Gold nanoparticle-streptavidin conjugates for rapid and efficient screening of aptamer function in lateral flow sensors using novel CD4-binding aptamers identified through Crossover-SELEX†
Tamika
Fellows
,
Lance
Ho
 ,
Shane
Flanagan
,
Shane
Flanagan
 ,
Ronen
Fogel
,
Ronen
Fogel
 ,
Dupe
Ojo
,
Dupe
Ojo
 and
Janice
Limson
and
Janice
Limson
 *
*
Rhodes University Biotechnology Innovation Centre, Grahamstown, Eastern Cape, South Africa. E-mail: j.limson@ru.ac.za
First published on 22nd June 2020
Abstract
To decrease the burden of laborious and reagent-intensive screening of modified aptamers, their binding function requires assessment in assay formats compatible with the end diagnostic application. Here, we report on the use of an alternative and cost-effective approach: a rapid lateral flow assay (LFA) utilising streptavidin-conjugated gold nanoparticles (AuNP) as reporter molecules to screen novel ssDNA aptamers for their ability to detect CD4. Crossover-SELEX was employed to identify CD4-targeting aptamers from a ssDNA library enriched against a recombinant human CD4 protein (hCD4) conjugated to magnetic-beads and to endogenous CD4 expressed by U937 cells. Counter-selection with IgG-conjugated beads and CD4-negative Ramos RA-1 cells was employed. Following SELEX, four sequences (U4, U14, U20 and U26) were selected for candidate screening. Fluorescence confocal microscopy showed comparable localization of the Cy5-labeled aptamer U26, compared to antibodies binding CD4's cytoplasmic domain. Aptamer-hCD4 binding kinetics were evaluated by a qPCR-based magnetic-bead binding assay to unmodified aptamers. U26 exhibited the highest binding affinity (Kd = 2.93 ± 1.03 nM) to hCD4-conjugated beads. Citrate-stabilized gold nanoparticles (mean particle diameter, 10.59 ± 1.81 nm) were functionalized with streptavidin to allow immobilization of biotin-labeled aptamers. Except for U4, the aptamer–gold nanoparticle conjugates (Apt–AuNP) remained stable under physiological conditions with their size (approx. 15 nm) appropriate for use in the LFAs. Lateral-flow based screening was used to evaluate the suitability of the Apt–AuNPs as CD4-detecting reporter molecules by immobilizing hCD4 and flowing the nanoparticle conjugates across the LFA. Using this approach, two novel sequences were identified as being suitable for the detection of hCD4: visual detection at 9 min was obtained using U20 or U26. After 20 min, equivalent colorimetric hCD4 responses were observed between anti-CD4 monoclonal antibody (ΔI = 94.19 ± 3.71), an existing CD4 aptamer F1-62 (ΔI = 90.31 ± 19.31) and U26 (ΔI = 100.14 ± 14.61) LFA's, each demonstrating high specificity to hCD4 compared to IgG. From the above, Crossover-SELEX allowed for the successful identification of ssDNA aptamers able to detect hCD4. Streptavidin-conjugated AuNPs, when bound to candidate aptamers, show potential application here as screening tools for the rapid evaluation of aptamer performance in low-cost lateral flow diagnostics.
Introduction
The CD4 (human Cluster of Differentiation 4) glycoprotein is mainly expressed on the surface of T-lymphocytes (T-cells) found in circulating blood1 and adipose tissue.2 CD4 functions as a co-receptor for peptides presented by the major histocompatibility complex (MHC) of antigen-presenting cells, used in T-cell activation of the immune response.3 Structurally, the extracellular region of CD4 is composed of four linked immunoglobulin-like domains that dictate its recognition function.3 Despite a critical role in immune function, CD4 is extensively researched for its role as a cellular portal of the Human Immunodeficiency Virus (HIV). As a consequence of HIV infection, the immune regulatory function is disrupted, leading to CD4-expressing (CD4+) T-cell depletion, maturation, and exhaustion of T-cells, chronic immune activation and inflammation.1 Consequently, the degree and progression of immunodeficiency are directly related to the decline of circulating T-cells as a result of cell rupture upon viral release.4Though a reciprocal trend exists between CD4+ cell counts and viral load,5 the correlation lacks linearity6 due to factors such as patients being late presenters of CD4+ cell decline,7 viral life cycle phases8 and viral rebound events.9 In practice, the evaluation of CD4+ cells has been established as a general indicator of HIV progression, care and management.10–12
Currently, clinical analysis of CD4+ cell counts mainly utilize fluorescently-labelled antibodies: immunochromatographic interactions between the antibody and CD4 antigen on T-cells in whole blood and subsequent measurement of fluorescent intensity by flow cytometry analysis.13–15 As most people infected with HIV live in low-income, resource-limited areas, flow cytometry remains out of reach as a routine diagnostic test within the public healthcare sector in those areas. As an expensive laboratory-confined technique, it also requires highly-trained personnel to operate and interpret data.13,16
Point-of-care (POC) diagnostic devices have emerged to meet the need for more cost-effective, accurate and sensitive means of CD4 antigen measurement for HIV monitoring.16–24 Existing POC devices utilize antibody-based recognition of the CD4 antigen coupled to either lateral flow sensors (LFS),16,20 impedance spectroscopy,15,21 microflow cytometry22,23 or other fluorescent imaging techniques to facilitate CD4+ cell detection.14,15,21
The choice of antibodies as biorecognition agents in commercial diagnostic devices monitoring CD4 levels is due to their high target selectivity, high affinity, and established immobilization strategies.16,25 However, antibodies are limited by high cost, thermal and chemical stability and their ability to only target antigenic molecules.25 The inability to utilize negative selection pressure during antibody generation further limits their specificity between a targeted antigen and homologous unrelated epitopes.26
Compared to antibodies, aptamers possess lower production costs, extended shelf-lives and are readily modified for immobilization and reporting purposes.26 These biorecognition agents are identified through Systematic Evolution of Ligands by EXponential enrichment (SELEX), a process that screens a library of oligonucleotides for the capability to recognize a specific target molecule with high affinity and selectivity.27 Several variations of SELEX exist, of which Cross-over SELEX combines aptamer enrichment against a purified target (recombinant protein, lipid, oligosaccharide, etc.) and whole-cells expressing the target in a native folding and post-translational modification state.28 In targeting cell-expressed protein, the advantage of Cross-over SELEX is to promote aptamer enrichment against conserved epitopes between the recombinant and native protein, with diminished interference from non-target cell surface markers.29
Aptamer sequences reported to bind recombinantly-produced30 or cell-expressed CD431–33 have found application in fluorescent detection of CD4+ cells, targeted CD4+ cell capture,31,34,35 and the delivery of siRNA to CD4+ cells for selective gene silencing in HIV therapy.36–38 Crossover-SELEX against both a CD4-IgG2 recombinant protein and CD4+ Karpas-299 cells identified the ssDNA aptamer, F1-62, which was shown to partially inhibit binding between HIV gp120 protein and CD4+ T-cells.31 Additionally, the CD4+ cell binding specificity of F1-62 was established against CD4-negative B-lymphoma and several other human cancer cell lines.31 Despite these promising studies, current literature reports no application of aptamers for CD4 detection in a POC device.32,33 The identification of additional CD4-targeting aptamers may also allow these biorecognition agents to become more applicable to POC devices, similar to the use of antibody pairs as target-capture and reporter components of commercial LFS.39
When combined with the simple, scalable and widely-accepted LFS format, aptamers present a viable alternative to antibodies for a rapid and cost-effective means to detect CD4, and other clinically-relevant biomarkers.40 For application in LFS, aptamers require modification to either signal target detection or to concentrate the target protein for signal enhancement. Various nanoparticles are conjugated to aptamers for colorimetric signal generation in LFS including latex beads,41 silica,42 silver,34 and most commonly gold nanoparticles.40,43–48 However, target recognition is dependent on the folded structure of aptamers often leading to disruption of their binding capability following modification or immobilization to solid supports.49,50 As the binding conformation of an aptamer is sequence-dependent, this further necessitates an evaluation of aptamer-target recognition on a case-by-case basis. Due to these limitations, the screening of aptamers requires assay formats with similar aptamer modifications and testing conditions to the intended sensing application. Lateral flow-based methods also present a standardised, rapid, and cost-effective means of functional screening.24
Here, we report on the use of a simple gold nanoparticle (AuNP)-based lateral flow assay (LFA) to screen novel CD4-targeting aptamers generated by Crossover-SELEX. The LFA proposed here shares the advantages of LFS, specifically the low cost, ease of assembly and accepted use. Similarly, AuNPs are established colorimetric reporters compatible with existing LFS technologies. As explored here, further modification of the AuNPs with streptavidin provides a generic reporter molecule for the assessment of aptamer-target binding. This approach allows for the rapid functionalization of streptavidin-AuNPs with a variety of biotin-labeled aptamers and the screening thereof for target recognition in the LFA.
