Double inner standard plot model of an electrophoresis titration chip for a portable and green assay of protein content in milk†
Cunhuai
Wang‡
ab,
Qiang
Zhang‡
ab,
Xiaoping
Liu
a,
Guoqing
Li
a,
Hao
Kong
b,
Muhammad Idrees
Khan
b,
Hua
Xiao
 b,
Yuxing
Wang
*c,
Weiwen
Liu
*a and
Chengxi
Cao
b,
Yuxing
Wang
*c,
Weiwen
Liu
*a and
Chengxi
Cao
 *ab
*ab
aDepartment of Instrument Science and Engineering, School of Electronic Information and Electrical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: wyx75@sjtu.edu.cn; weiwenliu@sjtu.edu.cn; cxcao@sjtu.edu.cn
bSchool of Life Sciences and Biotechnology, State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai 200240, China
cSchool of Physics and Astronomy, Shanghai Jiao Tong University, Shanghai 200240, China
First published on 3rd December 2018
Abstract
High portability and environmental safety (“green”) are two of the most important objectives pursued by microfluidic methods. However, there remain many challenges for the design of portable and visual microfluidic methods (e.g., chip electrophoresis) due to use of a cumbersome pump, power supply and detector. Herein, a facile double inner standard plot (DISP) model of electrophoresis titration (ET) was proposed for portable and visual assay of proteins in test milk samples without use of a pump, power supply or detector based on a moving reaction boundary (MRB) chip. The DISP-ET model predicted that: (i) by setting the upper limit (UL) and lower limit (LL) of double inner standard milk protein contents, points U and L were, respectively, achieved in the relationship D = −aC + b (D: MRB motion distance; C: protein content); and (ii) the two points divided both the C-axis and D-axis into “poor”, “eligible” and “superior” rulers scaled for quantitative assay of test samples. To demonstrate the model of DISP-ET, an original portable device (120 mm × 78 mm × 30 mm, 341 g) was designed, which had a chip (25 mm × 25 mm × 4 mm) of three channels (15 mm × 200 μm × 80 μm), platinum electrodes, a lithium cell and touch screen. A series of experiments were undertaken based on the developed portable device. The relevant experiments demonstrated systemically the validity of the DISP-ET model, theory and method. In particular, the experiments clearly showed the advantages of the DISP-ET chip: portability, visuality, green use, rapidity, and flexibility for real-life use. Finally, the device was applied for a portable and visual assay of fresh milk from a cow on a dairy farm. The DISP-ET model opens a window for designing portable and visual quantitative methods of food-safety control and clinical diagnoses.
1. Introduction
Owing to the importance of assays measuring protein content in food safety,1 diagnosis,2 proteomics3,4 and biomedical engineering,5 several methods have been developed for protein measurement: Kjeldahl method,6,7 biosensors,8–10 spectroscopy11,12 and Dumas method7,13 and the dye-binding method.14–16 Nevertheless, serious disturbance of non-protein nitrogen (NPN) often occurs.7,10,17To address NPN disturbance, electrophoresis titration (ET) was proposed for protein-content and double-protein ratio assays based on the moving reaction boundary (MRB, Fig. S1†).18–22 The specific advantage of protein ET is the non-disturbance of illegal NPN additives (e.g., urea, melamine),18–21 which greatly improves the selectivity of the protein-content assay of milk samples. However, these ET methods are based on cumbersome and expensive pumps,18,19 power supplies18,19 and detectors,20,21,23–27 which hamper the design of ET methods for portable and visual assays of protein content in food-safety control. In addition, Kjeldahl,6,7 Dumas7,14 and ET methods18–22 suffer from high consumption and high cost.
In particular, the portable ET sensor22 has been developed for on-spot detection of melamine in milk samples. However, portable sensing22 suffers from key issues stopping on-spot assays. First, a standard curve for sensing22 must be prepared based on a series of standard concentrations of analyte. An accurate standard curve can be prepared in a laboratory with closely controlled temperature (rather than in cowsheds) thanks to differences in environmental factors (e.g., temperature). Preparation of a standard curve with seven points22 requires more ≥4 h, which prohibits on-spot assays by ET sensors. However, the novel model of a facile double inner standard plot (DISP) developed herein could address this key issue. Second, the ET sensing platform for real on-spot assays requires a constant electric field, automatic timing, lights, touch screens and systemic integration as well as software. However, the ET sensing22 method does not have these key requirements.
Herein, the DISP model and theory were first advanced for portable and visual protein-content assays of test milk. To demonstrate the model and theory, a portable device was designed that contained a facile ET chip, platinum electrodes, lithium cell and touch screen as well as software. The relevant experiments were conducted and validated the DISP-ET model. In particular, the experiments displayed the merits of the DISP-ET chip: portable, visible, environmentally sound (“green”), low-cost and rapid. Finally, the device was used for portable and visual assays of fresh milk in a dairy farm.
