Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sampling, isolating and identifying microplastics ingested by fish and invertebrates†
A. L.
Lusher
 *a,
N. A.
Welden
b,
P.
Sobral
c and
M.
Cole
d
*a,
N. A.
Welden
b,
P.
Sobral
c and
M.
Cole
d
aDepartment of Animal Ecology I, University of Bayreuth, Universitaetsstr. 30, 95440 Bayreuth, Germany. E-mail: amy.lusher7@gmail.com
bFaculty of Science, Technology, Engineering and Maths, Open University, Milton Keynes, MK7 6AA, UK
cMARE – Marine and Environmental Sciences Centre, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, Campus da Caparica. 2829-516 Caparica, Portugal
dCollege of Life and Environmental Sciences: Biosciences, University of Exeter, Geoffrey Pope Building, Stocker Road, Exeter, EX4 4QD, UK
First published on 24th October 2016
Abstract
Microplastic debris (<5 mm) is a prolific environmental pollutant, found worldwide in marine, freshwater and terrestrial ecosystems. Interactions between biota and microplastics are prevalent, and there is growing evidence that microplastics can incite significant health effects in exposed organisms. To date, the methods used to quantify such interactions have varied greatly between studies. Here, we critically review methods for sampling, isolating and identifying microplastics ingested by environmentally and laboratory exposed fish and invertebrates. We aim to draw attention to the strengths and weaknesses of the suite of published microplastic extraction and enumeration techniques. Firstly, we highlight the risk of microplastic losses and accumulation during biotic sampling and storage, and suggest protocols for mitigating contamination in the field and laboratory. We evaluate a suite of methods for extracting microplastics ingested by biota, including dissection, depuration, digestion and density separation. Lastly, we consider the applicability of visual identification and chemical analyses in categorising microplastics. We discuss the urgent need for the standardisation of protocols to promote consistency in data collection and analysis. Harmonized methods will allow for more accurate assessment of the impacts and risks microplastics pose to biota and increase comparability between studies.
1 Introduction
Over the past century there has been an exponential increase in plastic demand and production.1 Concurrently, improper disposal, accidental loss, and fragmentation of plastic materials, have led to an increase in tiny plastic particles and fibres (microplastic, <5 mm) polluting the environment.2,3 Microplastics have been observed in marine,4 freshwater5,6 and terrestrial7 ecosystems across the globe, and biotic interactions are widely evidenced (Fig. 1). Microplastics can be consumed by a diverse array of marine organisms, across trophic levels, including protists,8 zooplankton,9–17 annelids,18–26 echinoderms,27–31 cnidaria,32 amphipods,19,26,33 decapods,34–41 isopods,42 bivalves,43–60 cephalopods,61 barnacles,62 fish,58,66–94 turtles,95 birds96 and cetaceans.97,98 Over 220 different species have been found to consume microplastic debris in natura. Of these, ingestion is reported in over 80% of the sampled populations of some invertebrate species.34,38,41 Interactions between microplastics and freshwater invertebrates, fish and birds are increasingly reported99–107 although some researchers are focussing on model species such as Daphnia magna.108–111 The consumption of microplastics by terrestrial organisms is poorly documented, however, laboratory studies indicate earthworms (Lumbricus terrestris) can consume plastic particles present in soil.112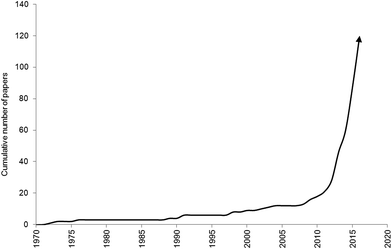 | ||
| Fig. 1 Publication trend of studies investigating biota interactions with microplastics until 30th June 2016. | ||
There are a number of exposure pathways by which organisms may interact with microplastic debris. Direct consumption of microplastic is prevalent in suspension feeders, including zooplankton,10 oysters59 and mussels,43–58,60–63 and deposit feeders, such as sea cucumbers,28 crabs35–37,39,40 and Nephrops,34,41 owing to their inability to differentiate between microplastics and prey. Predators and detritivores may indirectly ingest plastic while consuming prey (i.e. trophic transfer) or scavenging detrital matter (e.g. marine snows, faecal pellets, carcasses) containing microplastic.13,34,35,41,113–115 Micro- and nanoplastics can adhere to external appendages, including the gills of the shore crab (Carcinus maenas)37 and mussels (Mytilus edulis),62 and setae of copepod swimming legs and antennules.10 Other studies have identified that microplastics can bind to microalgae116–118 or macroalgae.119 Microplastic exposure has been associated with a suite of negative health effects, including increased immune response,49 decreased food consumption,20,22 weight loss,20 decreased growth rate,112 decreased fecundity,59 energy depletion22 and negative impacts on subsequent generations59,104. Microplastics have also been shown to readily accumulate waterborne persistent organic pollutants including pesticides, solvents and pharmaceuticals, which may pose further health effects such as endocrine disruption and morbidity.106,120,121
The United Nations Environment Programme (UNEP) has identified plastic pollution as a critical problem, the scale and degree of this environmental issue is comparable to that of climate change.3 There is currently much public and political debate surrounding the issue of microplastics as additives to household and industrial products, and the methods by which impacts of said microplastics on the environment are to be measured. Determining the degree to which biota consume microplastics is essential to determine and monitor ‘good environmental status’ for plastic pollution (e.g., EU Marine Strategy Framework Directive, 2008/56/EC; UNEA, US EPA). Equally, the development of robust environmental legislation is reliant on toxicological studies with ecological relevance, requiring an accurate measure of microplastic loads in natura.122 As such, it is imperative that researchers are able to accurately isolate, identify and enumerate microplastic debris consumed by or entangled with biota. Here we systematically and critically review methods employed in the extraction, identification and quantification of microplastic particles ingested by biota. We consider the effectiveness and limitations of a range of field sampling, laboratory exposure, extraction, and analytical techniques, and consider steps for mitigating contamination. Our review primarily focuses on peer-reviewed publications that have investigated the interactions between invertebrates and fish from the wild, and following controlled laboratory exposure. A review on extraction of microplastics from larger marine organisms has been conducted by Provencher et al. (this issue).
2 Methodological review
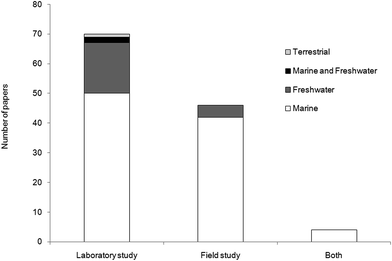
For this literature review, we examined original peer-reviewed research articles, grey literature and conference proceedings from the 1970s to July 2016. We identified literature referring to the extraction of microplastics from marine, freshwater and terrestrial biota using Web of Knowledge, Science Direct, Scopus and Google Scholar. We also mined the journals Marine Pollution Bulletin, Environmental Pollution and Environmental Science and Technology owing to the regularity with which they publish relevant material. Analysis of microplastic specific studies was expanded to include historical literature that did not necessarily have microplastics as the central theme of the research, such as studies which used fluorescent latex beads as a tracer for feeding and retention experiments. Of the 120 papers included in our meta analysis, 58.3% of studies were conducted in the laboratory, 38.3% focused on organisms collected from the wild and 3.4% involved both laboratory exposure and field collection (Fig. 2). There were 96 studies wholly focused on marine organisms, 21 on freshwater, two studies on both marine and freshwater organisms and one published study on a terrestrial species.2.1. Sampling
2.1.1.1. Microplastic losses during field sampling. Handling stress, physical movement, and the physiological and behavioural specificities of the sampled organism, may result in the loss of microplastics prior to animal preservation. Gut evacuation times for animals are varied, ranging from as little as 30 minutes for decapod crustaceans (N. Welden, personal observations), <2 hours for calanoid copepods,10 10 to 52 hours for fish68,124 to over 150 hours in larger lobsters.125 Therefore, some animals might egest microplastic debris prior to analysis.41 In such cases, the time between sample collection and the preservation of the animal must be as short as possible.
Care must also be taken to minimise handling stress or physical damage. This will reduce the potential for microplastic regurgitation; the frequency with which animals expel consumed plastics during sampling is unknown. The copepod Eurytemora affinis126 and some fish species have been observed regurgitating their stomach contents.127 The main cause of regurgitation in fish is thought to be related to the expansion of gas in the swim bladder: this causes the compression of the stomach and may, in extreme cases, result in total stomach inversion.128 Compression of a catch in the cod end might induce regurgitation in fish.129 The likelihood of regurgitation increases with depth of capture, and gadoids are more prone to regurgitation than flatfish. Piscivorous predators are prone to regurgitation due to their large distensive oesophagus and stomach.128,130 As such, regurgitation may bias the stomach content estimation, affecting consumption estimates and the presence of plastic debris.
2.1.1.2. Microplastic accumulation during field sampling. Laboratory studies have identified that nano- and microplastics can adhere to external appendages of marine copepods.10 Cataloguing such interactions in natura is complicated as determining whether the resulting accumulation has occurred naturally, or as a by-product of the sampling regimen, is prohibitive. While most studies focus on the consumption of plastic, any research considering external adherence of microplastics should be aware that observed entanglement may have occurred during sampling and may be unrepresentative of microplastic–biota interactions at large. A similar interaction may occur with organisms feeding on microplastics during capture in nets, this is particularly a concern when the mesh size of the net is capable of collecting microplastics, for example, in manta nets (common mesh size 0.33 mm.69 Control of microplastic contamination is discussed in Section 2.4.