Experimental
Materials and methods
The ssDNA library [5′-GCCTGTTGTGAGCCTCCTAAC-(49N)-CATGCTTATTCTTGTCTCCC-3′] and all aptamer sequences were purchased from Integrated DNA Technologies (Coralville, USA), reconstituted to 100 μM stock concentrations in 10 mM Tris-EDTA (TE) buffer pH 8.0, and stored at −20 °C until use. Human CD4 (hCD4) protein (recombinantly expressed in Human Embryonic Kidney 293 cells; >95% pure, ab167756) was obtained from Abcam (Cambridge, USA) and reconstituted in 10 mM HEPES, pH 7.4 at a concentration of 0.5 mg mL−1. The hCD4 protein corresponds to amino acids 26–390 of the extracellular domain of CD4 (Uniprot: P01730) and is fused to an Fc fragment of human IgG1 at the C-terminus.Rabbit anti-hCD4 (extracellular domain) monoclonal antibody (ab133622) denoted mAB A, mouse anti-hCD4 (cytoplasmic domain) monoclonal antibody (ab25804), goat anti-rabbit IgG (H + L) horseradish peroxidase (HRP)-conjugated polyclonal antibody (ab205718) and goat anti-mouse IgG (H + L) Cy3-labeled polyclonal antibody (ab97035) were sourced from Abcam. Rabbit anti-hCD4 (extracellular domain) monoclonal antibody (10400-R104) denoted mAB B, was obtained from Sino-Biological (CH). Human immunoglobulin G (IgG, 401114) was purchased from Calbiochem (Merck-Millipore, USA).
Unless specified, reagent-grade chemicals were obtained from Sigma-Aldrich (USA) and Merck (DE). All aqueous solutions used were prepared with Milli-Q water (>18.2 MΩ cm−1), prepared from a Direct-Q 3 Millipore (Merck-Millipore, USA). Phosphate-buffered saline (1× PBS) consisted of 137 mM NaCl, 2.7 mM KCI, 1.8 mM KH2PO4 and 10 mM Na2HPO4, pH 7.4. The PBS++ buffer consisted of a 1× PBS pH 7.4 solution, supplemented with 150 μM HSA, 0.45% (w/v) glucose, and 5 mM MgCl2. Once filter-sterilized (0.2 μm syringe filter, GVS Life Sciences, UK), PBS++ was used as the main buffer for aptamer generation via SELEX. Quantitative PCR (qPCR) analyses were performed using a QuantStudio™ 3 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, USA). Spectroscopy (UV-Vis) was performed using a FLUOstar Omega microplate reader (BMG Labtech, DE) and transmission electron microscopy (TEM) by a Zeiss Libra 120 TEM operating at 80 kV (DE). A description of additional materials and apparatus used in this study is described in the ESI.†
Preparation of mammalian cell lines
Non-adherent human histiocytic lymphoma cell lines U937 (ATCC® CRL-1593.2) and Ramos RA-1 (ATCC® CRL-1596) were sourced from ATCC (UK). The Ramos RA-1 cell line was used in SELEX as the counter-selection target due to its absence of CD4 expression. Both cell lines were cultured in RPMI growth media – supplemented with 5% (v/v) fetal calf serum and 1% (v/v) penicillin–streptomycin (P/S) solution – within vented culture flasks at 37 °C in 10% CO2. Cell lines were monitored by PCR for the absence of mycoplasma infection. Cell viability was monitored with trypan blue cell exclusion assays using a Brightline hemocytometer viewed under a Zeiss Axiovert A1 F1-LED epifluorescent microscope.51 For SELEX enrichment, cell populations exhibiting ≥95% viability were harvested, washed with PBS++ through centrifugation at 150g for 5 min, and incubated with 500 pmol sheared salmon sperm DNA for 5 min at 25 °C under slow tilt rotation.52 Harvesting and preparation of cells occurred immediately before use in SELEX.Preparation of ssDNA library
Initially, 10 μL of the 100 μM stock of the commercially obtained ssDNA library (corresponding to ∼6 × 1014 ssDNA oligonucleotides) was dissolved in PBS++ to create an aptamer candidate pool. For all subsequent SELEX rounds, approximately ∼1 μg of the enriched ssDNA library was dissolved in PBS++ and used for selection. Before use, all DNA pools were first resuspended in 1× PBS pH 7.4, and encouraged to form complex tertiary structures through heat denaturation and rapid cooling as detailed by:53 heating the pools to 95.5 °C for 10 min and cooled at −20 °C for 5 min. The ssDNA solution was then supplemented with HSA, MgCl2, and glucose to formulate PBS++, and allowed to thaw at room temperature for at least 15 min before use.Protein functionalization of M-270 epoxy magnetic-beads
The target (hCD4) and control (human IgG) proteins were conjugated to epoxy-functionalized M-270 DynaBead® superparamagnetic-beads (2.8 μm, Life Technologies) at ratios of 1 μg protein to 1 mg magnetic-beads, according to the manufacturer's instructions. Briefly, a 5 mg sample of magnetic-beads (∼3 × 108 beads) were resuspended in 1 mL of 100 mM potassium phosphate buffer pH 7.4, vortexed for 30 s and incubated under constant tilt-rotation for 10 min. The beads were then collected by a magnetic force for 1 min and the supernatant discarded; this procedure was repeated for a second wash. Resuspended beads were incubated with 5 μg protein dissolved in 1 mL of 100 mM potassium phosphate buffer pH 7.4 with 1 M ammonium sulphate overnight at 25 °C under constant tilt-rotation. Following functionalization, the protein-conjugated magnetic-beads were collected and washed twice by repeated resuspension-collection in 1 mL aliquots of 100 mM potassium phosphate buffer pH 7.4. Remaining unreacted epoxy groups were blocked by a 1 h incubation in 100 mM Tris-HCl pH 8.5 under tilt-rotation, followed by four washes in 1 mL 1× PBS, pH 7.4. Unless otherwise stated, the final protein-conjugated beads were stored at a concentration of 1 mg mL−1 in PBS++ at 4 °C until use.Crossover-SELEX
The Crossover-SELEX strategy combined selection against the CD4 target as a purified recombinant protein (hCD4) and as an endogenously expressed protein on the cell membrane surface (whole histiocytic lymphoma cells, U937). This was employed to enhance the aptamer specificity to CD4 in its native state and avoid generating aptamers to non-target cell surface constituents.31 A schematic and table of selection conditions for each SELEX round conducted in the Crossover-SELEX strategy to isolate CD4-targeting aptamer candidates are detailed in Fig. 1 and Table 1. For magnetic-bead protein SELEX counter selection, the snap-cooled ssDNA library was incubated with 5 mg of IgG-conjugated beads for 1 h at 25 °C. During counter selection in round 1, 0.75% (w/v) glycine and 0.61% (w/v) Tris-HCl were also added to PBS++ to account for the presence of Tris and glycine present in the commercial hCD4 protein solution. This amendment was not included in rounds 2 and 3.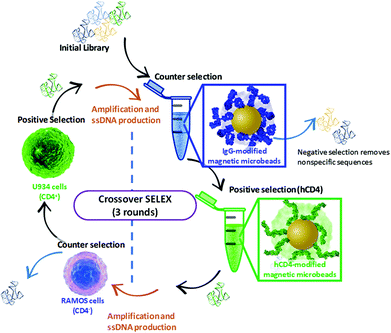 | ||
| Fig. 1 Schematic representation of the Crossover-SELEX strategy designed in this study. Clockwise, from top: enrichment of the ssDNA library first employed magnetic-bead protein SELEX using IgG beads and hCD4 beads for counter and positive selection, respectively. This method is previously described by ref. 54, with minor modifications. After PCR amplification and λ-exonuclease digestion, further enrichment of the ssDNA pool was performed using whole-cell SELEX against Ramos RA-1 and CD4 + U937 cells for counter and positive selection, conducted as described by ref. 52 and 55 with several modifications. Alternating protein and cell SELEX approaches were repeated for subsequent ssDNA pool enrichment as outlined in Table 1. | ||
| Round | ssDNA added | SELEX type | Selection type | Target | Selection binding time (min) |
|---|---|---|---|---|---|
| 1 | 6 × 1014 (1 nmol) | Protein | Counter | 5 mg IgG beads | 60 |
| Positive | 5 mg hCD4 beads | 30 | |||
| PCR amplification and production of 1 μg (3.3 pmol) ssDNA | |||||
| Cell | Counter | 5 × 106 Ramos RA-1 cells | 60 | ||
| Positive | 5 × 106 U937 cells | 30 | |||
| PCR amplification and production of 1 μg (3.3 pmol) ssDNA | |||||
| 2 | 2 × 1012 (3.3 pmol) | Protein | Counter | 5 mg IgG beads | 60 |
| Positive | 5 mg hCD4 beads | 15 | |||
| PCR amplification and production of 1 μg (3.3 pmol) ssDNA | |||||
| Cell | Counter | 5 × 106 Ramos RA-1 cells | 60 | ||
| Positive | 2.5 × 106 U937 cells | 15 | |||
| PCR amplification and production of 1 μg (3.3 pmol) ssDNA | |||||
| 3 | 2 × 1012 (3.3 pmol) | Protein | Counter | 5 mg IgG beads | 60 |
| Positive | 5 mg hCD4 beads | 10 | |||
Subsequently, for magnetic-bead protein SELEX positive selection, the IgG-modified beads were removed via magnetic force and the supernatant was immediately applied to 5 mg hCD4-conjugated beads with slow tilt rotation at 25 °C as specified in Table 1. Following exposure, the beads were collected and the supernatant was removed and reserved for analysis. ssDNA bound to hCD4-modified beads was eluted by resuspending the collected beads in 200 μL of elution buffer (40 mM Tris, 10 mM EDTA, 3.5 M urea, and 0.02% (v/v) Tween-20, adjusted to pH 8), heating the solution to 80 °C for 10 min.55 Eluted ssDNA was recovered through ethanol precipitation in the presence of sodium acetate and glycogen at −20 °C for 24 h. The resulting ssDNA was resuspended in 30 μL of ddH2O and used as a template for PCR amplification. Amplification of the ssDNA pools was optimized to avoid the inclusion of cross-primer and concatemer artefacts.56,57 Following PCR amplification, dsDNA was digested to ssDNA with λ-exonuclease (ESI†), purified and used in the next round of Crossover-SELEX against CD4 + U937 cells.