2. Model, device and method of DISP-ET
2.1 DISP-ET model
Fig. 1 shows the facile model of DISP-ET with quantitative readout of protein content by the naked eye. As shown in previous works,18–21 there is a linear relationship between the running time (t) of protein ET and MRB motion distance (D),| D = Vt | (1) |
| D = −aC + b | (2) |
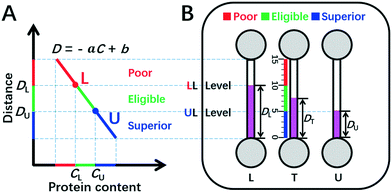 | ||
| Fig. 1 DISP-ET model (schematic). (A) Qualitative assay of protein content by the DISP model in the coordinate of MRB motion distance (D) vs. protein content (C) (eqn (2)). (B) Quantitative readout of protein content in the ET chip via the naked eye. In panel A, CU and CL denote the contents of UL and LL, respectively; point L and point U divide both C-axis and D-axis as poor (red), eligible (green) and superior (blue) rulers scaled. In panel B, DU, DT and DL indicate the MRB motion distances for UL, test and LL samples in channels U, T and L, respectively; the three scaled rulers and relevant colors created by UL and LL levels are poor, eligible and superior. For other symbols, see the figure and context. | ||
Herein, the double inner standard milk protein samples of the upper limit (UL, e.g., 3.6 mg mL−1 protein content for the national milk standard of New Zealand) and lower limit (LL, e.g., 2.8 mg mL−1 for the national milk standard of China) are applied for construction of a facile plot scale model of DISP (Fig. 1). According to eqn (1) and (2) and the DISP model in Fig. 1A, point U (CU, DU) and point L (CL, DL) can be obtained, respectively, from the simultaneous DISP-ET runs in channels U and L for the double inner milk samples of UL and LL (Fig. 1B). Simultaneously, the test sample can be run in channel T via the same DISP-ET chip, and the value of DT can be read out from channel T.
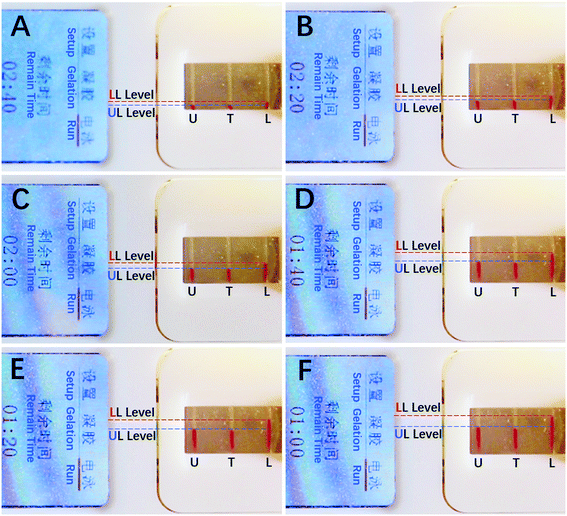
Using eqn (2) for the simultaneous DISP-ET runs of samples UL, test and LL in channels U, T and L (Fig. 1A and B), one can obtain eqn (3)–(5), respectively,
| DU = −aCU + b | (3) |
| DT = −aCT + b | (4) |
| DL = −aCL + b | (5) |
The difference of eqn (3) and (5) yields,
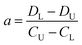 | (6) |
Inserting eqn (6) into eqn (3) or eqn (5), we have
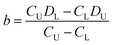 | (7) |
Thus, the two constants of a and b in eqn (2) can be calculated by using two points of (CU, DU) and (CL, DL), viz., the DISP-ET model, as shown in eqn (6) and (7).
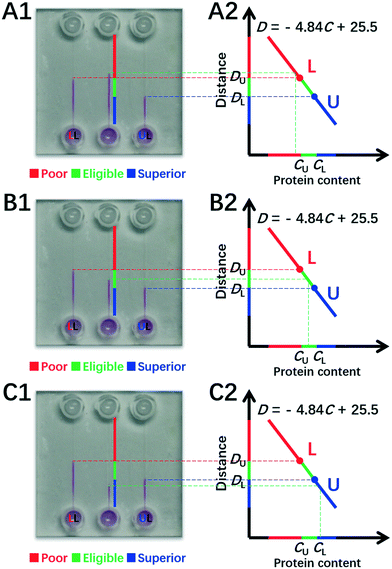
Finally, we obtain eqn (8) by re-expression of eqn (4)viaeqn (6) and (7),
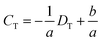 | (8) |
Obviously, (CU, DU) and (CL, DL) and DT are all available parameters via a single run of DISP-ET in Fig. 1B. Thus, eqn (6)–(8) indicate that the protein content (CT) of a test sample can be obtained visually by measuring the motion distance of MRB (DT) under the certain running time of DISP-ET. In other words, one can visually conduct portable quantitation of protein content with the DISP-ET model without a traditional pump,18,19 power supply18–21 or optical detector.20–27
Evidently, the two points of (CU, DU) and (CL, DL) divide the fitting curve of eqn (2)–(5) into the red, green and blue readouts scaled, thereby implying poor, eligible and superior milk samples, respectively (Fig. 1A). The range of protein content of eligible milk samples is highlighted as a green “ruler” between points U and L.