2.1.1.3. Sample storage. Consideration should be given to the storage of biotic samples. Choice of preservation technique will largely depend on the research question being considered; for example, will the fixative affect the structure, microbial surface communities, chemical composition, colour or analytical properties of any microplastics within the sample? 4% formaldehyde and 70% ethanol are commonly used fixatives, however, consultation of resistance tables suggests these preservatives, albeit at higher concentrations, can damage some polymers; for example, polyamide is only partially resistant to 10% formaldehyde solution, while polystyrene can be damaged by 100% alcohol (ESI, Table S1†). Alternative methods for storage of organisms include desiccation12 and freezing.41,77,83,89
2.2. Isolating microplastics
In recent years an increasing number of techniques have been developed to detect microplastics consumed by biota. Methods for extracting microplastics from biotic material include dissection, depuration, homogenisation and digestion of tissues with chemicals or enzymes. Here we consolidate a range of optimised methods, and evaluate their benefits, biases and areas of concern:| Treatment | Exposure | Organism | Author |
|---|---|---|---|
| HNO3 (22.5 M) | 20 °C (12 h) + 100 °C (2 h) | Blue mussels | Claessens et al. (2013)51 |
| HNO3 (22.5 M) | 20 °C (12 h) + 100 °C (2 h) | Blue mussels oysters | Van Cauwenberghe & Jansen (2014)54 |
| HNO3 (22.5 M) | 20 °C (12 h) + 100 °C (2 h) | Blue mussels lugworms | Van Cauwenberghe et al. (2013)23 |
| HNO3 (100%) | 20 °C (30 min) | Euphausids copepods | Desforges et al. (2015)16 |
| HNO3 (69–71%) | 90 °C (4 h) | Manilla clams | Davidson & Dudas (2016)61 |
| HNO3 (70%) | 2 h | Zebrafish | Lu et al. (2016)105 |
| HNO3 (22.5 M) | 20 °C (12 h) + 100 °C (15 min) | Brown mussels | Santana et al. (2016)63 |
| HNO3 (65%) | 20 °C (12 h) + 100 °C (10 min) | Blue mussels | De Witte et al. (2014)52 |
HClO4 (68%) (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| HNO3 (65%) | 20 °C (12 h) + 100 °C (10 min) | Brown shrimp | Devriese et al. (2015)38 |
HClO4 (68%) (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
|||
| CH2O2 (3%) | 72 h | Corals | Hall et al. (2015)32 |
| KOH (10%) | 2–3 weeks | Fish | Foekema et al. (2013)75 |
| KOH (10%) | 60 °C (12 h) | Fish | Rochman et al. (2015)58 |
| KOH (10%) | 2–3 weeks | Fish | Lusher et al. (2016)89 |
| H2O2 (30%) | 60 °C | Blue mussels | Mathalon & Hill (2014)53 |
| H2O2 (30%) | 20 °C (7 d) | Biogenic matter | Nuelle et al. (2015)137 |
| H2O2 (15%) | 55 °C (3 d) | Fish | Avio et al. (2015)81 |
| H2O2 (30%) | 65 °C (24 h) + 20 °C (<48 h) | Bivalves | Li et al. (2015)57 |
| NaClO (3%) | 20 °C (12 h) | Fish | Collard et al. (2015)82 |
NaClO3 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
20 °C (5 min) | ||
| Proteinase K | 50 °C (2 h) | Zooplankton copepods | Cole et al. (2014)12 |
2.2.3.1. Nitric acid. Nitric acid (HNO3) is a strong oxidizing mineral acid, capable of molecular cleavage and rapid dissolution of biogenic material.134 When tested against hydrochloric acid (HCl), hydrogen peroxide (H2O2) and sodium hydroxide (NaOH), HNO3 resulted in the highest digestion efficacies, with >98% weight loss of biological tissue.51 The optimised protocol involved digesting excised mussel tissue in 69% HNO3 at room temperature overnight, followed by 2 h at 100 °C. Desforges et al.16 also tested HNO3, HCl and H2O2 in digesting zooplankton, and similarly identified nitric acid as the most effective digestion agent based on visual observations; here the optimised digestion protocol consisted of exposing individual euphausids to 100% HNO3 at 80 °C for 30 minutes. Adaptations of nitric acid protocols have been successfully used to isolate fibres, films and fragments from a range of organisms.23,38,54,61,64,105 While largely efficacious in digesting organic material, a number of studies observed that oily residue and/or tissue remnants remained post-digestion,15,51,63 which have the potential to obscure microplastics. In response, De Witte et al.52 proposed using a mixture of 65% HNO3 and 68% perchloric acid (HClO4) in a 4
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v ratio (500 ml acid to 100 g tissue) to digest mussel tissue overnight at room temperature followed by 10 minutes boiling; this resulted in the removal of the oily residue. Recovery rates for 10 and 30 μm PS microspheres spiked into mussel tissue and subsequently digested with nitric acid range between 93.6–97.9%.51 However, the high concentrations of acid and temperatures applied resulted in the destruction of 30 × 200 μm Nylon fibres and melding of 10 μm polystyrene microbeads following direct exposure. Researchers have found that polymeric particles, including polyethylene (PE) and polystyrene (PS), dissolved following overnight exposure and 30 minutes boiling with 22.5 M HNO3.81,135 Polyamide (PA, Nylon), polyester (PET) and polycarbonate have low resistance to acids, even at low concentrations; furthermore, high concentrations of nitric, hydrofluoric, perchloric and sulphuric acid are likely to destroy or severely damage the majority of polymers tested, particularly at higher temperatures (ESI, Table S1†). The absence of synthetic fibres in biota digested using HNO3 is likely a reflection of the destructive power of the acid.57
1 v/v ratio (500 ml acid to 100 g tissue) to digest mussel tissue overnight at room temperature followed by 10 minutes boiling; this resulted in the removal of the oily residue. Recovery rates for 10 and 30 μm PS microspheres spiked into mussel tissue and subsequently digested with nitric acid range between 93.6–97.9%.51 However, the high concentrations of acid and temperatures applied resulted in the destruction of 30 × 200 μm Nylon fibres and melding of 10 μm polystyrene microbeads following direct exposure. Researchers have found that polymeric particles, including polyethylene (PE) and polystyrene (PS), dissolved following overnight exposure and 30 minutes boiling with 22.5 M HNO3.81,135 Polyamide (PA, Nylon), polyester (PET) and polycarbonate have low resistance to acids, even at low concentrations; furthermore, high concentrations of nitric, hydrofluoric, perchloric and sulphuric acid are likely to destroy or severely damage the majority of polymers tested, particularly at higher temperatures (ESI, Table S1†). The absence of synthetic fibres in biota digested using HNO3 is likely a reflection of the destructive power of the acid.57
2.2.3.2. Other acids. Formic and hydrochloric acid (HCl) have also been suggested as digestive agents. With scleractinian corals (Dipsastrea pallida), formic acid (3%, 72 h) has been used to decalcify polyps to assist in the visualisation of ingested blue polypropylene shavings.32 HCl has also be trialled as a digestant of microplastics from pelagic and sediment samples; however this non-oxidizing acid proved inconsistent and inefficient in digesting organic material.12
2.2.3.3. Alkalis. Strong bases can be used to remove biological material by hydrolysing chemical bonds and denaturing proteins.136 Excised fish tissues, including the oesophagus, stomach and intestines, have been successfully digested using potassium hydroxide (KOH, 10%) following a 2–3 week incubation.75,89 The protocol has been adapted for the dissolution of gastrointestinal tracts of fish and mussel, crab and oyster tissues, either directly or following baking (450 °C, 6 h), by incubating tissues in 10% KOH at 60 °C overnight.58,135 This latter method has proven largely efficacious in removing biogenic material, being well suited to the dissolution of invertebrates and fish fillets, but proving less applicable for fish digestive tracts owing to the presence of inorganic materials; as with HNO3, an oily residue and bone fragments may remain following digestion. Another strong base, sodium hydroxide (NaOH; 1 M and 10 M), has been successfully applied to remove biogenic material (e.g. zooplankton) from surface trawls, with 90% efficiency based on sample weight loss.12 Foekema et al.75 suggests polymers are resistant to KOH, and Dehaut et al.135 showed no demonstrable impact on polymer mass or form, except in the case of cellulose acetate (CA). Testing the rapid KOH digestion protocol achieved a 100% microplastic recovery rate.135 Tabulated data confirms PA, PE and polypropylene (PP) are resistant to 10% KOH, but polycarbonate (PC) and PET are degraded (ESI, Table S1†). Cole et al.12 tested 40% NaOH (60 °C) on a range of polymers, and observed deformation of PA fibres, yellowing of PVC granules and melding of polyethylene particles; similarly, Dehaut et al.135 noted PC, CA and PET were degraded using this protocol. Notably the compiled chemical resistance data indicates PE and polyvinyl chloride (PVC) are resistant to NaOH, even at concentrations of 50% at 50 °C (ESI, Table S1†). That Cole et al.12 observed changes in polymers supposedly resistant to the given treatment highlights the necessity for comprehensive testing of applied treatments prior to use on biota.