Similar to the above, SELEX enrichment to CD4 + U937 cells is divided between a counter-selection stage preceding the positive selection. During counter-selection of the cell stage of SELEX, ∼1 μg of ssDNA (2 × 1012 oligonucleotides) aptamer pool was incubated with 5 × 106 Ramos RA-1 cells for 1 h at 25 °C under slow tilt-rotation, the cells were subsequently pelleted by centrifugation at 800g for 5 min. The supernatant was removed, and the cells were washed once through resuspension in 1 mL of PBS++ and centrifuged at 800g for 5 min. The supernatants of the two wash steps were pooled and incubated with U937 cells at 25 °C under slow tilt-rotation, for round-dependent durations detailed in Table 1. Following exposure to the ssDNA pools, the U937 cells were washed 3× through centrifugation at 800g for 5 min and successive resuspensions in 1 mL of PBS++.
U937-bound ssDNA was collected by heat-induced denaturation of the U937 cell pellet. Briefly, cells were heated to 95 °C for 10 min, the cell debris collected through centrifugation at 2000g for 5 min and the remaining supernatant was column-purified to recover the enriched ssDNA sequences for PCR amplification and λ-exonuclease digestion (further details presented in ESI†). The SELEX cycle of protein and cell positive- and counter-selection was repeated with the enriched ssDNA pool of sequences. For each SELEX round, enrichment of ssDNA was quantified by qPCR as outlined in Fig. S1 and Table S2 (ESI).†
The enriched ssDNA pool attained from the final SELEX round was ligated into a pGEM®-T Easy vector, transformed into competent DH5α E. coli cells and plated for blue-white screening. Extracted DNA of the isolated colonies were prepared using a BigDye® Direct Cycle Sequencing Kit, column purified and sent for Sanger sequencing by the NRF-SAIAB Molecular Genetics Laboratory (South Africa). Details of the approach are outlined in the ESI.†
Selection of aptamer sequences
The sequenced oligonucleotides obtained from the final round of Crossover-SELEX were aligned using MUSCLE 6.0 Multiple Sequence Alignment tool to assign phylogeny. The following aptamer candidates were selected from bioinformatic analysis: U4, U14, U20 and U26. For each obtained sequence, secondary structure analyses were performed using the RNA fold server from the ViennaRNA Web Services, (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). Folding parameters were selected for linear DNA (Matthews model, 2004) at 25 °C allowing for dangling energies on both sides of a helix, using minimum free energy (MFE) and partition function fold algorithms. The structure prediction algorithm was set to incorporate G–Quadruplex formation. Two control sequences were used within this study: A previously reported CD4 binding aptamer, F1-6231 and a C27 negative control sequence containing a randomized variable region flanked by primer regions identical to the enriched aptamer candidates. The candidate aptamer and control sequences are described in Table 2.| Aptamer | Variable region sequence 5′ → 3′ | Length (bases) |
|---|---|---|
| U4 | CF-TTCTCTCTCTTTGCTTTCATGTCGGGTAGGTCACACCACTTTGTTGTTC-CR | 90 |
| U14 | CF-ACGTTAAAGTGAATTCTAACCTAGTGAGTTTTTCGTCTTGTATTATTGG-CR | 90 |
| U20 | CF-TTATATGATGCATCAGCGCGAGGGCGACACCGCTACTCGGGTCGATTTT-CR | 90 |
| U26 | CF-GATGTCGACGTGCAGCTTCCTTGAGCCTTACTGAAAATACTACCCAGTC-CR | 90 |
| C27 | CF-TAGCTCGTAGAAAAAAAATATAAAGGGCGTGTGCTGGGACTGCTCGGGATTGCGGACA-CR | 99 |
| F1-62 | ATCCAGAGTGACGCAGCACCACCACCGTACAATTCGCTTTCTTTTTTCATTACCTACTCTGGC | 63 |
Cy5 fluorescently labeled aptamer evaluation through confocal microscopy
Aptamer candidates were Cy5-labeled at the 5′ end and assessed for binding to U937 cells by confocal microscopy. Approximately 9 × 105 U937 cells were harvested, washed and resuspended in PBS++ containing 1 μM Cy5-labeled aptamers for 30 min at room temperature. After a 3× wash step, the cells were collected by centrifugation at 500g and resuspended in 100 μL PBS++. Once vortexed, 10 μL of each cell solution was transferred to a PLL coated coverslip, air-dried for 10 min and incubated with 100 μL 4% (v/v) paraformaldehyde in PBS++ for 30 min at RT. Excess paraformaldehyde was removed, and the fixed cells were stained with 1 μg mL−1 DAPI for 1 min and mounted with DAKO mounting medium. The mounted cells were visualized using the Zeiss LSM 780 Confocal Scanning Microscope using 355 nm UV excitation for DAPI, multiline 458/488/514 nm excitation for Cy3 and, 633 nm excitation for Cy5. Images were analyzed using Zen 2 software.Binding affinity determination using qPCR magnetic-bead assay
Before qPCR kinetic analysis, the 5′-biotin-labeled ssDNA aptamers were heat-denatured at 95 °C for 5 min in 1× PBS pH 7.4, thereafter rapidly cooled and maintained at 4 °C for 30 min. For each aptamer, separate 50 μg hCD4-conjugated bead amounts were incubated with aptamer concentrations of 1 μM, 0.5 μM, 0.1 μM, 0.05 μM, 0.01 μM, 0.001 μM, 0.0001 μM in 100 μL 1× PBS pH 7.4 at 25 °C for a period of 2 h. Following incubation, the supernatant (unbound aptamer fraction) was discarded from the hCD4-conjugated beads collected by magnetic force. The magnetic-beads were washed twice by 200 μL 1× PBS pH 7.4 containing 0.001% Tween-20, once with 200 μL 1× PBS pH 7.4, and resuspended to a final volume of 50 μL in 1× PBS pH 7.4. Aptamer sequences were amplified directly off the hCD4-conjugated beads using a reaction mixture utilizing a proprietary SYBR Green PCR master mix (qMax Green No Rox qPCR Mix, Accuris, USA). A single qPCR reaction included 1 μg magnetic-bead ssDNA template, 0.5 μM SELEX library forward primer (5′-GCCTGTTGTGAGCCTCCTAAC-3′), 0.5 μM 5′-phosphorylated SELEX library reverse primer (5′-PO3-GGGAGACAAGAATAAGCATG-3′) and 1× propriety master mix (containing reaction buffer, nucleotides, fluorogenic dye and HotStart DNA polymerase) made up to 10 μL with ddH2O. The PCR cycle parameters were as follows: initial hold at 30 °C for 30 s followed by 95 °C for 2 min; 40 cycles of denaturation at 95 °C for 5 s, annealing at 54 °C for 20 s and extension at 72 °C for 10 s.Minor modifications of the qPCR procedure were necessary for the F1-62 aptamer, utilizing forward and reverse primers of 5′-ATCCAGAGTGACGCAGCA-3′ and 5′-GCCAGAGTAGGTAATGAA-3′. All qPCR amplification was performed in triplicate. The derived Cq values of the hCD4-conjugate bead-bound ssDNA were normalized to those obtained from 5 fmol, 1 fmol, 0.5 fmol, 0.1 fmol, 0.01 fmol, 0.001 fmol, 0.0001 fmol standard additions of each corresponding aptamer amplified by qPCR as described above. For affinity analysis, normalized Cq values were modeled to the Langmuir isotherm, using the least-squares regression algorithm in Statistica 13, by eqn (1).58
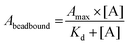 | (1) |
Preparation of the lateral flow assay screening platforms
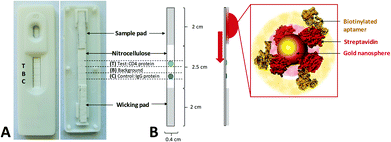
The Lateral Flow Assay (LFA) was composed of 3 primary components: sample pad, nitrocellulose strip and wicking pad, as detailed in Fig. 2. Protein solutions (0.5 mg mL−1) of the target hCD4 (test protein) and control IgG proteins were separately drop-dried in 2 × 0.25 μL aliquots onto the nitrocellulose strip at their designated zones (Fig. 2), using a 37 °C drying time of 10 min between each aliquot addition. To remove nonspecific binding sites on the material, the entire nitrocellulose strip was then blocked for 20 min by immersion in 1× PBS containing 5% (w/v) milk powder. The blocked strip was subsequently dried at 37 °C for 2 h and stored in an airtight container at 4 °C, for a maximum of 7 days until use. The sample pad was similarly blocked in 5% (w/v) milk powder in 1× PBS and dried. The wicking pad, prepared nitrocellulose strip and sample pad were assembled on the surface of adhesive cardboard and mounted into a plastic cassette. The LFA was stored at room temperature until use.AuNP synthesis and characterization
Colloidal nanosphere gold particles (AuNP) were synthesized using conventional citrate reduction with minor modifications.59 A solution of 50 mL of water containing 400 μM dissolved HAuCl4·3H2O was heated to boiling under reflux and 2 mL of 1% (w/v) trisodium citrate was added slowly, in increments. After 30 min, the solution turned characteristically deep red in color and was subsequently cooled to 4 °C in the dark for storage and later use. Samples of the synthesized AuNPs were characterized using UV–Vis spectrophotometry and Transmission Electron Microscopy (TEM, Zeiss Libra 120). For TEM analysis, 5 μl of the citrate-stabilized AuNP solution was drop-dried on a carbon-coated copper grid before TEM analysis. Citrate-stabilized particle size distribution was evaluated over 390 individual counts using high-resolution TE micrographs.Aptamer functionalization of AuNPs for application in lateral flow assays
Citrate-stabilized AuNPs were used as colorimetric reporter molecules in the aptamer-based LFA. For aptamer functionalization, 1 μg streptavidin (1 mg mL−1 in PBS) was first incubated in 1 mL citrate stabilized AuNP solution for 2 h at room temperature under slow tilt-rotation.60 For each test, 200 μL of the AuNP solution was collected by centrifugation at 7000g for 5 min and the supernatant was removed. To immobilize the aptamer sequences, streptavidin-coated nanoparticles (SA-AuNP) were incubated with 250 nM heat-treated aptamer in 100 μL 1× PBS for 1 h at RT under slow tilt-rotation. The aptamer–functionalized AuNP conjugates (Apt–AuNP) were collected at 7000g for 5 min and the supernatant was removed. The Apt–AuNP were resuspended in 50 μL 1× PBS and directly added to the sample well of the prepared LFA. To serve as a positive control, two rabbit (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution) monoclonal antibodies targeting the extracellular domain of CD4 were each separately incubated in citrate stabilized AuNPs for 2 h at RT under slow tilt-rotation. For each test, 200 μL of the antibody-coated AuNPs were collected by centrifugation at 7000g for 5 min, resuspended in 50 μL 1× PBS and directly added to the LFA sample well.