According to the DISP-ET model shown in Fig. 1A, a facile ET chip can be designed, which has channels U, T and L, the relevant anodic wells, and cathodic wells (Fig. 1B and 2). Correspondingly, the DISP milk samples of UL (CU) and LL (CL) are added, respectively, into channels U and L (Fig. 1B), and the test sample is injected in channel T for assaying the protein content of test milk (CT). By running the DISP-ET chip simultaneously (Video SI†), the distances DU, DT and DL can be obtained for the three samples of UL, test and LL, respectively.
Levels LL and UL divide the ET channels as three readable rulers scaled (Fig. 1A and B): (i) the “poor” ruler (in red) of milk quality above level LL; (ii) the “eligible” ruler (in green) between levels LL and UL; and (iii) the “superior” ruler (in blue) under level UL. Based on the DISP-ET model and eqn (8), the three theoretical predictions can be drawn below:
Prediction 1: if there is the inequality of DT > DL, then CT < CL, implying a poor milk sample tested in channel T;
Prediction 2: if there exists the inequality of DU ≤ DT ≤ DL, then CU ≥ CT ≥ CL, indicating an eligible milk sample tested in channel T;
Prediction 3: if there is the inequality of DT < DU, then CT > CU, suggesting a superior milk tested in channel T.
Thus, the protein level of a milk sample tested in channel T can be assayed directly via the readable rulers of Fig. 1B designed from the DISP-ET model of Fig. 1A without use of a bulk pump,18,19 power supply18–21 or optical detector.18–27 Evidently, the model of DISP-ET is prerequisite for portable and visual detection of protein content in milk samples.
2.2 DISP-ET chip
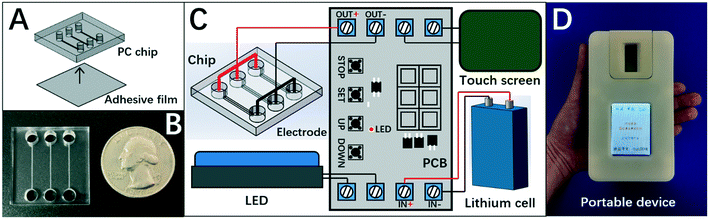
Herein, the chip of DISP-ET (Fig. 2A) was designed in our laboratory and fabricated by Shanghai BioChemAn Biotechnology (Shanghai, China) (Fig. 2B). The chip consisted of a polycarbonate plate (25 mm × 25 mm × 4 mm) and an adhesive film (25 mm × 25 mm × 1 mm). The chip had three anodic wells (i.d. 4 mm × 3 mm), three cathodic wells (i.d. 4 mm × 3 mm), and three channels (15 mm × 200 μm × 80 μm) (Fig. 2A).The chip device comprised the DISP-ET chip, ternary platinum electrodes, lithium cell, touch screen, LED and printed circuit board (Fig. 2C). The touch screen could control the entire ET process for various ET conditions. The LED was used for real-time observation of protein titration. The device (Fig. 2D) was designed to implement on-chip gel polymerization, ET and visual protein-content assay.
2.3 DISP-ET procedure
There were four steps for a whole run of DISP-ET.18–21 First, 100 μL of gel solution was prepared, which consisted of 62.5 μL of PAG solution (20% T and 3% C), 0.5 μL of phenolphthalein (0.5%), 6.75 μL of KCl solution (2 M), 1.25 μL of APS (10%, w/v), 0.05 μL of TEMED and 28.95 μL of ultrapure water. Then, 20 μL of a test milk sample and two DISP milk samples (UL and LL) were mixed uniformly with the 80 μL of prepared gel solution in three Eppendorf tubes, respectively. Then, the three mixed milk gel solutions of UL, test and LL were injected correspondingly into channels U, T and L for protein immobilization.Second, after injection of the gel solution, the prepared chip was assembled into the portable device for gel polymerization for 7 min (Video SI†). Third, after gel polymerization, 55 μL of the anodic and cathodic solutions were loaded into the anode and cathode wells, respectively (Video SI†). Fourth, after electrolyte loading, DISP-ET was started by pressing the “Start” button. When the running time reached the set time of 3 min, the circuit system stopped the chip ET run automatically, and the protein content of the test sample could be assayed based on the DISP-ET model shown in Fig. 1.
3. Results and discussion
3.1 Validation of DISP-ET portability and visuality
High portability and visuality are two of the most important objectives pursued by microfluidic techniques. However, many challenges for mini design of microfluidic methods (including chip electrophoresis) remain due to use of a cumbersome pump, power supply and detector.23–27 For the chip ET of protein content in milk,18–21 a bulk pump,18,19 power supply20,21 and fluorescent detector20,21,23–27 must be used for a sensitive ET assay, resulting in great challenges for the mini design on chip ET of visual protein-content assays.The model and device of DISP-ET addressed the issues of mini design of visual chip electrophoresis. As shown in Fig. 2, the lithium cell rather than a cumbersome power supply18–21 was used to yield a 20 V electric field. In addition, a bulk pump18,19 was not applied in the DISP-ET chip. In particular, a bulk pump or expensive detector20,21,23–27 were not used in the chip device due to direct readout via the naked eye or mobile-phone camera (if necessary). Clearly, the three prerequisites of a portable and visual assay (viz., no pump,18,19 no power supply18,19 and no optical detector20,21,23–27) were achieved via the design shown in Fig. 2.