2.2.3.4 Oxidizing agents. Hydrogen peroxide (H2O2) and peroxodisulfate potassium (K2S2O8) are oxidizing agents. Mathalon and Hill53 used H2O2 (30%) at 55–65 °C to digest mussel soft tissue, and although largely effective, the authors noted “flakes of debris” remained. Li et al.57 also applied this method, but incubated samples in an oscillating incubator, and then at room temperature for 24–48 h. Avio et al.81 similarly tested alternate treatments in digesting intestinal tracts of mullet (Mugil cephalus); while, H2O2 was an efficacious digestant, Avio et al.81 identified that direct application of H2O2 resulted in only a 70% retrieval of spiked microplastics, with losses linked to H2O2 foaming. A number of studies noted excessive foaming might obscure samples or lead to sample loss.51,53,81 A density separation of stomach contents with hypersaline (NaCl) solution followed by digestion of isolated material with 15% H2O2 resulted in a much improved 95% recovery rate for spiked microplastics. Dehaut et al.135 trialled 0.27 M K2S2O8 with 0.24 M NaOH in digesting biological tissues; while largely efficient in digesting biogenic material (<99.7% mass reduction), the authors noted its expense and highlighted issues with crystallisation of the digestive solutions and incomplete digestion causing blockages during filtration. Avio et al.81 observed that 15% H2O2 had no visible impact on PE or PS microspheres, although a slight modification to FTIR spectra was observed. Conversely, Nuelle et al.137 identified some visual deformities to exposed plastic, and quantified a 6.2% loss in size for PP and PE particles (<1 mm). Resistance data indicates 30% H2O2 should have little or no effect on PE or PP, again highlighting the importance of thorough testing of protocol applicability. Tabulated data indicates PA and PE are also prone to damage or dissolution from 30% H2O2 (ESI, Table S1†). K2S2O8 resulted in no changes in the mass or appearance in the majority of exposed polymers, but caused complete dissolution of cellulose acetate;135 chemical resistance data is currently unavailable and requires further testing.
2.2.3.5. Sodium hypochlorite. Sodium hypochlorite (NaClO) is used as an endodontic irrigant, with a near linear dose-dependent dissolution efficiency for biological tissue.138 Collard et al.82 digested fish stomach contents, in an overnight exposure with ∼3% NaClO; filtered digestants were subsequently washed with 65% HNO3 and digested in a 10
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 NaClO and HNO3 solution for 5 minutes. The technique caused no visible degradation of a range of polymers (PET, PVC, PE, PP, PS, PC or PA). Resistance chart data indicates 15% NaClO would degrade PA, although no data is provided for the ∼3% NaClO concentration applied by Collard et al.82
1 NaClO and HNO3 solution for 5 minutes. The technique caused no visible degradation of a range of polymers (PET, PVC, PE, PP, PS, PC or PA). Resistance chart data indicates 15% NaClO would degrade PA, although no data is provided for the ∼3% NaClO concentration applied by Collard et al.82
2.2.3.6. Enzymes. Enzymatic digestion has been mooted as a biologically specific means of hydrolysizing proteins and breaking down tissues.12 To remove biological material from field-collected samples, Cole et al.12 developed a digestion protocol employing a serine protease (Proteinase K). Material was desiccated (60 °C, 24 h), ground, and homogenised by repeatedly drawing samples through a syringe. Next, samples were mixed with homogenizing solution (400 mM Tris–HCl buffer, 60 mM EDTA, 105 mM NaCl, 1% SDS), acclimated to 50 °C, enzymatically digested with Proteinase K (500 mg mL−1 per 0.2 g DW sample), and mixed with sodium perchlorate (NaClO4, 5 M). Ultrasonication was demonstrated to have a deleterious effect on digestion efficiency, owing to protein precipitation in the media. With marine samples, the Proteinase K method proved to have a digestion efficacy of >97%, and the method was used to isolate fluorescent polystyrene microspheres (20 μm) ingested by marine copepods. The authors note that additional enzymes could be used depending on the chemical make-up of the organism or samples in question (e.g. chitinase with chitinous invertebrates). The enzyme Pepsin causes no damage to polymers, but proved only partially effective at digesting biogenic material.137 It has recently been reported that enzymes have been successfully applied in the isolation of microplastics from: intestinal tracts of turtles with Proteinase K,139 mussel tissue with Corolase® 7089 (AB enzymes),140 and herring digestive tracts with Proteinase K and H2O2.141 In contrast to chemical digestion techniques, enzymes ensure no loss, degradation or surface change to plastics present,12 and are less hazardous to human health. The trade-off is a protracted method, necessitating increased researcher time when considering large-scale field sampling and monitoring.
2.2.3.7. Filtering digestants. Following digestion, chemical agents can be filtered to retain any recalcitrant materials (e.g. undigested tissue, inorganic residue, microplastics). Viable filters include 0.2 and 0.7 μm glass fibre filters,61,63 5 μm cellulose nitrate filters,54 5 μm cellulose acetate membranes,82 50 μm mesh,12 and 250 μm mesh.77 Larger pore size facilitates rapid filtering but will result in the loss of smaller plastics.12 Glass fibre filters can shed and might be considered a source of contamination; smoother filters (e.g. membrane filters) are typically easier to scrape and less prone to fragment (personal observations of the authors). Microplastics on filters can be visualised directly (see Section 2.3), transferred to slides,63 or extracted. Collard et al.82 suggests placing filters in methanol solution, ultrasonicating (50 Hz), centrifuging (5000 rpm, 5 min, 20 °C) and then removing pelleted plastic by pipette; while this method was suitable for a range of polymers, the methanol caused a 25% weight reduction in tested PVC particles.
2.3. Microplastic identification
Following the preparation of target tissues, the quantity and types of microplastics should be ascertained. Of the methods currently employed, visual identification is most widely utilized; often in combination with one or more follow-up analytical techniques. Researchers can use characteristics, including morphology and density, to identify the presence of microplastics. Visual identification is based on the morphological and physical characteristics of particles whereas chemical characteristics are determined by more advanced analytical techniques.Visual identification is rapid, relatively cheap and can be conducted without the need for additional technical staff and consumables; however, accurately differentiating microplastics, particularly in the smaller size ranges, requires training and experience. Criteria for visually identifying microplastics include: the absence of cellular or organic structures; a homogenous thickness across the particles; and, homogenous colors and gloss.77,123 Manually sorting plastics under a microscope is most effective for particles >500 micrometers; the effort and accuracy required for sorting increases with decreasing particle size. Owing to the difficulties in handling and differentiating microplastics from organic and inorganic matter,146 error rates could be as high as 70%, increasing with decreasing particle size,61 with incorrect identification most prevalent with microfibres.123,147 To gauge the accuracy of visual discrimination, sub-samples of potential plastics should be chemically analysed.77,123,147–150 It has been observed that training and experience can significantly lower the error rates and misidentification stemming from visual identification.83
Plastics are largely classified by their morphological characteristics: size, shape, and colour. Size is typically based on the longest dimension of a particle; size categories can be used where appropriate. When reporting microplastic shape, researchers tend to use five main categories, although the nomenclature used varies between research groups (Table 2). Finally, colours are often reported across a wide spectrum; colour differentiation is subjective, and visual identification of microplastics cannot be based on colour alone. Caution should be given to categorising microplastics suffering embrittlement, fragmentation or bleaching, or encrusted with biota, as this may skew results.
| Shape classification | Other terms used |
|---|---|
| Fragments | Irregular shaped particles, crystals, fluff, powder, granules, shavings |
| Fibres | Filaments, microfibres, strands, threads |
| Beads | Grains, spherical microbeads, microspheres |
| Foams | Polystyrene, EPS |
| Pellets | Resin pellets, nurdles, pre-production pellets, nibs |
Confirming the identity of suspected plastics may be carried out in a number of ways depending on the funds and equipment available to the researcher. Perhaps the simplest technique is the use of a hot needle to observe melting points.38,52,60,86 While both cheap and fast, this method does not allow for the accurate identification of the polymer; however, the temperature range at which melting occurs does provide a specific range of potential plastics. A converse method is to exclude non-plastics rather than identifying the plastics present, oven and freeze drying removes water from organic material causing it to wither. This increases the likelihood of non-plastic material being identified and removed from mixed samples.151,152 Combining these two techniques provides a cheap, if laborious method of plastic identification.
Another low cost technique involves the examination of microplastics under a polarised light microscope to observe the birefringent properties of the suspected polymer. The birefringence of a polymer is the result of its chemical structure and manufacturing methods which results in unique anisotropic properties; by passing polarised light through a sample, unique spectra are created, from which it is possible to confirm the identity of plastic materials.153 As with the hot needle technique, this method require plastics to be viewed individually; whilst initial costs are low, the time taken makes it prohibitive for large samples. More complex – and costly – methods can also be used to infer resin constituents, plastic additives and dyes. Often, these techniques require the purification of potential microplastic prior to analysis. The removal of biofilms, organic and inorganic matter adhered to the surface will avoid impeding polymer identification and the removal of non-plastic particles.146 Following purification, suspected plastics are submitted to analytical techniques including: Fourier Transformed Infra-Red spectrometry (FT-IR) in transmittance or reflectance; Attenuated Total Reflectance (ATR); Raman spectrometry for colour pigment spectra; and, Pyrolysis-Gas Chromatography combined with Mass Spectroscopy (Pyr-GC-MS), which analyzes particles using their thermal degradation properties and can be used to analyse polymer type and organic plastic additives simultaneously.154 Alternate analytical methods include: high temperature gel-permeation chromatography (HT-GPC) with IR detection; SEM-EDS and thermoextraction; and, desorption coupled with GC/MS.150,155,156
If coupled with microscopy, FTIR and Raman can be used to identify microplastics with a size >20 μm.123,149 Raman spectroscopy combined with microscopy has a higher resolution (approx. 1–2 μm)100,149 and can be used to locate particles within biological tissues.10 FT-IR and Raman have been recommended for determining resin constituents.123,149 There is minimal sample preparation, other than clean up, required for FT-IR. However, FT-IR and PYr-GS-MS are both destructive. Raman is non-destructive as it does not require the sample to be flattened or manipulated. The disadvantage of PYr-GS-MS is the manual placement of the particle in the instrument, which can incur size limitations and only one particle can be run per sample. However, qualitative and quantitative analysis are being developed.141,157 A drawback of chemical analysis is that the isolation of small, highly degraded samples increases the chances of misidentification and producing noisy spectra in which the vital fingerprint areas are obscured, although this can be improved by the use of microscope aided instrumentation (micro-FTIR and micro-Raman), which is designed to target and read responses from samples of a smaller size.