1000 dilution) monoclonal antibodies targeting the extracellular domain of CD4 were each separately incubated in citrate stabilized AuNPs for 2 h at RT under slow tilt-rotation. For each test, 200 μL of the antibody-coated AuNPs were collected by centrifugation at 7000g for 5 min, resuspended in 50 μL 1× PBS and directly added to the LFA sample well.
Image and video footage were captured for 15 min upon initial addition of the Apt–AuNPs to the LFA using a Samsung Galaxy Note 10+ primary camera (12MP sensor with 1.4 μm pixels, 26 mm (equivalent) variable-aperture f/1.5–2.4 lens, dual-pixel AF, OIS). A still image captured every minute of the LFA run was evaluated for colorimetric signal intensity using the color histogram function of the Fiji image-processing package (https://fiji.sc/). Here, the sum of square differences in individual RGB color components of a 30 × 30 pixel sample between the target protein and blank regions of the LFA was returned as a measure of colorimetric signal intensity in eqn. (2).61
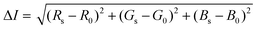 | (2) |
Where the colorimetric signal intensity is represented by ΔI, the RGB color of the target protein Rs, Gs and Bs and corresponding R0, G0 and B0 as the RGB of the LFA blank region.
Statistical analysis
Where stated, experimental reproducibility was reported as the arithmetic mean between individual measurements ± standard error of the mean. The number of individual measurements (n) is reported for each study. Statistical difference in the means of normally distributed measurements between sensor zones on the same sensors was identified using two-tailed, equal-variance Student's t-tests. Unless otherwise stated, significant differences in the means of three or more normally-distributed groups were assigned using one-way ANOVA; significantly-different groups were identified from their counterparts using Tukey's Honest Significant Difference post hoc test. All statistical analyses were performed using Statistica® 13.5. For all statistical analyses, the significance threshold, α, was set to 0.05.Results and discussion
Bioinformatic characterization of aptamer candidates
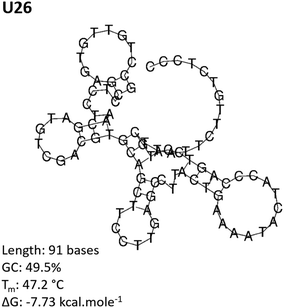
The aptamer pool obtained after five selection conditions was cloned into competent E. coli and sequenced using the Sanger method. Bioinformatic analysis of the sequences was conducted to identify potential aptamer candidates and their secondary structures. A summary of all the sequences obtained from the SELEX process is presented in the ESI, Table S1.† Sequences U4, U14, U20 and U26 were selected as representative aptamer candidates from phylogenetic analysis. From Fig. 3 and Fig. S2,† these sequences show several unique conformational stem and loop secondary structures.62–64Cy5-aptamer localization via confocal microscopy
Fluorescence confocal microscopy was used to determine the localization of the Cy5-labeled aptamer candidates upon exposure to U937 cells, Fig. 4. Candidate localization was compared to a previously reported CD4 binding aptamer, F1-62, and a C27 negative control sequence containing a randomized variable region flanked by primer binding sites identical to the enriched aptamer candidates.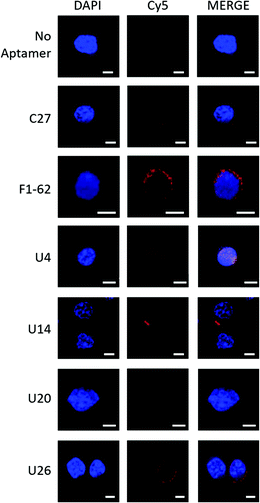 | ||
| Fig. 4 Fluorescence confocal microscopy demonstrating localization of Cy5-labeled aptamer candidates to U937 cells. Cy5-aptamer localization following a 30 min incubation with CD4-expressing U937 cells fixed onto PLL coated coverslips with 4% paraformaldehyde in PBS++ and mounted with DAKO mounting medium. Slides were visualized for the presence of Cy5 fluorescence with the Zeiss LSM 780 Confocal Scanning Microscope using three illuminating lasers (UV Laser 355 nm for DAPI excitation (355/461) and Argon Laser 633 nm for Cy5 excitation (633/666)). Images were processed with Zen 2 microscopy software. Unlabeled cells (‘No aptamer’) exposed to both the UV and Argon lasers presented a fluorescent blue nucleus stained by DAPI. Similarly, Cy5-C27 control staining demonstrated no presence of the Cy5 fluorescence localized to the U937 cells (‘C27’). In contrast, the mouse anti-hCD4 (cytoplasmic domain) monoclonal antibody and F1-62 aptamer demonstrated cell membrane-localized fluorescence signal at 568 nm (Fig. S3,† 1° and 2° Ab) and 666 nm (‘F1-62’), respectively. Scale bar: 5 μm. | ||
The Cy5-labeled U26 aptamer (Fig. 4, U26) presented clusters of fluorescence around the cell membrane periphery and along the peri-nuclear cell membrane defined by DAPI staining. Similar membrane clustering was observed for the Cy5-labeled F1-62 positive control. The small clustering effect may be due to the enrichment of CD4 in lipid rafts which plays a role in enhanced immune response.65,66 Cell staining was also observed for the U4 aptamer candidate (Fig. 4, U4). However, the fluorescent profile of this aptamer indicates binding localized to the nuclear region which was attributed to non-specific cellular uptake of the aptamer due to loss of cell membrane integrity.51,67,68 Aptamers U14 and U20, and the C27 control failed to elicit an observable Cy5 fluorescent response (Fig. 4, U14, U20 and C27).
Binding affinity characterization via magnetic-bead-based qPCR
Dissociation constants of the selected candidates were determined using a qPCR-based hCD4-conjugated magnetic-bead assay. Antibody magnetic-bead based ELISA validated both bead immobilization of hCD4 and exposure of the CD4 extracellular domain to solvent-accessible areas (Fig. S4†). As hCD4 was randomly orientated onto the magnetic-beads, evident exposure of the hCD4 extracellular domain proved varied epitope presentation to aptamer candidates during the qPCR-based binding assay.From Fig. 5, the successive increase in sequence retention to hCD4-conjugate beads was indicative of candidate aptamer binding affinity to hCD4 in a concentration-dependent manner. Among the novel sequences generated in this study, U26 showed the highest affinity for hCD4 with a Kd value of 2.93 ± 1.03 nM. The dissociation constants of the sequences U20, U14, U4, C27 and F1-62 were evaluated as 457.61 ± 126.67 nM, 118.26 ± 46.19 nM, 19.04 ± 9.18 nM, 287.61 ± 72.48 nM and 221.07 ± 24.49 nM respectively.