Fig. 2 and Table 1 show that the size of the DISP-ET device was only 120 × 78 × 30 mm (slightly larger than an iPhone 6) and the total weight decreased <350 g (Table 1), making a portable assay of DISP-ET become reality. Obviously, the size (120 × 78 × 30 mm), volume (0.28 dm3) and weight (<350 g) of the DISP-ET chip device (Table 1) were much less than those of the Kjeldahl instrument (1185 × 790 × 610 mm, 571 dm3, >48 kg) and MRB ET instrument (300 × 300 × 270 mm, 24.3 dm3, >20 kg), strongly implying the high portability of our DISP-ET chip device.
| Method | Kjeldahl instrument | MRB ET instrument | Portable device |
|---|---|---|---|
| Portability | No | No | Yes |
| Size | 1185 × 790 × 610 mm | 300 × 300 × 270 mm | 120 × 78 × 30 mm |
| Equipment volume | 571 dm3 | 24.3 dm3 | 0.28 dm3 |
| Weight | >48 kg | >20 kg | <350 g |
| Visuality | No | No | Yes |
| Green | No | No | Yes |
| Voltage | 220 V | 220 V | 20 V |
| Rated power | 3760 W | 100 W | 5 W |
| Consumed power | 11.5 kW h | 0.1 kW h | 0.001 kW h |
| Sample volume | 10–20 mL | 200 μL | 2 μL |
| Total time | >200 min | >30 min | About 10 min |
| Anti-NPN | No | Yes | Yes |
| Cost | $40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
$15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
$500 |
| Safety | Explosive and corrosion | Fair safety | Good safety |
The DISP-ET device was used further for a portable and visual protein-content assay of fresh milk (Video SI,†Fig. 3). As shown in Video S1,† the test milk sample was taken on-spot from cow number 140132 at Shanghai Yinan Dairy Farming. The fresh milk was added directly to the gel solution and, after a short period of mixing by hand, the mixture of gel–milk solution was loaded directly into a channel for the DISP-ET assay (see section 3.5). The experiments in Video SI† and Fig. 3 as well as real application in section 3.5 demonstrated the portability and visuality of the DISP-ET device and method clearly.
 | ||
| Fig. 3 Photographs of runs of superior milk at (A) 20 s, (B) 40 s, (C) 60 s, (D) 80 s, (E) 100 s and (F) 120 s via the portable device on-spot. | ||
Evidently, the mini design in Fig. 2 and the experiments in Fig. 3 and Video SI† as well as application in section 3.5 showed the portability and visuality of the DISP ET model, device and method.
3.2 Validation of a green DISP-ET model
The DISP-ET method was green and low cost. First, the cost of a lithium cell shown in Fig. 2 was ∼$3, much less than the one from a traditional power supply (∼$1000).18–21 Second, the total cost of a DISP-ET device was ∼$500 (Table 1), much less than the one from a fluorescent detector (∼$40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) used in our previous works,20,21 the one from a MRB ET instrument ($15
000) used in our previous works,20,21 the one from a MRB ET instrument ($15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and one from the Kjeldahl instrument ($40
000) and one from the Kjeldahl instrument ($40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) (Table 1).
000) (Table 1).
Third, the rated power of the DISP-ET device was only ∼5 W (Table 1), which was much less than that for the Kjeldahl instrument (3760 W),6,7 MRB ET instrument (100 W),18,19 and MRB ET chip device with a fluorescent detector (200 W).20,21 The consumed power of a portable device for a test was 0.001 kW h (Table 1), which was evidently less than that for the Kjeldahl instrument (11.5 kW h),6,7 MRB ET instrument (0.1 kW h),18,19 and chip ET system (0.2 kW h).20,21
Fourth, the consumed sample volume of the ET chip was 2 μL (Table 1), which was obviously more than the one for the chip ET method (∼1 mL),20,21 but much less than the one for the Kjeldahl instrument (10–20 mL)6,7 and MRB ET instrument (200 μL).18,19 The portable device used regular low-cost microfluidic materials and the ET chip could be reused >30 times. Fifth, the total detection time of the DISP-ET chip was ∼10 min (Table 1), which was rapid in contrast to that for the Kjeldahl method (200 min)6,7 and MRB ET method (20 min),18,19 which saved a great deal of running time.