2.4. Contamination
At all stages care must be taken to prevent the contamination or cross-contamination of samples. Airborne contamination of samples with synthetic fibres stemming from clothing75,158 or atmospheric fallout159 is a recurrent issue within the literature.52,53,58,61,63 Sources of contamination should be eliminated where possible, and otherwise quantified using environmental filters or procedural blanks. Here we highlight sources of contamination during sampling and sample processing, and consider protocols for contamination mitigation.2.5. Data analysis
The varied methods by which microplastic uptake by biota is measured understandably results in differing levels of recording. At the highest level, researchers record the number of items, often in relation to organism size. This may be recorded simply, as the percentage of individuals seen to ingest microplastic,36,66,71,76,77,79,84,89 the number of microplastics per individual,16,53,54,57,58,65,74,78,80,85,88–91,102 or as the number of microplastic items by length or weight.38,54,60,61,73,77,89 Many types of plastic, for example microfibre boluses, do not lend themselves to the enumeration of individual plastic items. In addition, mastication and peristaltic action may break down plastic items within the gut; as a result, the number of items in the gut may exceed that originally ingested. In such cases, researchers have reported the weight of plastic aggregations,41 descriptions of the aggregation of microplastic observed,34 or a combination of the two. Such issues in enumeration are more often observed in studies of wild caught organisms, where the initial level of microplastic exposure is not known and the type of microplastic recovered being susceptible to tangling. A similar issue may be observed in the study of microplastic uptake in laboratory experiments; here concentrations of introduced microplastic may be recorded solely by number or mass per individual,19,161 or as a value in relation to mass of food33,34,92,99,161–163 or volume of water.10,26,164The use of multiple methods to quantify the level of microplastic uptake by fish and invertebrates is also an issue in the reporting of environmental plastic levels. Inconsistency in the use of units can mask or inflate the apparent impact of microplastic on a species or location. This increases the likelihood of errors arising when comparing multiple studies carried out by unrelated researchers. The manner in which plastic abundance and concentration is recorded influences the range of statistical analyses available; for example, grouping aggregations into specific classifications reduces the power of the available tests. In field experiments, a range of techniques have been used to determine the relationship between microplastic uptake and both biological and environmental factors. Many of these methods combine continuous and categorical variables in linear models of varying complexity, and require careful structuring in statistical software such as R statistical software.165
3 Discussion
Techniques used to isolate and enumerate microplastics in fish and invertebrates have largely been adapted from studies focussed on large vertebrates (e.g. marine mammals, seabirds; Provencher, this issue), or have derived from traditional biology methods; as these fields advance, it is vital that we continue to observe their progress and incorporate relevant methods. Owing to the challenges in sampling, isolating and identifying microplastics, and the diverse physiology of taxonomic groups under investigation, a degree of flexibility, innovation and ingenuity on the part of the researcher is clearly required.Of the numerous studies investigating microplastic uptake in fish and invertebrates, it is analysis of wild-caught animals that presents the most issues. In these studies, potential sources of error are numerous, including microplastic losses or contamination during sampling; furthermore, there are substantial challenges in drawing links between exposure and effect. These studies, however, are essential for establishing ecologically relevant data, which ultimately provides researchers with a clear view of the quantity and types of plastic (and associated contaminants) experienced by biota in the natural environment. In this section, we address the need for standardising protocols for microplastic quantification,166 outlining preferred methods, best practice, and steps for mitigating contamination. It is anticipated that compiling standardized methodologies will provide researchers with a grounding in developing future experimental design.
3.1. Controlling sampling bias
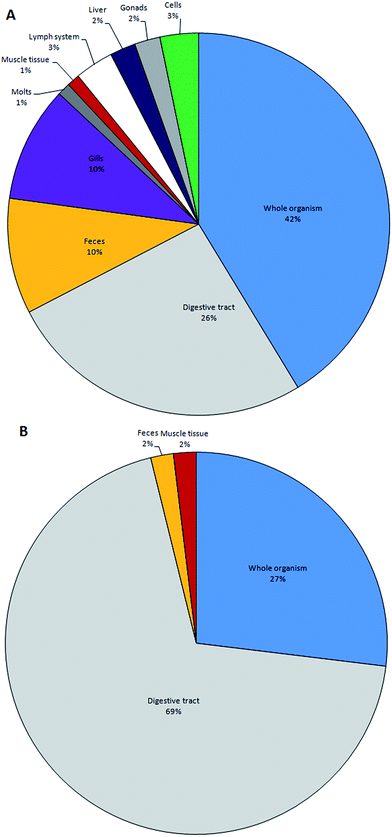
Throughout the sampling process, utmost care must be taken to prevent the artificial inflation or loss of microplastics. In Table 3 we outline the commonly used methods for sampling and isolating microplastics across a range of taxa. In all cases, the least damaging sampling gear is preferable, and sampling periods should be kept as short as practically realistic. Organisms that spend longer in nets are subject to additional stress that increases the likelihood of regurgitation or stomach inversion and artificially increases contact time between microplastics and biota; this could facilitate microplastic ingestion and adherence to external appendages. Individuals should be rinsed following capture to remove adhered particles, and samples of fishing gear should be taken to exclude material ingested as a result of capture. We recommend researchers over-sample where practical, so that individuals with recently emptied stomachs, or otherwise damaged during sampling, should be omitted from the dataset; this will help reduce the bias caused by regurgitation, and enable more robust comparisons between animals sampled from different sites or collected using alternate sampling methods. Specimens should be rapidly transported to the laboratory or preserved promptly to avoid microplastic egestion in transit. We note that collection from commercial fish markets or artisanal fishers is not ideal, as the researcher will have less, if any, control on the method of capture and the handling conditions on transport. Where applicable, we suggest researchers work closely with fishers to ensure animals are sampled appropriately and sufficient information on the capture procedure is collected.| Ecosystem | Life strategy | Subphylum | Size range | Sampling method | Initial plastic separation |
|---|---|---|---|---|---|
| Aquatic | Benthic | Annelida | — | Grabs | Digestion |
| Aquatic | Benthic | Crustacea | >50 mm | Trawls/creels | Dissection |
| Aquatic | Benthic | Crustacea | <50 mm | Otter-/beam-trawls | Digestion |
| Aquatic | Benthic | Echindodermata | — | Grab/trawls | Dissection |
| Aquatic | Benthic | Mollusca | >30 mm | Grabs | Dissection |
| Aquatic | Benthic | Mollusca | <30 mm | Grabs | Digestion |
| Aquatic | Benthic | Flatfish | — | Otter-/beam-trawls | Dissection |
| Aquatic | Nektonic | Crustacea (juv.) | <50 mm | Mid-water trawls | Digestion |
| Aquatic | Nektonic | Gadids | Otter-/mid-water trawls | Dissection | |
| Aquatic | Nektonic | Echindodermata (juv.) | <2 cm | Trawls | Digestion |
| Aquatic | Nektonic | Mollusca (juv.) | <2 cm | Trawls | Digestion |
| Aquatic | Nektonic | Fish | <10 cm | Mid-water trawls | Dissection |
| Aquatic | Planktonic | Annelida | <2 cm | Trawls | Digestion |
| Aquatic | Planktonic | Cnidaria | <10 cm | Trawls | Digestion |
| Aquatic | Planktonic | Crustacea | <2 cm | Trawls | Dissection |
| Terrestrial | — | Annelida | — | Sediment collection | Digestion |
| Terrestrial | — | Arachnida | — | Trapping/hand gathering | Digestion |
| Terrestrial | — | Crustacea | — | Trapping/hand gathering | Digestion |
| Terrestrial | — | Insecta | — | Trapping/hand gathering | Digestion |
| Terrestrial | — | Mollusca | — | Trapping/hand gathering | Digestion |
3.2. Effective plastic isolation
Researchers are presented with a range of techniques for isolating microplastics from biota, including: dissection, depuration, digestion and density separation. Determining the appropriate method will largely depend on the research question (e.g. risks of human consumption, total body burden, localised accumulation). Digestion of whole organisms or excised tissues is widely used (Table 2), however caution must be given in selecting an appropriate digestive agent due to the potential destruction of contaminants. In Table S1 (ESI†) we highlight the damage that a range of digestive agents can cause to polymers. For example, >50% formic acid, >35% HClO4, >40% hydrofluoric acid, >80% H2O2, >50% HNO3, >70% HClO4, >50% KOH and >95% sulphuric acid can be particularly damaging to specific polymers. Some digestive agents, including H2O and HClO4, are simply ineffective in breaking down tissues. Our analysis found that some recent studies used high percentages of acids to which several polymers are not resistant. In particular, high percentages of and HClO4 have been utilised in several studies.16,23,38,51,52,54,60,61,105 For example, only PET and PVC are resistant to 50% HNO3, whereas, PE and PP are partially resistant and PA, PC and PS are not resistant. Typically a balance will need to be struck between finding a cost-effective digestive agent with the capacity to effectively break down tissue, without losing microplastics. Based on our analyses, the rapid 10% KOH (60 °C overnight)58,135 and enzymatic digestion protocols12,140 appear, on balance, to be among the most widely tested and effective digestive treatments currently available; in all cases, the costs, strengths and weaknesses, and applicability of each method to the study organism in question should be carefully considered. As with sampling, steps for mitigating and accounting for external contamination are paramount.3.3. Polymer verification