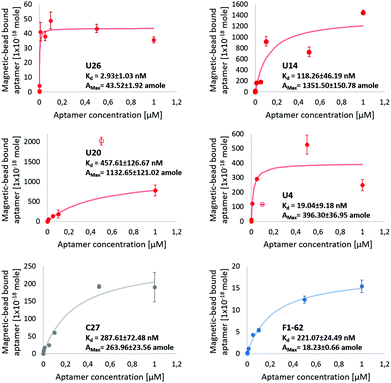 | ||
| Fig. 5 Screening of aptamer binding to hCD4-conjugated magnetic-beads, via qPCR. The U4, U14, U20, U26 aptamer and control C27 and F1-62 sequences at initial concentrations of 1 μM, 0.5 μM, 0.1 μM, 0.05 μM, 0.01 μM, 0.001 μM, 0.0001 μM and 0 μM exposed to 50 μg hCD4-conjugated magnetic-beads. Data points excluded from the modeled kinetic curve are shown as empty circles. The amount of ssDNA retained to hCD4-conjugated beads was determined relative to qPCR calibration curves of each respective aptamer, Fig. S5.† | ||
Variation in the apparent maximal binding capacities of the aptamer candidates to 50 μg hCD4-conjugated beads (Amax) was noted to two orders of magnitude, similar to the variation in Kd. Variation in Amax was not expected to arise from sequence-specific variation in amplification efficiencies of the aptamers (controlled by comparing bead-bound responses of each aptamer through standards generated using each respective sequence (Fig. S5†), nor from differences in the available protein sites between each aptamer (as bead masses were maintained between samples). Rather, observed differences in Amax between candidates may relate to differences in the rate constants by which the complexes assemble and dissociate.69 Aptamer candidate binding kinetics to immobilised hCD4 require further investigation by surface plasmon resonance to account for variation in binding capacity.
The evaluated binding affinity of F1-62 was lower than the previously reported Kd of 1.59 nM, determined through flow cytometry analysis of Cy5-labeled F1-62 binding to the CD4+ T-cell lymphoma cells, Karpas-299.31 This may be attributed to differences in the ligand-analyte pair and interaction environment between cell and magnetic-bead based affinity assays. Association of the C27 control provides evidence of nonspecific affinity to the hCD4-conjugated bead matrix by the selected aptamer candidates in the nM range. Despite potential nonspecific interactions, comparison of binding affinities tended to favor hCD4 binding in order of the sequences U26, U4, U14, F1-62 and U20.
Characterization of gold nanoparticle synthesis
Citrate-stabilized colloidal nanosphere gold particles (AuNP) were synthesized as described by Turkevich et al. with minor modification.59 TEM was used to validate synthesis and provide a physical measurement of the AuNP size range, Fig. S6A.† Fractions of the synthesized AuNPs were modified with streptavidin via physical adsorption (SA-AuNP), resuspended in PBS and subsequently functionalized with biotin-labeled aptamers (Apt–AuNP). In screening aptamer function for use in a lateral flow sensor configuration, it was necessary to apply the Apt–AuNPs at equivalent loadings. Thus, to evaluate the dispersion, size range and the apparent concentration of AuNPs modified with the various aptamers, UV–Vis spectroscopic characterization of each sample was conducted, Fig. S6B and C.† Nanoparticle-conjugate absorbance profiles estimated size and concentrations, as summarized in Table S3.†70Comparable sizing of the AuNPs was observed between TEM (10.59 ± 1.81 nm) and spectrophotometric (10.12 ± 1.12 nm) methods, despite experimental differences in AuNP synthesis of this study and that of W. Haiss et al.70 Resuspension of the synthesized AuNPs in 1× PBS caused salt-induced destabilization and aggregation of the nanoparticles, evidenced by their increased plasmon wavelength (from 523 to 646 ± 5 nm), Fig. S6B.†71 Attachment of streptavidin to the surface of the AuNPs via physical adsorption appears to prevent aggregation, with AuNPs exhibiting a small change in apparent size (12.64 ± 2.45 nm) compared to unmodified AuNPs.72 Streptavidin coating increased the hydration shell extending around the nanoparticles, shifting the plasmon resonance wavelength and the peak absorbance73 and stabilising the AuNPs in 1× PBS.
Immobilization of the biotin-labeled aptamers U14, U20, U26, C27 or F1-62 to the streptavidin-modified AuNPs (Apt–AuNP) increased the apparent particle diameter (average 15.36 ± 0.33 nm), as evident from the shift in spectral peaks in Fig. S6C.† However, no significant difference in particle diameter was found when compared to the streptavidin-modified AuNPs (Dunn's multiple comparison post hoc test, p > 0.05). In AuNP-based LFAs, the core particle size is indirectly proportional to detection sensitivity.45 Similarly, a large hydration shell inversely affects the diffusion speed of particles in solution.74 Hence, the small particle size of the Apt–AuNP conjugates, and associated hydration shell, were favorable for use in LFA screening.
Separately, a significant increase in particle size occurred upon immobilization of biotin-labeled U4 (d = 63.06 ± 1.28 nm) evident by the shift in the peak wavelength, λmax from 523 to 537 ± 1 nm. As U4 is of equal length, GC content and flanked by identical primer binding sites of the aptamer candidates, destabilization and subsequent aggregation of the U4 SA-AuNP conjugate remains unclear. Despite the promising binding affinity of U4 to hCD4-conjugated beads in qPCR assays, aggregation of the associated SA-AuNP conjugate excluded the application of the U4 aptamer in AuNP-based LFAs.
Screening of the Apt–AuNP conjugates for suitability in labeling hCD4 in an LFA format
Each Apt–AuNP conjugate was applied to an LFA screening platform to both evaluate their ability to bind hCD4, and potential use as reporter molecules in low-cost CD4 LFS. For LFA-based screening, SA-AuNPs were used as a conjugate negative control while F1-62 and C27 AuNP conjugates served as positive and negative hCD4 binding controls, respectively. Fig. 6 presents the multiple time point study of the various AuNP conjugates towards 250 ng of hCD4. As the time-dependent responses across the same sensors were examined in this study, a repeated measures variant of ANOVA was used to assign significant differences in the sensor responses, compared to the measured signal intensities at t = 0.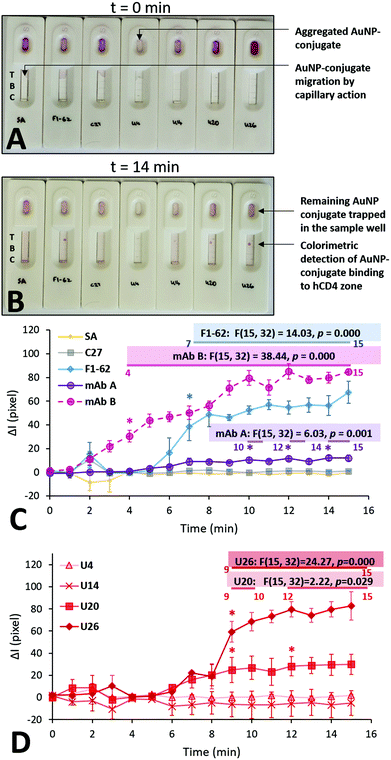 | ||
| Fig. 6 Multiple time point study evaluating the rate of colorimetric signal generation by the aptamer gold-nanoparticle conjugates on the LFA screening platforms. (A) Initial addition of 50 μL of the various AuNP conjugates; t = 0 min represents the time after the immediate addition of the last U26-AuNP conjugate. (B) Colorimetric detection of hCD4 by the various AuNP conjugates; t = 14 min represents the time after the immediate addition of the last U26-AuNP conjugate. (C) and (D) Comparison of the background-corrected quantified signal intensities at the CD4 zones, caused by the accumulation of the SA-AuNPs functionalized with U4, U14, U20, U26 aptamer and control C27 and F1-62 sequences at a concentration of 0.25 μM. Annotations show the relevant calculated statistics determined using repeated measures ANOVA. The asterisks and the numbered horizontal bars beneath the presented statistics show the times which have been determined to be statistically-higher than the baseline (t = 0) responses, as determined using Tukey HSD post hoc tests (n = 3 separate sensors). Recorded footage of the time point study is shown at 4× speed in Videos S7 and S8.† | ||
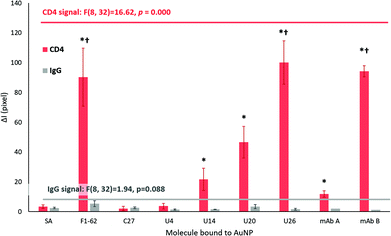
Upon addition to the Apt–AuNP screening platforms, the conjugates began migrating towards the test and control zones of the nitrocellulose via passive capillary action.39 Notably, the U4 Apt–AuNP conjugate was not recoverable following centrifugation (Fig. 6A) potentially due to factors leading the conjugate to an aggregated state (Fig. S6C†). The average migration rate of the various conjugates was consistent and rapid. For all stable Apt–AuNP conjugates, slight fluctuations were observed in ΔI for the initial 1–3 min due to discoloration of the nitrocellulose strip by the Apt–AuNP solution when compared to the dry nitrocellulose membrane (Fig. 6A and C). The ΔI of each sensor achieved a consistent baseline response following saturation of the nitrocellulose strip with the Apt–AuNP solution upon 4 min (Fig. 6B).
As shown in Fig. 6C, F1-62 produced a statistically significant (p = 0.000) increase in ΔI compared to the baseline response 7 min (p = 0.000) after the addition of the Apt–AuNP conjugate. Similarly, U26 produced a statistically significant response after 9 min (p = 0.000) and the U20 Apt–AuNP conjugate at 9 min (p = 0.029).
Comparison of aptamer hCD4 binding selectivity against human IgG
As for a rapid response, sensor selectivity is a crucial aspect to consider for Apt–AuNP application in LFS. For the screening platforms presented in Fig. 7, IgG was used as a measure of selectivity due to the similar domains shared by the extracellular region of CD4.75 Additionally, IgG forms a major component of blood plasma in combination with human serum albumin, transferrin, and fibrinogen.76The ΔI responses of the U14, U20 and U26 Apt–AuNP conjugates produced significant colorimetric responses to hCD4 in comparison to the IgG control (Fig. 7). The high selectivity of the aptamers for hCD4 may be attributed to stringent counter selection procedures against both IgG and the protein-complex membrane surface of Ramos RA-1 cells during Crossover-SELEX. After 20 min, a comparable colorimetric response to hCD4 was observed between U26 (ΔI = 100.14 ± 14.61) and the F1-62 control (ΔI = 90.31 ± 19.31) Apt–AuNP conjugate. Within the reported literature, this is the first time that F1-62 has been tested for its applicability towards CD4 LFS.