3.3 Demonstration of the DISP-ET model
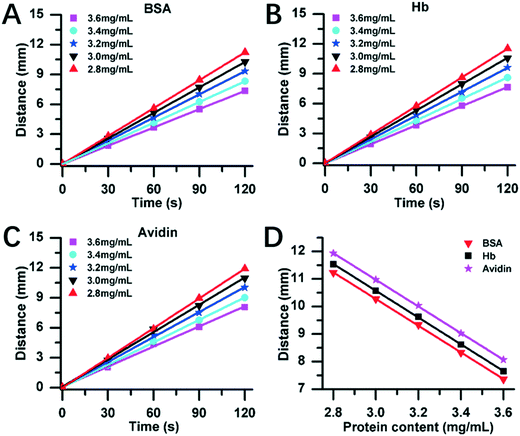
Fig. 4 exhibits the DISP-ET experiments on the MRB motion distance versus protein content. Three model proteins (bovine serum albumin (BSA), hemoglobin (Hb) and avidin) with five content levels (2.8, 3.0, 3.2, 3.4 and 3.6 mg mL−1) were chosen for DISP-ET experiments. The higher the protein content, the shorter was the MRB motion distance in a given running time (Fig. 4A–C). The experiments in Table S1† show that there were good fitting curves between the MRB motion distance and running time of DISP-ET, and the coefficients (R2) ranged from 0.9986 to 0.9997. Based on the data of Fig. 4A–C, the relationship between the motion distance and protein content was achieved (Fig. 4D and Table S2†). The three relationships were the curve of D = −4.84C + 25.5 (R2 = 0.9985) for BSA, D = −4.87C + 25.2 (R2 = 0.9989) for Hb, and D = −4.85C + 24.8 (R2 = 0.9998) for avidin. All these experiments in Fig. 4D and Table S2† demonstrated the validity of eqn (3)–(8).In Fig. 4A–C, the 3.4 and 3.0 mg mL−1 of protein content were, respectively, supposed to be the standard samples of UL and LL, and the test protein of 3.6, 3.2 and 2.8 mg mL−1 was simulated to be the superior, eligible and poor protein samples, respectively. Fig. 4A revealed three experimental results:
(i) there was a relationship of DT,BSA,2.8 > DL,BSA,3.0, hence the test (2.8 mg mL−1) showed the relationship of CT,BSA,2.8 < CL,BSA,3.0, thereby unveiling a poor BSA sample (viz., 2.8 mg mL−1 of the test <3 mg mL−1 of the LL);
(ii) there existed an inequality of DU,BSA,3.4 ≤ DT,BSA,3.2 ≤ DL,BSA,3.0, so CU,BSA,3.4 ≥ CT,BSA,3.2 ≥ CL,BSA,3.0, thereby demonstrating an eligible BSA sample (viz., 3 mg mL−1 of the LL < 3.2 mg mL−1 of the test < 3.4 mg mL−1 of the UL);
(iii) there was a relationship of DT,BSA,3.6 < DU,BSA,3.4, hence the test of 3.6 mg mL−1 confirmed the result of CT,BSA,3.6 > CU,BSA,3.4, demonstrating the superior BSA sample (viz., 3.4 mg mL−1 of the UL <3.6 mg mL−1 of the test).
Evidently, the three experiments of Fig. 4A were all in accordance with the three predictions based on eqn (3)–(8), thus demonstrating the DISP-ET model in Fig. 1. The same experimental conclusions could be also drawn from the DISP-ET runs in Fig. 4B and C, further showing the model of DISP-ET.
To demonstrate the DISP-ET chip device, experiments were undertaken for direct readouts of poor, eligible and superior protein samples via the LL (3.0 mg mL−1) and UL (3.4 mg mL−1) samples of DISP (Fig. 5). In Fig. 5A1–2, 2.8 mg mL−1 of BSA was simulated as the test sample in channel T, and the distance of DT was evidently longer than the one of DL for sample LL (3.0 mg mL−1). In Fig. 5B1–2, 3.2 mg mL−1 of BSA was chosen as the test sample, and its distance of DT was clearly longer than the one of DU for sample UL (3.4 mg mL−1) but shorter than the one of DL for sample LL (3.0 mg mL−1). In Fig. 5C1–2, 3.6 mg mL−1 of BSA was used as the test, and the distance of DT was obviously shorter than the one of DU for sample UL (3.4 mg mL−1). The three readouts of poor, eligible and superior BSA samples were in good agreement with the real qualities (2.8, 3.2 and 3.6 mg mL−1 of BSA), thereby validating the readout of the DISP-ET chip in Fig. 1B. Readouts in Fig. S3 and S4† further showed the validity of the DISP-ET chip readout. Obviously, the systemic data in Table S3† showed the validity of the DISP-ET chip readout of Fig. 1B quantitatively.