Methods for verifying isolated microplastics vary in complexity and expense. The method used is dependent not only on resources available to the research group, but also the degree of information required by the study. Studies examining the total body burden or the rate of uptake may only require the most cursory identification to confirm that the particles recovered are indeed plastic; whereas research examining the potential origins of plastics or the presence of adsorbed contaminants and additives require more rigorous testing. We concur with the European standard of polymeric identification of a 5–10% subsample of isolated microplastics. Whilst it has been suggested that this level is insufficient to accurately determine the ability of the researcher to accurately identify plastics, this can be improved when selecting subsamples for verification. Researchers must ensure a representative sample encompassing all categories of recovered microplastics, and particular consideration should be given to commonly misidentified forms, such as fibres and small size fractions. Where possible, researchers should include the weight and number of plastic items, and the mass of sampled organism should be recorded in cases where animals are grouped prior to analysis. Such standardisation will improve comparability between studies.3.4. Mitigating contamination
Contamination, cross-contamination and loss of plastics are a major challenge for microplastics research. It is recommended that all laboratory processing should include steps for preventing or limiting airborne contamination, and procedural blanks used to account for this.124 Additional processing may be utilised during microplastic isolation to improve the detectability or identification of microplastic, however each additional step increases the opportunity for contamination. Where possible plastic consumables should be avoided.167 All samples should be preserved by freezing, desiccation, or in filtered ethanol or formalin, although the latter may result in loss of some plastics. On research vessels, glassware may not be feasible, so plastic may be used following sufficient cleaning with filtered water.3.5. Data analysis
Researchers should be aware of biases in sampling environmentally exposed individuals. Firstly, the condition of organisms prior to capture is unknown, and linking microplastic (and co-contaminant) burden with condition is prohibitive. Secondly, sampling may lead to an underestimation of the microplastic burden in a population because highly contaminated individuals are dead or dying, or remain in shelters and burrows owing to reduced functionality. Thirdly, it is vital that sampling is spatially and/or temporally broad to ensure that observed levels of contamination are representative of the wider population. For example, Welden et al.41 observed significant spatial variation in fibre contamination in three Scottish populations of Nephrops norvegicus.3.6. Recommendations for future work
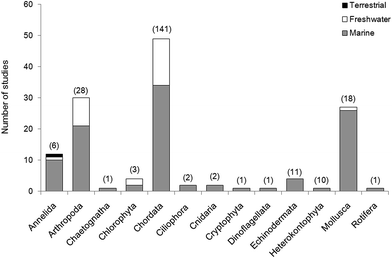
In reviewing the relevant literature, it is apparent that research is currently skewed towards vertebrates (Fig. 4). The range of ecological functions carried out by invertebrates and the diverse niches which they occupy all suggest that the impacts of microplastics on these groups may have a marked effect on the environment. We recommend further assessment of the uptake and impact of microplastics in these groups as this is essential if we are to predict the extent of the effects on biodiversity, ecosystems and ecological processes. A comparison of the relative uptake and retention of the different categories and shapes of microplastics is also required to determine which are the most harmful. Many laboratory studies of microplastic ingestion rely solely on pre-produced plastics which are easily purchased from suppliers (e.g. polystyrene microbeads10–15,17,26,47,105,117); however, these are not representative of the diverse forms currently present in the environment. Understanding which plastics are readily retained is necessary to determine the threat posed by the relative levels of environmental microplastics and to link these to evidence of negative effects in physiology and behaviour which may impair function at the ecosystem level.In fact, a number of studies have considered the observable impacts of microplastic on organisms. In 11/120 studies reviewed here, researchers examined the relationship between microplastic uptake and changes in physiology and body chemistry. Endpoints have included: organism behaviour,104,168 lysosomal response,56 lipid content,169,170 protein content,170 population fitness,161 cellular population growth171 and individual growth.15,170 They also utilised ecotoxicological assays to monitor embryonic development31,172 and the uptake of metals94,173 and chemicals.162,163,174 Laboratory studies must use environmentally realistic concentrations of micro particles, to allow evaluation of potential harm to the individual as a result of field exposure. In wild populations, where the presence of confounding factors make it difficult to attribute biological responses and condition directly to plastic exposure, direct observation of plastic type and abundance remains the most reliable method of determining microplastic impacts.
Microplastics may be selectively or non-selectively ingested or acquired through trophic transfer. Again, the artificially inflated levels of non representative plastics used in a number of currently available studies greatly increases the potential for chance transfer of plastics. The result of such studies, whilst useful in indicating the potential for transfer, display transfer in a way not feasible at normal contamination levels. In studies not focusing on the ecotoxicological impacts, we recommend inflated levels of microplastic should only be used in the presence of an ecologically valid control determined by reference to rigorous environmental sampling.
Lastly, few studies have addressed the movement of microplastic within ecosystems. Many rely on seeding plastic into non-natural food sources, in mesocosm experiments with no alternate food sources, and whilst these experiments have shown the transfer of plastic between food and organisms, there is a clear need for more robust information on the validity of these results in natura.175
The need for rapid, accurate assessment of the levels of microplastic in wild populations is essential for determining baseline levels of contamination, and assessing the risk of microplastic to organisms and ecosystems. The diverse physiology of the organisms covered in this review has necessitated the analysis of a range of protocols for microplastic extraction and enumeration which could limit comparability between studies. As such, we recommend the development of a standard methodology per subphylum or class of organism which will combine efficiency in digestion and recovery of microplastic with the use of the least toxic chemicals to preserve plastic polymers.
Particular attention should be given to harmonising the way in which data is recorded (e.g. mass and number of isolated microplastics per mass of organism) to promote comparability. Prevention of overestimation of plastic contamination must be controlled by confirming the identity of a proportion of the recovered materials. This may be carried out either by chemical analysis, density separation, birefringent microscopy or other physical examination.
4 Concluding remarks
In this review various methods for sampling, isolating and identifying microplastics ingested by fish and invertebrates were examined. As research progresses the need for method standardisation becomes clear, so that a finer picture of the threat of microplastic to organisms emerges and ecological and environmental risk can be assessed. Such standardised methodologies must take into account the numerous potential sources of error and contamination, as outlined above, and also the general need for monitoring which demands that a great number of samples be processed hastily.Many of the studies covered in this review have focused on the issue of whether microplastic uptake occurs, and only addresses the level of plastic contamination in a single species or group and do not allow the assess of risk and disturbance at the ecosystem level. These studies have resulted in numerous further questions regarding the uptake and transfer of microplastics within ecosystems: is plastic uptake selective, or passive? To what extent does trophic transfer occur? And, are the negative effects of plastic ingestion observed in laboratory experiments valid in the environment? There is a strong need to design studies in such a way that their results contribute to clarify these issues, for example contrasting microplastic loads with environmental contamination or diet, to give a more holistic approach to the study of microplastic pollution.
Acknowledgements
MC is funded by the Natural Environment Research Council (NERC UK: NE/L007010). Thanks to S. Nelms for her useful comments on an earlier version of the manuscript. Thanks are extended to C. Rochman and R. Thompson for inviting us to contribute to this special issue.References
- Plastics – the Facts 2014/2015, http://issuu.com/plasticseuropeebook/docs/final_plastics_the_facts_2014_19122, accessed August 2015.
- M. Cole, P. Lindeque, C. Halsband and T. S. Galloway, Mar. Pollut. Bull., 2011, 62(12), 2588–2597 CrossRef CAS PubMed.
- UNEP, Marine Plastic Debris and Microplastic Technical Report, United Nations Environmental Programme, Nairobi, 2016 Search PubMed.
- A. Lusher, in Marine Anthropogenic Litter, ed. M. Bergmann, L. Gutow and M. Klages, Springer, Berlin, 2015, ch. 10, pp. 245–308 Search PubMed.
- M. Wagner, C. Scherer, D. Alvarez-Muñoz, N. Brennholt, X. Bourrain, S. Buchinger, E. Fries, C. Grosbois, J. Klasmeier, T. Marti, S. Rodriguez-Mozaz, R. Urbatazka, D. Vertaak, M. Winther-Nielsen and G. Reifferscheid, Environ. Sci. Eur., 2014, 26(12), 1–9 Search PubMed.
- D. Eerkes-Medrano, R. C. Thompson and D. C. Aldridge, Water Res., 2015, 75, 63–82 CrossRef CAS PubMed.
- K. Duis and A. Coors, Environ. Sci. Eur., 2016, 28(1), 1–25 CrossRef CAS PubMed.