Using the LFA platform, hCD4 detection by two different monoclonal antibody–AuNP conjugates occurred after 4 min (mAB B, p = 0.001) and 10 min (mAB A, p = 0.000) with no apparent cross-reactivity to human IgG (Fig. 6C). Response time in commercial CD4 POC tests such as the Pima™ and Visitect® CD4 were interpretable in 20 min17 and 40 min20,77 respectively. After 20 min, the novel U26 aptamer produced a comparable hCD4 detection response when compared to the anti-CD4 antibody mAb B (ΔI = 94.19 ± 3.71, p = 1.000). This response was significantly greater than the mAb A (ΔI = 11.93 ± 2.19, p = 0.000).
AuNP-streptavidin conjugates were shown to be viable reporter molecules for the screening of aptamer binding to their target. From the response time, sensitivity and specificity of hCD4 detection, the LFA screening platform identified three different aptamers (U20, U26 and F1-62) for potential application in rapid, low-cost and selective aptamer-based CD4 lateral-flow sensors. Despite possessing notably different nucleotide sequences, the rapid detection of hCD4 by both F1-62 and U26 indicates potential application of the aptamers in sandwich-based LFS for use as target-capture and reporter sequences.
Conclusions
Novel CD4-targeting aptamers were successfully identified from a Crossover-SELEX approach combining magnetic-bead protein selection against recombinant hCD4, and whole-cell selection against CD4 + U937 cells. Aptamer candidate binding to CD4 was validated by fluorescent localization of Cy5-labeled aptamers to the U937 cell membrane periphery, and from aptamer binding kinetics towards hCD4-conjugated magnetic-beads. The aptamer U26 showed the highest affinity for hCD4-conjugated magnetic-beads showing a Kd value of 2.93 ± 1.03 nM.To evaluate the applicability of the aptamer candidates as biorecognition agents in low-cost, rapid diagnostics for CD4, the aptamers were biotin-labeled and immobilized to streptavidin-conjugated AuNPs for use as colorimetric reporters in a simple lateral-flow assay (LFA). Streptavidin-conjugation protected the AuNPs from salt-induced nucleation and aggregation. However, destabilization occurred for U4 aptamer-conjugated AuNPs excluding the potential use of this aptamer in similar lateral-flow sensors. Of the identified aptamers, both U20- and U26-AuNP conjugates showed rapid detection (9 min) of hCD4 and high selectivity compared to human IgG within the LFAs.
Proven hCD4 detection by the aptamers in the LFA allows for a more analogous transition towards their intended use as commercial CD4 AuNP-based lateral flow sensors. When incorporated into the LFA platform, the combination of biotin–streptavidin chemistry and AuNPs present an interchangeable screening method for identifying prospective aptamer-target interactions and their applicability to lateral-flow based diagnostics.
Conflicts of interest
The C27 control sequence is stated in a South African patent titled “Analysis of human immune status” with patent number: 2014/01017. A patent application regarding the novel CD4-targeting aptamers described in this work has been filed in South Africa.Acknowledgements
This research was supported in part by UNICEF Innovation. This work is also based on the research supported wholly or in part by the National Research Foundation of South Africa (Grant Number 95319). T. F. acknowledges the Mandela Rhodes Foundation for a postgraduate scholarship. The authors thank Dr Gwynneth F. Matcher of the National Research Foundation (NRF) – South African Institute of Aquatic Biodiversity (SAIAB) Molecular Genetics Laboratory (Rhodes University, South Africa) for performing Sanger DNA sequencing on the enriched SELEX library. Additional acknowledgment to Ms Shirley Pinchuck of the Electron Microscopy Unit at Rhodes University for assistance with TEM imaging.Notes and references
- B. Levast, L. Barblu, M. Coutu, J. Prévost, N. Brassard and A. Peres,
et al., HIV-1 gp120 envelope glycoprotein determinants for cytokine burst in human monocytes, PLoS One, 2017, 12(3), e0174550 CrossRef PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28346521.
- R. L. Travers, A. C. Motta, J. A. Betts, A. Bouloumié and D. Thompson, The impact of adiposity on adipose tissue-resident lymphocyte activation in humans, Int. J. Obes., 2015, 39(5), 762–769 CrossRef CAS
. Available from: http://www.nature.com/articles/ijo2014195.
- D. Glatzová and M. Cebecauer, Dual Role of CD4 in Peripheral T Lymphocytes, Front. Immunol., 2019, 10, 618 CrossRef PubMed
. Available from: https://www.frontiersin.org/article/10.3389/fimmu.2019.00618/full.
- K. K. Vidya Vijayan, K. P. Karthigeyan, S. P. Tripathi and L. E. Hanna, Pathophysiology of CD4+T-Cell Depletion in HIV-1 and HIV-2 Infections, Front. Immunol., 2017, 8, 580 CrossRef CAS
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28588579.
- M. Javaid, R. Nawaz, M. Natiq, S. Haider, S. Mazhar and M. Ghazanfar,
et al., Relationship of CD4+Count with Viral Load in HIV/AIDS Patients, Biomedica, 2019, 35(1), 41 Search PubMed
.
- C. J. Gill, J. L. Griffith, D. Jacobson, S. Skinner, S. L. Gorbach and I. B. Wilson, Relationship of HIV Viral Loads, CD4 Counts, and HAART Use to Health-Related Quality of Life, JAIDS, J. Acquired Immune Defic. Syndr., 2002, 30(5), 485–492 CrossRef
. Available from: http://journals.lww.com/00126334-200208150-00004.
- H.B Krentz, M.J Auld and M.C Gill, The high cost of medical care for patients who present late (CD4 <200 cells/microL) with HIV infection, HIV Med., 2004, 5(2), 93–98 CrossRef CAS
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15012648.
- J. M. Mabuka, A.-S. Dugast, D. M. Muema, T. Reddy, Y. Ramlakhan and Z. Euler,
et al., Plasma CXCL13 but Not B Cell Frequencies in Acute HIV Infection Predicts Emergence of Cross-Neutralizing Antibodies, Front. Immunol., 2017, 8, 1104 CrossRef PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28943879.
- M. Robertson, F. Laraque, H. Mavronicolas, S. Braunstein and L. Torian, Linkage and retention in care and the time to HIV viral suppression and viral rebound - New York City, AIDS Care, 2015, 27(2), 260–267 CrossRef PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25244545.
- E. P. G. Coakley, M. H. Samore, J. M. Gillis, M. D. Hughes and S. M. Hammer, The values of quantitative serum HIV-1 RNA levels and CD4 cell counts for predicting survival time among HIV-positive individuals with CD4 counts of ≤50×106 cells/l, AIDS, 2000, 14(9), 1147–1153 CrossRef CAS PubMed
. Available from: http://journals.lww.com/00002030-200006160-00011.
- E. L. Korenromp, B. G. Williams, G. P. Schmid and C. Dye, Clinical Prognostic Value of RNA Viral Load and CD4 Cell Counts during Untreated HIV-1 Infection—A Quantitative Review, ed. N. P. Pai, PLoS One, 2009, 4(6), e5950 CrossRef
. Available from: http://dx.plos.org/10.1371/journal.pone.0005950.
- WHO, Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV, 2015 [cited 2020 Mar 20]. Available from: http://www.who.int/hiv/pub/guidelines/earlyrelease-arv/en/.
- S. Thorslund, R. Larsson, F. Nikolajeff, J. Bergquist and J. Sanchez, Bioactivated PDMS microchannel evaluated as sensor for human CD4+cells—The concept of a point-of-care method for HIV monitoring, Sens. Actuators, B, 2007, 123(2), 847–855 CrossRef CAS
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0925400506007143.
- Z. Wang, S. Y. Chin, C. D. Chin, J. Sarik, M. Harper and J. Justman,
et al., Microfluidic CD4+T-Cell Counting Device Using Chemiluminescence-Based Detection, Anal. Chem., 2010, 82(1), 36–40 CrossRef CAS PubMed
. Available from: https://pubs.acs.org/doi/10.1021/ac902144w.
- D. S. Boyle, K. R. Hawkins, M. S. Steele, M. Singhal and X. Cheng, Emerging technologies for point-of-care CD4 T-lymphocyte counting, Trends Biotechnol., 2012, 30(1), 45–54 CrossRef CAS
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0167779911001193.
- M. T. Glynn, D. J. Kinahan and J. Ducrée, CD4 counting technologies for HIV therapy monitoring in resource-poor settings – state-of-the-art and emerging microtechnologies, Lab Chip, 2013, 13(14), 2731 RSC
. Available from: http://xlink.rsc.org/?DOI=c3lc50213a.
- G. Wu and M. H. Zaman, Low-cost tools for diagnosing and monitoring HIV infection in low-resource settings, Bull. W. H. O., 2012, 90(12), 914–920 CrossRef PubMed
. Available from: http://www.who.int/entity/bulletin/volumes/90/12/12-102780.pdf.