 | ||
| Fig. 5 Quantitative assay of BSA content by the DISP model. Direct readouts of poor (A1–2), eligible (B1–2) and superior (C1–2) protein samples via the DISP-ET chip. The other conditions are the same as those shown in Fig. 4. | ||
Furthermore, experiments were conducted for real milk samples (Fig. 6). Herein, 2.8 mg mL−1 and 3.6 mg mL−1 of milk protein contents were used as DISP samples of LL and UL, respectively (section 3.5), and the test milk had protein content of 3.2 mg mL−1. As shown in Fig. 6A, the distance of DT for 3.2 mg mL−1 of test milk was between the one of DU for the UL sample of 3.6 mg mL−1 and the one of DL for the LL sample of 2.8 mg mL−1 during the whole running time of DISP-ET. From the experiments in Fig. 6C, the boundary positions of UL, tested and LL samples were obtained after 120 s, and the two horizontal lines of UL and LL were coincident with the DISP-ET assays completely (Fig. 6C and D), briefly validating the model of Fig. 1. Furthermore, fitting curves of 2.8 mg mL−1 (LL) and 3.6 mg mL−1 (UL) were obtained, the spiked contents of 3.0, 3.2 and 3.4 mg mL−1 could be detected via the DISP-ET method (Fig. 6A and B), and the relevant measurements are given in Table S3.† Clearly, the detected values of 3.09, 3.25 and 3.46 mg mL−1 were in agreement with the spiked contents (Table S3†), thereby systemically demonstrating the validity of the PSM-ET model. The relative differences were, in general, <3% for milk samples. The results shown in Fig. 6D and the application (section 3.5) not only showed the validity of DISP-ET readouts qualitatively, but also validated the chip readout model quantitatively.
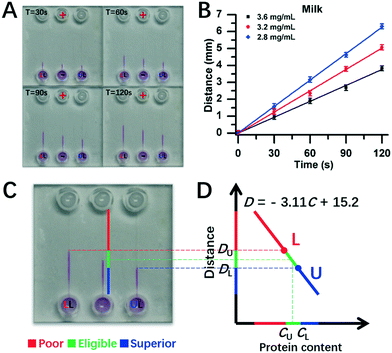 | ||
| Fig. 6 Assay for protein content in milk via chip DISP-ET. (A) Photograph on a chip DISP-ET run of milk at different running times. (B) Calibration curves of boundary motion distance vs. running time by milk ET with three content levels in panel A. (C) Photograph of ET chip at 120 s running time. (D) Readout of milk quality in a ET chip run for panel C (schematic). The other conditions are the same as those shown in Fig. 4. Each point was measured thrice. | ||
3.4 Analytical performance
The good anti-interference of the conventional MRB-ET method to NPN and discussion of indicators has been demonstrated in previous works,18–20 and was paid little attention here (S-5, ESI†). Displacement of the ET boundary of 100 μm was the minimum distinguishable scale of the human naked eye under the DISP-ET system. Thus, the sensitivity of the DISP-ET method was defined as the difference in protein concentration of a dilute sample of a model protein, which corresponded to motion of the ET boundary distance of 100 μm (viz., the minimum distinguishable scale of the human naked eye) over the same period of 120 s. We calculated that a relevant sensitivity of 0.2 mg mL−1 of BSA could be clearly distinguished via the DISP-ET readout, and the relevant absolute sensitivity was ∼2 μg. The relative and absolute sensitivities of DISP-ET were obviously worse than those of the MRB ET chip with a fluorescent detector (0.2 μg mL−1 and 0.2 ng).20,21 Considering that only the naked eye was used for readouts, the sensitivity of the DISP-ET method was quite good. In addition, the sensitivity was sufficient for portable assay of protein content in milk.The intra-day values of RSD were calculated based on ten replicated group runs of 2.8 mg mL−1 of BSA every 3 h in 1 day. The inter-day values were calculated by using ten group runs from the same BSA solutions obtained on 3 continuous days. The intra-day RSD values were <3.71% and the inter-day RSD values were <4.93%. The recovery was calculated using the ratio of the protein content detected via the chip DISP-ET to the precise content measured by an electronic balance. The recoveries were from 99% to 103% (Tables S3 and S4†), which indicated fair accuracy.
3.5 Application
Eligible protein content in a milk standard has different critical values for different countries. For example, the eligible protein content is 2.8 mg mL−1 in China, 3.2 mg mL−1 in Europe and USA, and 3.6 mg mL−1 in New Zealand. Herein, two milk samples of 2.8 mg mL−1 and 3.6 mg mL−1 from China and New Zealand were simulated as the two DISP milk levels of LL and UL, respectively.The LL sample of 2.8 mg mL−1 and the UL sample of 3.6 mg mL−1 were loaded into channels L and U, respectively, and the test sample was injected into channel T for the protein-content assay (Fig. 7). The three readouts were: (i) the boundary of channel T was under the level of UL and the detected value was 3.78 mg mL−1 (spiked 3.75 mg mL−1), suggesting a superior milk (panel A); (ii) one was between LL and UL and the detected value was 3.36 mg mL−1 (spiked 3.35 mg mL−1), indicating an eligible milk (panel B); and (iii) one was higher than the level of LL and the assayed value was 2.61 mg mL−1 (spiked 2.60 mg mL−1), implying a poor milk (panel C). Evidently, one could quickly read the milk quality and protein content based on the DISP-ET readout, and the assay values were are the spiked ones shown in Fig. 7.