- U. Christaki, J. R. Dolan, S. Pelegri and F. Rassoulozadegan, Limnol. Oceanogr., 1998, 43(3), 458–464 CrossRef.
- D. S. Wilson, Ecology, 1973, 54(4), 909–914 CrossRef.
- M. Cole, P. Lindeque, E. Fileman, C. Halsband, R. Goodhead, J. Moger and T. S. Galloway, Environ. Sci. Technol., 2013, 47(12), 6646–6655 CrossRef CAS PubMed.
- K.-W. Lee, W. J. Shim, O. Y. Kwon and J.-H. Kang, Environ. Sci. Technol., 2013, 47(19), 11278–11283 CrossRef CAS PubMed.
- M. Cole, H. Webb, P. K. Lindeque, E. S. Fileman, C. Halsband and T. S. Galloway, Sci. Rep., 2014, 4, 4528, DOI:10.1038/srep04528.
- O. Setälä, V. Flemming-Lehtinen and M. Lehtiniemi, Environ. Pollut., 2014, 185, 77–83 CrossRef PubMed.
- M. Cole, H. Lindeque, E. S. Fileman, C. Halsband and T. S. Galloway, Environ. Sci. Technol., 2015, 49(2), 1130–1137 CrossRef CAS PubMed.
- M. Cole and T. S. Galloway, Environ. Sci. Technol., 2015, 49(24), 14625–14632 CrossRef CAS PubMed.
- J. P. W. Desforges, M. Galbraith and P. S. Ross, Arch. Environ. Contam. Toxicol., 2015, 69(3), 320–330 CrossRef CAS PubMed.
- M. Cole, P. Lindeque, E. S. Fileman, C. Halsband and T. S. Galloway, Environ. Sci. Technol., 2016, 50(6), 3239–3246 CrossRef CAS PubMed.
- T. F. Bolton and J. N. Havenhand, J. Plankton Res., 1998, 20(11), 2153–2164 CrossRef.
- R. C. Thompson, Y. Olsen, R. P. Mitchell, A. Davis, S. J. Rowland, A. W. John, D. McGonigle and A. E. Russell, Science, 2004, 304(5672), 838 CrossRef CAS PubMed.
- E. Besseling, A. Wegner, E. M. Foekema, M. J. Van Den Heuvel-Greve and A. A. Koelmans, Environ. Sci. Technol., 2013, 47(1), 593–600 CrossRef CAS PubMed.
- M. A. Browne, S. J. Niven, T. S. Galloway, S. J. Rowland and R. C. Thompson, Curr. Biol., 2013, 23(23), 2388–2392 CrossRef CAS PubMed.
- S. L. Wright, D. Rowe, R. C. Thompson and T. S. Galloway, Curr. Biol., 2013, 23(23), R1031–R1033 CrossRef CAS PubMed.
- L. Van Cauwenberghe, M. Claessens, M. B. Vandegehuchte and C. R. Janssen, Environ. Pollut., 2015, 199, 10–17 CrossRef CAS PubMed.
- D. S. Green, B. Boots, J. Sigwart, S. Jiang and C. Rocha, Environ. Pollut., 2016, 208, 426–434 CrossRef CAS PubMed.
- F. Gusmão, M. Di Domenico, A. C. Z. Amaral, A. Martinez, B. C. Gonzalez, K. Worsaae, J. A. I. do Sul and P. da Cunha Lana, Environ. Pollut., 2016, 216, 584–590 CrossRef PubMed.
- O. Setälä, J. Norkko and M. Lehtiniemi, Mar. Pollut. Bull., 2016, 102(1), 95–101 CrossRef PubMed.
- M. W. Hart, Biol. Bull., 1991, 180(1), 12–27 CrossRef.
- E. R. Graham and J. T. Thompson, J. Exp. Mar. Biol. Ecol., 2009, 368(1), 22–29 CrossRef.
- C. Della Torre, E. Beergami, A. Salvati, C. Faleri, P. Cirino, K. A. Dawson and I. Corsi, Environ. Sci. Technol., 2014, 48(20), 12302–12311 CrossRef CAS PubMed.
- K. L. Kaposi, B. Mos, B. P. Kelaher and S. A. Dworjanyn, Environ. Sci. Technol., 2014, 48(3), 1638–1645 CrossRef CAS PubMed.
- C. R. Nobre, M. F. M. Santana, A. Maluf, F. S. Cortez, A. Cesar, C. D. S. Pereira and A. Turra, Mar. Pollut. Bull., 2015, 92(1–2), 99–104 CrossRef CAS PubMed.
- N. M. Hall, K. L. E. Berry, L. Rintoul and M. O. Hoogenboom, Mar. Biol., 2015, 162(3), 725–732 CrossRef CAS.
- A. Ugolini, G. Ungherese, M. Ciofini, A. Lapucci and M. Camaiti, Estuarine, Coastal Shelf Sci., 2013, 129, 19–22 CrossRef CAS.
- F. Murray and P. R. Cowie, Mar. Pollut. Bull., 2011, 62(6), 1207–1217 CrossRef CAS PubMed.
- A. J. Watts, C. Lewis, R. M. Goodhead, D. J. Beckett, J. Moger, C. Tyler and T. S. Galloway, Environ. Sci. Technol., 2014, 48(15), 8823–8830 CrossRef CAS PubMed.
- D. Brennecke, E. C. Ferreira, T. M. M. Costa, D. Appel, B. A. P. da Gama and M. Lenz, Mar. Pollut. Bull., 2015, 96, 491–495 CrossRef CAS PubMed.
- A. J. Watts, M. A. Urbina, S. Corr, C. Lewis and T. S. Galloway, Environ. Sci. Technol., 2015, 49(24), 14597–14604 CrossRef CAS PubMed.
- L. I. Devriese, M. D. van der Meulen, T. Maes, K. Bekaert, I. Paul-Pont, L. Frère, J. Robbens and A. D. Vethaak, Mar. Pollut. Bull., 2015, 98(1–2), 179–187 CrossRef CAS PubMed.
- G. Stasolla, G. Innocenti and B. S. Galil, Isr. J. Ecol. Evol., 2016, 61(3–4), 130–134 Search PubMed.
- A. J. Watts, M. A. Urbina, R. Goodhead, J. Moger, C. Lewis and T. S. Galloway, Environ. Sci. Technol., 2016, 50(10), 5364–5369 CrossRef CAS PubMed.
- N. A. Welden and P. R. Cowie, Environ. Pollut., 2016, 214, 859–865 CrossRef CAS PubMed.
- J. Hämer, L. Gutow, A. Köhler and R. Saborowski, Environ. Sci. Technol., 2012, 48(22), 13451–13458 CrossRef PubMed.
- J. E. Ward and N. M. Targett, Mar. Biol., 1989, 101(3), 313–321 CrossRef.
- M. G. S. Brilliant and B. A. MacDonald, J. Exp. Mar. Biol. Ecol., 2000, 253(2), 211–227 CrossRef.
- M. Brillant and B. MacDonald, Mar. Biol., 2002, 141(3), 457–465 CrossRef CAS.
- J. E. Ward, J. S. Levinton and S. E. Shumway, J. Exp. Mar. Biol. Ecol., 2003, 293(2), 129–149 CrossRef.
- M. A. Browne, A. Dissanayake, T. S. Galloway, D. M. Lowe and R. C. Thompson, Environ. Sci. Technol., 2008, 42(13), 5026–5031 CrossRef CAS PubMed.
- J. E. Ward and D. J. Kach, Mar. Environ. Res., 2009, 68(3), 137–142 CrossRef CAS PubMed.
- N. Von Moos, P. Burkhardt-Holm and A. Köhler, Environ. Sci. Technol., 2012, 46(20), 11327–11335 CrossRef CAS PubMed.
- A. Wegner, E. Besseling, E. M. Foekema, P. Kamermans and A. A. Koelmans, Environ. Toxicol. Chem., 2012, 31, 2490–2497 CrossRef CAS PubMed.
- M. Claessens, L. Van Cauwenberghe, M. B. Vandegehuchte and C. R. Janssen, Mar. Pollut. Bull., 2013, 70(1–2), 227–233 CrossRef CAS PubMed.
- B. De Witte, L. Devriese, K. Bekaert, S. Hoffman, G. Vandermeersch, K. Cooreman and J. Robbens, Mar. Pollut. Bull., 2014, 85(1), 146–155 CrossRef CAS PubMed.
- A. Mathalon and P. Hill, Mar. Pollut. Bull., 2014, 81(1), 69–79 CrossRef CAS PubMed.
- L. Van Cauwenberghe and C. R. Janssen, Environ. Pollut., 2014, 193, 65–70 CrossRef CAS PubMed.
- C. G. Avio, S. Gorbi, M. Milan, M. Benedetti, D. Fattorini, G. d'Errico, M. Pauletto, L. Bargelloni and F. Regoli, Environ. Pollut., 2015, 198, 211–222 CrossRef CAS PubMed.
- L. Canesi, C. Ciacci, E. Bergami, M. P. Monopoli, K. A. Dawson, S. Papa, B. Canonico and I. Corsi, Mar. Environ. Res., 2015, 111, 34–40 CrossRef CAS PubMed.
- J. Li, D. Yang, L. Li, K. Jabeen and H. Shi, Environ. Pollut., 2015, 207, 190–195 CrossRef CAS PubMed.
