is © The Royal Society of Chemistry 2020 |