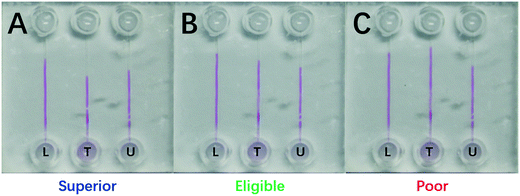 | ||
| Fig. 7 Photograph of a 210 s DISP-ET run for (A) superior (3.78 mg mL−1), (B) eligible (3.36 mg mL−1) and (C) poor (2.61 mg mL−1) milk sampled. ‘U’, ‘T’ and ‘L’ represent channels U, T and L, respectively. The other conditions are the same as those shown in Fig. 4. | ||
The portable device was also used for on-spot protein content assays of fresh milk (Video SI†). As shown in Video S1,† the test milk sample was taken on-spot from superior dairy cow number 140132 in Shanghai Yinan Dairy Farming. After milk addition, gel-solution loading, device assembly, chip gel polymerization, electrolyte injection and chip DISP-ET run, the result of superior quality of fresh milk could be read out via the portable chip device and was in accordance with the one obtained by the MRBET instrument (Fig. 3 and Table S5†). The entire process took ∼10 min. Fig. 3 shows the runs of milk from 20 to 120 s in the portable device on-spot. The MRB of channel T was under the one of channel U during the run of chip DISP-ET. The observations indicated that DT < DU and CT > CU, denoting a superior milk. The result showed, very clearly, real use of the DISP-ET device for on-spot milk sample assays.
3.6 Merits of DISP-ET
The developed DISP-ET method had six main advantages compared with the Kjeldahl,6,7 MRB ET18,19 and chip ET20,21 methods (Table 1, and Fig. 1–3 and 7). First, the DISP-ET method had high portability (section 3.1). Second, the DISP-ET method had visuality for the protein assay (Fig. 3, 4 and 6 and S3 and S4, Video SI†). Third, the method of DISP-ET was green and low cost. A total running time of 10 min of the DISP-ET method was much less than that of Kjeldahl (200 min) and MRB ET (30 min) methods, indicating fair rapidity.Fourth, the developed DISP-ET method had a wide range of quantitation.18,20 Herein, the portable device used a limited range from 2.8 mg mL−1 of LL to 3.6 mg mL−1 of UL due to application for on-spot milk protein assays. The quantitation range might not just restrict the sharp window from 2.8 mg mL−1 to 3.6 mg mL−1, but could be expanded by setting a wider one flexibly between the UL and LL values for real needs (up to 2 orders linearity of in Fig. S9 in ref. 20). In addition, quantitation of the ET chip could be adapted for different demands by designing longer/or shorter and wider/or narrower channels.
Fifth, even though the naked eye was used instead of an optical detector, the DISP-ET method had fair sensitivity (0.2 μg mL−1 and 0.2 ng), good stability (RSD of 3.71–4.93%) and high recoveries (99–103%). Finally, the portable device had high safety (sulfuric acid was not used) in contrast to the poor safety of the Kjeldahl method (use of high-concentration sulfuric acid).
4. Conclusions
We proposed a facile DISP-ET model. This model allowed qualitative readout of protein quality by the naked eye and quantitative assay of protein content by the DISP-ET method without a cumbersome and expensive pump, power supply or fluorescent detector used in our previous works. Accordingly, an original, portable and visual chip device was designed based on the DISP-ET model. The DISP-ET chip greatly reduced the size, volume and weight of the device, and the time of ET run, and was especially suited to a portable and visual assay of protein content in milk. The relevant experiments demonstrated that the original chip device had the advantages of high portability, visuality, green use, rapidity and flexibility for real-life use. Finally, the DISP-ET chip was used for on-spot protein content assays of fresh milk from a dairy farm. The DISP-ET model and device have great importance for portable assays in food-safety control.Notes
All fresh-milk samples were collected from a superior dairy cow (number 140132) from Shanghai Yinan Dairy Farming according to the approved protocol (IRB#201702014) by the Institutional Review Board of Shanghai Jiao Tong University. The methods were carried out in accordance with approved guidelines. All experimental protocols were approved by Institutional Animal Care and Use Committee of Shanghai Jiao Tong University.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank Shanghai Yinan Dairy Farming for the opportunity to use their cows and record videos. The authors are grateful for financial support from the NSFC (21475086, 21675067 and 31727801), National Research and Development Program (2017YFC1200204) and the National High-tech R&D Program of China (863 Program, 2014AA020545).References
- S. Maass, S. Sievers, D. Zühlke, J. Kuzinski, P. K. Sappa, J. Muntel, B. Hessling, J. Bernhardt, R. Sietmann and U. Völker, Anal. Chem., 2011, 83, 2677–2684 CrossRef CAS.