- C. M. Rochman, A. Tahir, S. L. Williams, D. V. Baxa, R. Lam, J. T. Miller, F. C. Teh, S. Werorilangi and S. J. Teh, Sci. Rep., 2015, 5, 14340, DOI:10.1038/srep14340.
- R. Sussarellu, M. Suquet, Y. Thomas, C. Lambert, C. Fabioux, M. E. J. Pernet, N. Le Goïc, V. Quillien, C. Mingant, Y. Epelboin and C. Corporeau, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(9), 2430–2435 CrossRef CAS PubMed.
- G. Vandermeersch, L. Van Cauwenberghe, C. R. Janssen, A. Marques, K. Granby, G. Fait, M. J. Kotterman, J. Diogène, K. Bekaert, J. Robbens and L. Devriese, Environ. Res., 2015, 143, 46–55 CrossRef CAS PubMed.
- K. Davidson and S. E. Dudas, Arch. Environ. Contam. Toxicol., 2016, 71(2), 147–156 CrossRef CAS PubMed.
- I. Paul-Pont, C. Lacroix, C. Gonzalez Fernandez, H. Hegaret, C. Lambert, N. Le Goic, L. Frere, A.-L. Cassone, R. Sussarellu, C. Fabioux, J. Guyomarch, M. Albentosa, A. Huvet and P. Soudant, Environ. Pollut., 2016, 216, 724–737 CrossRef CAS PubMed.
- M. F. M. Santana, L. G. Ascer, M. R. Custódio, F. T. Moreira and A. Turra, Mar. Pollut. Bull., 2016, 106(1–2), 183–189 CrossRef CAS PubMed.
- H. E. Braid, J. Deeds, S. L. DeGreasse, J. J. Wilson, J. Osborne and R. H. Hanner, Mar. Biol., 2012, 159(1), 25–31 CrossRef CAS.
- M. C. Goldstein and D. S. Goodwin, PeerJ, 2014, 1, e841 Search PubMed.
- E. J. Carpenter, S. J. Anderson, G. R. Harvey, H. P. Miklas and B. B. Peck, Science, 1972, 178, 749–750 CAS.
- S. Kartar, R. A. Milne and M. Sainsbury, Science, 1976, 79(3), 52 Search PubMed.
- J. Dos Santos and M. Jobling, ICES J. Mar. Sci., 1992, 49(2), 145–154 CrossRef.
- C. M. Boerger, G. L. Lattin, S. L. Moore and C. J. Moore, Mar. Pollut. Bull., 2010, 60(12), 2275–2278 CrossRef CAS PubMed.
- P. Davison and R. G. Asch, Mar. Ecol.: Prog. Ser., 2011, 432, 173–180 CrossRef.
- F. E. Possatto, M. Barletta, M. F. Costa, J. A. I. do Sul and D. A. Dantas, Mar. Pollut. Bull., 2011, 62(5), 1098–1102 CrossRef CAS PubMed.
- D. V. Dantas, M. Barletta and M. F. Costa, Environ. Sci. Pollut. Res., 2012, 19(2), 600–606 CrossRef PubMed.
- J. A. A. Ramos, M. Barletta and M. F. Costa, Aquat. Biol., 2012, 17, 29–34 CrossRef.
- C. A. Choy and J. C. Drazen, Mar. Ecol.: Prog. Ser., 2013, 485, 155–163 CrossRef.
- E. M. Foekema, C. De Gruijter, M. T. Mergia, J. A. van Franeker, A. J. Murk and A. A. Koelmans, Environ. Sci. Technol., 2013, 47(15), 8818–8824 CrossRef CAS PubMed.
- M. Gassel, S. Harwani, J.-S. Park and A. Jahn, Mar. Pollut. Bull., 2013, 73, 231–242 CrossRef CAS PubMed.
- A. L. Lusher, M. McHugh and R. C. Thompson, Mar. Pollut. Bull., 2013, 67(1–2), 94–99 CrossRef CAS PubMed.
- K. K. Saji Kumar, N. Ragesh, R. Remya and K. S. Mohamed, Marine Fisheries Information Service; Technical and Extension Series, 2013, vol. 217, p. 13 Search PubMed.
- V. Kripa, P. G. Nair, A. M. Dhanya, V. P. Pravita, K. S. Abhilash, A. A. Mohamed, D. Vijayan, P. G. Vishnu, G. Morhan, P. S. Anilkkumar and L. R. Khambadkar, Marine Fisheries Information Service; Technical and Extension Series, 2014, vol. 219, pp. 27–28 Search PubMed.
- B. Sulochanan, G. S. Bhat, S. Lavanya, A. P. Dineshbabu and P. Kaladharan, Indian J. Geo-Mar. Sci., 2013, 43(9), 1–6 Search PubMed.
- C. G. Avio, S. Gorbi and F. Regoli, Mar. Environ. Res., 2015, 111, 18–26 CrossRef CAS PubMed.
- F. Collard, B. Gilbert, G. Eppe, E. Parmentier and K. Das, Arch. Environ. Contam. Toxicol., 2015, 69(3), 331–339 CrossRef CAS PubMed.
- D. Neves, P. Sobral, J. L. Ferreira and T. Pereira, Mar. Pollut. Bull., 2015, 101, 119–126 CrossRef CAS PubMed.
- M. B. Phillips and T. H. Bonner, Mar. Pollut. Bull., 2015, 100, 264–269 CrossRef CAS PubMed.
- T. Romeo, B. Pietro, C. Peda, P. Consoli, F. Andaloro and M. C. Fossi, Mar. Pollut. Bull., 2015, 95, 358–361 CrossRef CAS PubMed.
- J. Bellas, J. Martínez-Armental, A. Martínez-Cámara, V. Besada and C. Martínez- Gómez, Mar. Pollut. Bull., 2016, 109(1), 55–60 CrossRef CAS PubMed.
- S. M. E. Cannon, J. L. Lavers and B. Figueiredo, Mar. Pollut. Bull., 2016, 107, 286–291 CrossRef CAS PubMed.
- D. D. A. Miranda and G. F. de Carvalho-Souza, Mar. Pollut. Bull., 2016, 103, 109–114 CrossRef CAS PubMed.
- A. L. Lusher, C. O'Donnell, R. Officer and I. O'Connor, ICES J. Mar. Sci., 2016, 73(4), 1214–1225 CrossRef.
- M. A. Nadal, C. Alomar and S. Deudero, Environ. Pollut., 2016, 214, 517–523 CrossRef CAS PubMed.
- T. Naidoo, A. J. Smit and D. Glassom, Afr. J. Mar. Sci., 2016, 38(1), 145–149 CrossRef.
- C. Pedà, L. Caccamo, M. C. Fossi, F. Gai, F. Andaloro, L. Genovese, A. Perdichizzi, T. Romeo and G. Maricchiolo, Environ. Pollut., 2016, 212, 251–256 CrossRef PubMed.
- C. D. Rummel, M. G. Löder, N. F. Fricke, T. Lang, E. M. Griebeler, M. Janke and G. Gerdts, Mar. Pollut. Bull., 2016, 102, 134–141 CrossRef CAS PubMed.
- P. Ferreira, E. Fonte, M. E. Soares, F. Carvalho and L. Guilhermino, Aquat. Toxicol., 2016, 170, 89–103 CrossRef CAS PubMed.
- P. S. Tourinho, J. A. I. do Sul and G. Fillmann, Mar. Pollut. Bull., 2010, 60(3), 396–401 CrossRef CAS PubMed.
- J. A. Van Franeker, C. Blaize, J. Danielsen, K. Fairclough, J. Gollan, N. Guse, P. L. Hansen, M. Heubeck, J. K. Jensen, G. Le Guillou and B. Olsen, Environ. Pollut., 2011, 159(10), 2609–2615 CrossRef CAS PubMed.
- E. Besseling, E. M. Foekema, J. A. van Franeker, M. F. Leopold, S. Kühn, E. B. Rebolledo, E. Heße, L. Mielke, J. IJzer, P. Kamminga and A. A. Koelmans, Mar. Pollut. Bull., 2015, 95(1), 248–252 CrossRef CAS PubMed.
- A. L. Lusher, G. Hernández-Milián, J. O'Brien, S. Berrow, I. O'Connor and R. Officer, Environ. Pollut., 2015, 199, 185–191 CrossRef CAS PubMed.
- P. Rosenkranz, Q. Chaudhry, V. Stone and T. F. Fernandes, Environ. Toxicol. Chem., 2009, 28(10), 2142–2149 CrossRef CAS PubMed.
- H. K. Imhof, J. Schmid, R. Niessner, N. P. Ivleva and C. Laforsch, Limnol. Oceanogr.: Methods, 2012, 10(7), 524–537 CrossRef CAS.
- W. Sanchez, C. Bender and J. M. Porcher, Environ. Res., 2014, 128, 98–100 CrossRef CAS PubMed.
- F. Faure, C. Demars, O. Wieser, M. Kunz and L. F. De Alencastro, Environ. Chem., 2015, 12(5), 582–591 CrossRef CAS.
- F. J. Biginagwa, B. S. Mayoma, Y. Shashoua, K. Syberg and F. R. Khan, J. Great Lakes Res., 2016, 42(1), 146–149 CrossRef CAS.
- O. M. Lönnstedt and P. Eklöv, Science, 2016, 352(6290), 1213–1216 CrossRef PubMed.