- N. Cassim, L. M. Coetzee, K. Schnippel and D. K. Glencross, Estimating Implementation and Operational Costs of an Integrated Tiered CD4 Service including Laboratory and Point of Care Testing in a Remote Health District in South Africa, ed. D. W. Dowdy, PLoS One, 2014, 9(12), e115420 CrossRef PubMed
. Available from: http://dx.plos.org/10.1371/journal.pone.0115420.
- M. W. Tenforde, A. S. Walker, D. M. Gibb and Y. C. Manabe, Rapid antiretroviral therapy initiation in low- and middle-income countries: A resource-based approach, PLoS Med., 2019, 16(1), e1002723 CrossRef PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30645592.
- S. Luchters, K. Technau, Y. Mohamed, M. F. Chersich, P. A. Agius and M. D. Pham,
et al., Field Performance and Diagnostic Accuracy of a Low-Cost Instrument-Free Point-of-Care CD4 Test (Visitect CD4) Performed by Different Health Worker Cadres among Pregnant Women, ed. A. M. Caliendo, J. Clin. Microbiol., 2018, 57(2), e01277-18 CrossRef
. Available from: http://jcm.asm.org/lookup/doi/10.1128/JCM.01277-18.
- X. Cheng, D. Irimia, M. Dixon, J. C. Ziperstein, U. Demirci and L. Zamir,
et al., A Microchip Approach for Practical Label-Free CD4+T-Cell Counting of HIV-Infected Subjects in Resource-Poor Settings, JAIDS, J. Acquired Immune Defic. Syndr., 2007, 45(3), 257–261 Search PubMed
. Available from: http://journals.lww.com/00126334-900000000-99751.
- H. Yun, H. Bang, J. Min, C. Chung, J. K. Chang and D.-C. Han, Simultaneous counting of two subsets of leukocytes using fluorescent silica nanoparticles in a sheathless microchip flow cytometer, Lab Chip, 2010, 10(23), 3243 RSC
. Available from: http://xlink.rsc.org/?DOI=c0lc00041h.
-
X. Mao, S.-C. S. Lin and T. J. Huang, High-throughput on-chip flow cytometry system using “microfluidic drifting” based three-dimensional (3D) hydrodynamic focusing, in TRANSDUCERS 2009–2009 International Solid-State Sensors, Actuators and Microsystems Conference, IEEE, 2009, pp. 425–428. Available from: http://ieeexplore.ieee.org/document/5285473/ Search PubMed
.
- G. A. Posthuma-Trumpie, J. Korf and A. van Amerongen, Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats, A literature survey, Anal. Bioanal. Chem., 2009, 393(2), 569–582 CrossRef CAS
. Available from: http://link.springer.com/10.1007/s00216-008-2287-2.
- P. J. Conroy, S. Hearty, P. Leonard and R. J. O'Kennedy, Antibody production, design and use for biosensor-based applications, Semin. Cell Dev. Biol., 2009, 20(1), 10–26 CrossRef CAS
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1084952109000147.
- A. D. Keefe, S. Pai and A. Ellington, Aptamers as therapeutics, Nat. Rev. Drug Discovery, 2010, 9(7), 537–550 CrossRef CAS PubMed
. Available from: http://www.nature.com/articles/nrd3141.
- S. D. Jayasena, Aptamers: an emerging class of molecules that rival antibodies in diagnostics, Clin. Chem., 1999, 45(9), 1628–1650 CrossRef CAS
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10471678.
- C. Pestourie, L. Cerchia, K. Gombert, Y. Aissouni, J. Boulay and V. De Franciscis,
et al., Comparison of Different Strategies
to Select Aptamers Against a Transmembrane Protein Target, Oligonucleotides, 2006, 16(4), 323–335 CrossRef CAS PubMed
. Available from: http://www.liebertpub.com/doi/10.1089/oli.2006.16.323.
- B. J. Hicke, C. Marion, Y.-F. Chang, T. Gould, C. K. Lynott and D. Parma,
et al., Tenascin-C Aptamers Are Generated Using Tumor Cells and Purified Protein, J. Biol. Chem., 2001, 276(52), 48644–48654 CrossRef CAS PubMed
. Available from: http://www.jbc.org/lookup/doi/10.1074/jbc.M104651200.
- K. A. Davis, Y. Lin, B. Abrams and S. D. Jayasena, Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry, Nucleic Acids Res., 1998, 26(17), 3915–3924 CrossRef CAS PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9705498.
- N. Zhao, S. Pei, P. Parekh, E. Salazar and Y. Zu, Blocking interaction of viral gp120 and CD4-expressing T cells by single-stranded DNA aptamers, Int. J. Biochem. Cell Biol., 2014, 51, 10–18 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1357272514000831.
- A. Nozari, Berezovski M V. Aptamers for CD Antigens: From Cell Profiling to Activity Modulation, Mol. Ther. – Nucleic Acids, 2017, 6, 29–44 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2162253116303699.
- J. Bala, S. Chinnapaiyan, R. K. Dutta and H. Unwalla, Aptamers in HIV research diagnosis and therapy, RNA Biol., 2018, 15(3), 327–337 CrossRef PubMed
. Available from: https://www.tandfonline.com/doi/full/10.1080/15476286.2017.1414131.
- J. Liu, J. Zeng, Y. Tian and N. Zhou, An aptamer and functionalized nanoparticle-based strip biosensor for on-site detection of kanamycin in food samples, Analyst, 2018, 143(1), 182–189 RSC
. Available from: http://xlink.rsc.org/?DOI=C7AN01476G.
- P. Zhang, N. Zhao, Z. Zeng, C.-C. Chang and Y. Zu, Combination of an Aptamer Probe to CD4 and Antibodies for Multicolored Cell Phenotyping, Am. J. Clin. Pathol., 2010, 134(4), 586–593 CrossRef CAS PubMed
. Available from: https://academic.oup.com/ajcp/article-lookup/doi/10.1309/AJCP55KQYWSGZRKC.
- Q. Zhu, T. Shibata, T. Kabashima and M. Kai, Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA, Eur. J. Med. Chem., 2012, 56, 396–399 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0223523412004710.
- L. A. Wheeler, V. Vrbanac, R. Trifonova, M. A. Brehm, A. Gilboa-Geffen and S. Tanno,
et al., Durable Knockdown and Protection From HIV Transmission in Humanized Mice Treated With Gel-formulated CD4 Aptamer-siRNA Chimeras, Mol. Ther., 2013, 21(7), 1378–1389 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1525001616319621.
- P. Song, Y. K. Chou, X. Zhang, R. Meza-Romero, K. Yomogida and G. Benedek,
et al., CD4 aptamer–RORγt shRNA chimera inhibits IL-17 synthesis by human CD4+T cells, Biochem. Biophys. Res. Commun., 2014, 452(4), 1040–1045 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0006291X14016532.
- K. M. Koczula and A. Gallotta, Lateral flow assays, ed. P. Estrela, Essays Biochem., 2016, 60(1), 111–120 CrossRef PubMed
. Available from: https://portlandpress.com/essaysbiochem/article/60/1/111/78237/Lateral-flow-assays.
- A. Chen and S. Yang, Replacing antibodies with aptamers in lateral flow immunoassay, Biosens. Bioelectron., 2015, 71, 230–242 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0956566315300427.
- E. M. Linares, L. T. Kubota, J. Michaelis and S. Thalhammer, Enhancement of the detection limit for lateral flow immunoassays: Evaluation and comparison of bioconjugates, J. Immunol. Methods, 2012, 375(1–2), 264–270 CrossRef CAS
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0022175911003085.
- V. C. Özalp, D. Çam, F. J. Hernandez, L. I. Hernandez, T. Schäfer and H. A. Öktem, Small molecule detection by lateral flow strips via aptamer-gated silica nanoprobes, Analyst, 2016, 141(8), 2595–2599 RSC
. Available from: http://xlink.rsc.org/?DOI=C6AN00273K.
- J. H. Lee, M. V. Yigit, D. Mazumdar and Y. Lu, Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates, Adv. Drug Delivery Rev., 2010, 62(6), 592–605 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0169409X10000724.
- S. C. B. Gopinath, T. Lakshmipriya, Y. Chen, W.-M. Phang and U. Hashim, Aptamer-based ‘point-of-care testing’, Biotechnol. Adv., 2016, 34(3), 198–208 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0734975016300106.
- D. Kim, Y. Kim, S. Hong, J. Kim, N. Heo and M.-K. Lee,
et al., Development of Lateral Flow Assay Based on Size-Controlled Gold Nanoparticles for Detection of Hepatitis B Surface Antigen, Sensors, 2016, 16(12), 2154 CrossRef PubMed
. Available from: http://www.mdpi.com/1424-8220/16/12/2154.
- E. Frohnmeyer, N. Tuschel, T. Sitz, C. Hermann, G. T. Dahl and F. Schulz,
et al., Aptamer lateral flow assays for rapid and sensitive detection of cholera toxin, Analyst, 2019, 144(5), 1840–1849 RSC
. Available from: http://xlink.rsc.org/?DOI=C8AN01616J.
- R. Reid, B. Chatterjee, S. J. Das, S. Ghosh and T. K. Sharma, Application of aptamers as molecular recognition elements in lateral flow assays, Anal. Biochem., 2020, 593, 113574 CrossRef CAS
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0003269719311534.
- E. B. Bahadır and M. K. Sezgintürk, Lateral flow assays: Principles, designs and labels, TrAC, Trends Anal. Chem., 2016, 82, 286–306 CrossRef
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0165993616300668.