- M. Drayson, L. X. Tang, R. Drew, G. P. Mead, H. Carrsmith and A. R. Bradwell, Blood, 2001, 97, 2900–2902 CrossRef CAS.
- A. Dupuis, J. A. Hennekinne, J. Garin and V. Brun, Proteomics, 2008, 8, 4633–4636 CrossRef CAS PubMed.
- B. V. D. Waterbeemd, G. P. M. Mommen, J. L. A. Pennings, M. H. Eppink, R. H. Wijffels, L. A. V. D. Pol and A. P. J. M. D. Jong, J. Proteome Res., 2013, 12, 1898–1908 CrossRef PubMed.
- M. Scott and A. Knight, J. Agric. Food Chem., 2009, 57, 4545–4551 CrossRef CAS PubMed.
- J. Kjeldahl, Z. Anal. Chem., 1883, 22, 366–382 CrossRef.
- J. C. Moore, J. W. Devries, M. Lipp, J. C. Griffiths and D. R. Abernethy, Compr. Rev. Food Sci. Food Saf., 2010, 9, 330–357 CrossRef CAS.
- C. Chen, X. Xiang, Y. Liu, G. Zhou, X. Ji and Z. He, Biosens. Bioelectron., 2014, 58, 205–208 CrossRef CAS.
- S. Wustoni, S. Hideshima, S. Kuroiwa, T. Nakanishi, M. Hashimoto, Y. Mori and T. Osaka, Biosens. Bioelectron., 2015, 67, 256–262 CrossRef CAS PubMed.
- T. L. Fodey, C. S. Thompson, I. M. Traynor, S. A. Haughey, D. G. Kennedy and S. R. Crooks, Anal. Chem., 2011, 83, 5012–5016 CrossRef CAS PubMed.
- J. Dziuba, D. N. j. m. cz and P. Minkiewicz, Anal. Chim. Acta, 2001, 449, 243–252 CrossRef CAS.
- F. Jin, J. J. Hickman, K. Lenghaus and R. K. Marcus, Anal. Bioanal. Chem., 2004, 380, 204–211 CrossRef CAS PubMed.
- A. C. Fieldner and C. A. Taylor, Ind. Eng. Chem., 1915, 7, 106–112 CrossRef CAS.
- V. F. Colenbrander and T. G. Martin, J. Dairy Sci., 1971, 54, 531–533 CrossRef CAS PubMed.
- D. L. Park and R. L. King, J. - Assoc. Off. Anal. Chem., 1974, 57, 42–47 CAS.
- J. W. Sherbon, J. - Assoc. Off. Anal. Chem., 1974, 57, 1338–1341 CAS.
- K. Ai, Y. Liu and L. Lu, J. Am. Chem. Soc., 2009, 131, 9496–9497 CrossRef CAS PubMed.
- H. Y. Wang, C. Y. Guo, C. G. Guo, L. Y. Fan, L. Zhang and C. X. Cao, Anal. Chim. Acta, 2013, 774, 92–99 CrossRef CAS PubMed.
- H. Y. Wang, S. Li, Y. Y. Tang, J. Y. Dong, L. Y. Fan and C. X. Cao, Analyst, 2013, 138, 3544–3551 RSC.
- H. Wang, Y. Shi, J. Yan, J. Dong, S. Li, H. Xiao, H. Xie, L. Y. Fan and C. X. Cao, Anal. Chem., 2014, 86, 2888–2894 CrossRef CAS PubMed.
- L. X. Zhang, Y. R. Cao, H. Xiao, X. P. Liu, S. R. Liu, Q. H. Meng, L. Y. Fan and C. X. Cao, Biosens. Bioelectron., 2016, 77, 284–291 CrossRef CAS PubMed.
- W. L. Li, F. Z. Kong, Q. Zhang, W. W. Liu, H. Kong, X. P. Liu, M. I. Khan, A. Wahid, S. Saud, H. Xiao, C. X. Cao and L. Y. Fan, Anal. Chem., 2018, 90, 6710–6717 CrossRef CAS PubMed.
- A. Marcello, D. Sblattero, C. Cioarec, P. Maiuri and P. Melpignano, Biosens. Bioelectron., 2013, 46, 44–47 CrossRef CAS PubMed.
- M. A. Kapil and A. E. Herr, Anal. Chem., 2014, 86, 2601–2609 CrossRef CAS PubMed.
- R. Bei, Y. Xianyu, M. Dong, Y. Chen, Z. Qian and X. Jiang, Anal. Chem., 2017, 89, 6113–6119 CrossRef PubMed.
- Y. Song, Y. Zhang, P. E. Bernard, J. M. Reuben, N. T. Ueno, R. B. Arlinghaus, Y. Zu and L. Qin, Nat. Commun., 2012, 3, 1283 CrossRef PubMed.
- T. Tian, J. Li, Y. Song, L. Zhou, Z. Zhu and C. J. Yang, Lab Chip, 2016, 16, 1139–1151 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8lc01015c |
| ‡ The first two authors have equal contribution to this work. |
| This journal is © The Royal Society of Chemistry 2019 |