- V. Ostatná, H. Vaisocherová, J. Homola and T. Hianik, Effect of the immobilisation of DNA aptamers on the detection of thrombin by means of surface plasmon resonance, Anal. Bioanal. Chem., 2008, 391(5), 1861–1869 CrossRef PubMed
. Available from: http://link.springer.com/10.1007/s00216-008-2133-6.
- C. Acquah, M. K. Danquah, J. L. S. Yon, A. Sidhu and C. M. Ongkudon, A review on immobilised aptamers for high throughput biomolecular detection and screening, Anal. Chim. Acta, 2015, 888, 10–18 CrossRef CAS PubMed
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0003267015008016.
- M. Avci-Adali, M. Metzger, N. Perle, G. Ziemer and H. P. Wendel, Pitfalls of Cell-Systematic Evolution of Ligands by Exponential Enrichment (SELEX): Existing Dead Cells During In Vitro Selection Anticipate the Enrichment of Specific Aptamers, Oligonucleotides, 2010, 20(6), 317–323 CrossRef CAS PubMed
. Available from: http://www.liebertpub.com/doi/10.1089/oli.2010.0253.
- K. Sefah, D. Shangguan, X. Xiong, M. B. O'Donoghue and W. Tan, Development of DNA aptamers using Cell-SELEX, Nat. Protoc., 2010, 5(6), 1169–1185 CrossRef CAS
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20539292.
- E Baldrich, A Restrepo and CK O'Sullivan, Aptasensor Development: Elucidation of Critical Parameters for Optimal Aptamer Performance, Anal. Chem., 2004, 76(23), 7053–7063 CrossRef CAS PubMed
. Available from: https://pubs.acs.org/doi/10.1021/ac049258o.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, FluMag-SELEX as an advantageous method for DNA aptamer selection, Anal. Bioanal. Chem., 2005, 383(1), 83–91 CrossRef CAS PubMed
. Available from: http://link.springer.com/10.1007/s00216-005-3388-9.
- D. Shangguan, Y. Li, Z. Tang, Z. C. Cao, H. W. Chen and P. Mallikaratchy,
et al., Aptamers evolved from live cells as effective molecular probes for cancer study, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(32), 11838–11843 CrossRef CAS PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16873550.
- K.-A. Frith, R. Fogel, J. P. D. Goldring, R. G. E. Krause, M. Khati and H. Hoppe,
et al., Towards development of aptamers that specifically bind to lactate dehydrogenase of Plasmodium falciparum through epitopic targeting, Malar. J., 2018, 17(1), 191 CrossRef
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29724225.
- L. S. J. Ho, R. Fogel and J. L. Limson, Generation and screening of histamine-specific aptamers for application in a novel impedimetric aptamer-based sensor, Talanta, 2020, 208, 120474 CrossRef PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31816738.
-
R. Joshi, Surface plasmon resonance, in Biosensors, Gyan Publishing House, Delhi, 2006, pp. 38–40 Search PubMed
.
- J. Turkevich, P. C. Stevenson and J. Hillier, A study of the nucleation and growth processes in the synthesis of colloidal gold, Discuss. Faraday Soc., 1951, 11, 55 RSC
. Available from: http://xlink.rsc.org/?DOI=df9511100055.
- F. Di Nardo, S. Cavalera, C. Baggiani, C. Giovannoli and L. Anfossi, Direct vs Mediated Coupling of Antibodies to Gold Nanoparticles: The Case of Salivary Cortisol Detection by Lateral Flow Immunoassay, ACS Appl. Mater. Interfaces, 2019, 11(36), 32758–32768 CrossRef CAS PubMed
. Available from: https://pubs.acs.org/doi/10.1021/acsami.9b11559.
- R. C. Murdock, L. Shen, D. K. Griffin, N. Kelley-Loughnane, I. Papautsky and J. A. Hagen, Optimization of a Paper-Based ELISA for a Human Performance Biomarker, Anal. Chem., 2013, 85(23), 11634–11642 CrossRef CAS PubMed
. Available from: https://pubs.acs.org/doi/10.1021/ac403040a.
- W. T. Horn, The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: Further challenges in the modeling of ligand-RNA interactions, RNA, 2004, 10(11), 1776–1782 CrossRef CAS PubMed
. Available from: http://www.rnajournal.org/cgi/doi/10.1261/rna.7710304.
- F. Nishikawa, K. Funaji, K. Fukuda and S. Nishikawa, In Vitro Selection of RNA Aptamers Against the HCV NS3 Helicase Domain, Oligonucleotides, 2004, 14(2), 114–129 CrossRef CAS PubMed
. Available from: http://www.liebertpub.com/doi/10.1089/1545457041526335.
- S. K. Lee, M. W. Park, E. G. Yang, J. Yu and S. Jeong, An RNA aptamer that binds to the β-catenin interaction domain of TCF-1 protein, Biochem. Biophys. Res. Commun., 2005, 327(1), 294–299 CrossRef CAS
. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0006291X04027871.
- Y. Percherancier, B. Lagane, T. Planchenault, I. Staropoli, R. Altmeyer and J.-L. Virelizier,
et al., HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains, J. Biol. Chem., 2003, 278(5), 3153–3161 CrossRef CAS PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12431990.
- R. Fragoso, D. Ren, X. Zhang, M. W.-C. Su, S. J. Burakoff and Y.-J. Jin, Lipid Raft Distribution of CD4 Depends on its Palmitoylation and Association with Lck, and Evidence for CD4-Induced Lipid Raft Aggregation as an Additional Mechanism to Enhance CD3 Signaling, J. Immunol., 2003, 170(2), 913–921 CrossRef CAS PubMed
. Available from: http://www.jimmunol.org/lookup/doi/10.4049/jimmunol.170.2.913.
- N. Li, J. N. Ebright, G. M. Stovall, X. Chen, H. H. Nguyen and A. Singh,
et al., Technical and Biological Issues Relevant to Cell Typing with Aptamers, J. Proteome Res., 2009, 8(5), 2438–2448 CrossRef CAS PubMed
. Available from: https://pubs.acs.org/doi/abs/10.1021/pr801048z.
- M. L. Raddatz, A. Dolf, E. Endl, P. Knolle, M. Famulok and G. Mayer, Enrichment of Cell-Targeting and Population-Specific Aptamers by Fluorescence-Activated Cell Sorting, Angew. Chem., Int. Ed., 2008, 47(28), 5190–5193 CrossRef CAS PubMed
. Available from: http://doi.wiley.com/10.1002/anie.200800216.
- G. Schreiber, G. Haran and H.-X. Zhou, Fundamental Aspects of Protein−Protein Association Kinetics, Chem. Rev., 2009, 109(3), 839–860 CrossRef CAS PubMed
. Available from: https://pubs.acs.org/doi/10.1021/cr800373w.
- W. Haiss, N. T. K. Thanh, J. Aveyard and D. G. Fernig, Determination of size and concentration of gold nanoparticles from UV-vis spectra, Anal. Chem., 2007, 79(11), 4215–4221 CrossRef CAS PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17458937.
- R. Pamies, J. G. H. Cifre, V. F. Espín, M. Collado-González, F. G. D. Baños and J. G. De La Torre, Aggregation behaviour of gold nanoparticles in saline aqueous media, J. Nanopart. Res., 2014, 16, 2376 CrossRef
.
- R. D'Agata, P. Palladino and G. Spoto, Streptavidin-coated gold nanoparticles: critical role of oligonucleotides on stability and fractal aggregation, Beilstein J. Nanotechnol., 2017, 8(1), 1–11 Search PubMed
. Available from: https://www.beilstein-journals.org/bjnano/articles/8/1.
- N. G. Khlebtsov, Determination of Size and Concentration of Gold Nanoparticles from Extinction Spectra, Anal. Chem., 2008, 80(17), 6620–6625 CrossRef CAS PubMed
. Available from: https://pubs.acs.org/doi/10.1021/ac800834n.
- A. Komalam, L. G. Muraleegharan, S. Subburaj, S. Suseela, A. Babu and S. George, Designed plasmonic nanocatalysts for the reduction of eosin Y: absorption and fluorescence study, Int. Nano Lett., 2012, 2(1), 26 CrossRef
. Available from: http://link.springer.com/10.1186/2228-5326-2-26.
- A. N. Barclay, R. L. Brady, S. J. Davis and G. Lange, CD4 and the immunoglobulin superfamily, Philos. Trans. R. Soc., B, 1993, 342(1299), 7–12 CrossRef CAS PubMed
. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7904350.
-
F. Putnam, Progress in Plasma Proteins, in The Plasma Proteins Structure, Function and Genetic Control, ed. F. W. Putnam, Academic Press, 1984, pp. 1–44 Search PubMed
.
- F. Scorgie, Y. Mohamed, D. Anderson, S. M. Crowe, S. Luchters and M. F. Chersich, Qualitative assessment of South African healthcare worker perspectives on an instrument-free rapid CD4 test, BMC Health Serv. Res., 2019, 19(1), 123 CrossRef PubMed
. Available from: https://bmchealthservres.biomedcentral.com/articles/10.1186/s12913-019-3948-x.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed SELEX methodology, complete aptamer sequences, antibody controls (fluorescence confocal microscopy and magnetic bead ELISA), aptamer qPCR calibration curves, LFA time point study video footage. See DOI: 10.1039/d0an00634c |
| This journal is © The Royal Society of Chemistry 2020 |