- Y. Lu, Y. Zhang, Y. Deng, W. Jiang, Y. Zhao, J. Geng, L. Ding and H. Ren, Environ. Sci. Technol., 2016, 50(7), 4054–4060 CrossRef CAS PubMed.
- P. Wardrop, J. Shimeta, D. Nugegoda, P. D. Morrison, A. Miranda, M. Tang and B. O. Clarke, Environ. Sci. Technol., 2016, 50(7), 4037–4044 CrossRef CAS PubMed.
- C. A. Peters and S. P. Bratton, Environ. Pollut., 2016, 210, 380–387 CrossRef CAS PubMed.
- E. Besseling, B. Wang, M. Lurling and A. A. Koelmans, Environ. Sci. Technol., 2014, 48(20), 12336–12343 CrossRef CAS PubMed.
- M. Ogonowski, C. Schur, A. Jarsen and E. Gorokhova, PLoS One, 2016, 11(5), e0155063 Search PubMed.
- F. Nasser and I. Lynch, J. Proteomics, 2016, 137, 45–51 CrossRef CAS PubMed.
- S. Rehse, W. Kloas and C. Zarfl, Chemosphere, 2016, 153, 91–99 CrossRef CAS PubMed.
- E. Huerta Lwanga, H. Gertsen, H. Gooren, P. Peters, T. Salánki, M. van der Ploeg, E. Besseling, A. A. Koelmans and V. Geissen, Environ. Sci. Technol., 2016, 50(5), 2685–2691 CrossRef CAS PubMed.
- T. Cedervall, L. A. Hansson, M. Lard, B. Frohm and S. Linse, PLoS One, 2012, 7(2), e32254 CAS.
- P. Farrell and K. Nelson, Environ. Pollut., 2013, 177, 1–3 CrossRef CAS PubMed.
- A. Batel, F. Linti, M. Scherer, L. Erdinger and T. Braunbeck, Environ. Toxicol. Chem., 2016, 35(7), 1656–1666 CrossRef CAS PubMed.
- P. Bhattacharya, J. P. Turner and P.-C. Ke, J. Phys. Chem. C, 2010, 114(39), 16556–16561 CAS.
- M. Long, B. Moriceau, M. Gallinari, C. Lambert, A. Huvet, J. Raffray and P. Soudant, Mar. Chem., 2015, 175, 39–46 CrossRef CAS.
- F. Lagarde, O. Olivier, M. Zanella, P. Daniel, S. Hiard and A. Caruso, Environ. Pollut., 2016, 215, 331–339 CrossRef CAS PubMed.
- L. Gutow, A. Eckerlebe, L. Gimenez and R. Saborowski, Environ. Sci. Technol., 2016, 50, 915–923 CrossRef CAS PubMed.
- A. Bakir, S. J. Rowland and R. C. Thompson, Environ. Pollut., 2014, 185, 16–23 CrossRef CAS PubMed.
- C. M. Rochman, in Marine Anthropogenic Litter, ed. M. Bergmann, L. Gutow and M. Klages, Springer, Berlin, 2015, ch. 5, pp. 117–140 Search PubMed.
- C. M. Rochman, Science, 2016, 352(6290), 1172 CrossRef CAS PubMed.
- V. Hidalgo-Ruz, L. Gutow, R. C. Thompson and M. Thiel, Environ. Sci. Technol., 2012, 46(6), 3060–3075 CrossRef CAS PubMed.
- B. S. Sæther and M. Jobling, Aquat. Living Resour., 1997, 10(6), 359–364 CrossRef.
- I. J. McGaw and D. L. Curtis, J. Comp. Physiol., B, 2013, 183(4), 443–465 CrossRef PubMed.
- M. D. Powell and A. J. Berry, Estuarine, Coastal Shelf Sci., 1990, 31(6), 763–773 CrossRef.
- P. J. Bromley, Rev. Fish Biol. Fish., 1994, 4(1), 36–66 CrossRef.
- R. E. Bowman, Environ. Biol. Fishes, 1986, 16(1), 171–181 CrossRef.
- S. H. Bowen, in Fisheries Techniques, ed. B. R. Murphy and D. W. Willis, American Fisheries Society, Bethesda, MD, 2nd edn, 1996, pp. 513–532 Search PubMed.
- N. Daan, Neth. J. Sea Res., 1973, 6(4), 479–517 CrossRef.
- D. Barltrop and F. Meek, Arch. Environ. Health, 1979, 34(4), 280–385 CrossRef CAS PubMed.
- J. J. Reineke, D. Y. Cho, Y.-T. Dingle, A. P. Morello, J. Jacob, C. G. Thanos and E. Mathiowitz, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(34), 13803–13808 CrossRef CAS PubMed.
- A. L. Andrady, in Marine Anthropogenic Litter, ed. M. Bergmann, L. Gutow and M. Klages, Springer, Berlin, 2015, ch. 3, pp. 57–72 Search PubMed.
- S. Nakashima, R. E. Sturgeon, S. N. Willie and S. S. Berman, Analyst, 1988, 113, 159–163 RSC.
- A. Dehaut, A.-L. Cassone, L. Frere, L. Hermabessiere, C. Himber, E. Rinnert, G. Riviere, C. Lambert, P. Soudant, A. Huvet, G. Duflos and I. Paul-Pont, Environ. Pollut., 2016, 215, 223–233 CrossRef CAS PubMed.
- Y. Jin, H. Li, R. B. Mahar, Z. Wang and Y. Nie, J. Environ. Sci., 2009, 21, 279–284 CrossRef CAS.
- M. T. Nuelle, J. H. Dekiff, D. Remy and E. Fries, Environ. Pollut., 2014, 184, 161–169 CrossRef CAS PubMed.
- S. Stojicic, S. Zivkovic, W. Qian, H. Zhang and M. Haapasalo, J. Endod., 2010, 36(9), 1558–1562 CrossRef PubMed.
- E. Duncan, presented in part at Micro 2016, Lanzarote, Canary Islands, Spain, May, 2016 Search PubMed.
- A. I. Catarino, R. Thompson, W. Sanderson and T. B. Henry, Environ. Toxicol. Chem., 2016 DOI:10.1002/etc.3608 , accepted author manuscript.
- B. Scholz-Böttcher, presented in part at Micro 2016, Lanzarote, Canary Islands, Spain, May, 2016 Search PubMed.
- A. A. Horton, C. Svendsen, R. J. Williams, D. J. Spurgeon and E. Lahive, Mar. Pollut. Bull., 2016 DOI:10.1016/j.marpolbul.2016.09.004.
- L. R. Gilfillan, M. D. Ohman, M. J. Doyle and W. Watson, California Cooperative Oceanic and Fisheries Investigations, 2009, vol. 50, pp. 123–133 Search PubMed.
- L. Sgier, R. Freimann, A. Zupanic and A. Kroll, Nat. Commun., 2016, 7, 11587, DOI:10.1038/ncomms11587.
- J. Reisser, J. Shaw, G. Hallegraeff, M. Proietti, D. A. Barnes, M. Thums, C. Wilcox, B. D. Hardesty and C. Pattiaratchi, PLoS One, 2014, 9(6), e100289 Search PubMed.
- M. Löder and G. Gerdts, in Marine Anthropogenic Litter, ed. M. Bergmann, L. Gutow and M. Klages, Springer, Berlin, 2015, ch. 8, pp. 201–227 Search PubMed.
- F. Remy, F. Collard, B. Gilbert, P. Compère, G. Eppe and G. Lepoint, Environ. Sci. Technol., 2015, 49(18), 11158–11166 CrossRef CAS PubMed.
- T. Rocha-Santos and A. C. Duarte, TrAC, Trends Anal. Chem., 2015, 65, 47–53 CrossRef CAS.
- Y. K. Song, S. H. Hong, M. Jang, G. M. Han, M. Rani, J. Lee and W. J. Shim, Mar. Pollut. Bull., 2015, 93(1–2), 202–209 CrossRef CAS PubMed.
- M. Eriksen, S. Mason, S. Wilson, C. Box, A. Zellers, W. Edwards, H. Farley and S. Amato, Mar. Pollut. Bull., 2013, 77(1–2), 177–182 CrossRef CAS PubMed.
- A. R. A. Lima, M. F. Costa and M. Barletta, Environ. Res., 2014, 132, 146–155 CrossRef CAS PubMed.
- A. R. A. Lima, M. Barletta and M. F. Costa, Front. Environ. Sci., 2016, 4, 56 Search PubMed.
- A. A. Hamza, T. Z. N. Sokkar and W. A. Ramadan, Pure Appl. Opt., 1992, 1(6), 321 CrossRef CAS.
- E. Fries, J. H. Dekiff, J. Willmeyer, M. T. Nuelle, M. Ebert and D. Remy, Environ. Sci.: Processes Impacts, 2013, 15(10), 1949–1956 CAS.
- E. Dumichen, A.-K. Barthel, U. Braun, C. G. Bannick, K. Brand, M. Jekel and R. Senz, Water Res., 2015, 85, 451–457 CrossRef PubMed.
- I. Hintersteiner, M. Himmelsbach and W. W. Buchberger, Anal. Bioanal. Chem., 2015, 407, 1253–1259 CrossRef CAS PubMed.
- M. Fischer, presented in part at Micro 2016, Lanzarote, Canary Islands, Spain, May, 2016 Search PubMed.
- M. A. Browne, P. Crump, S. J. Niven, E. Teuten, A. Tonkin, T. Galloway and R. C. Thompson, Environ. Sci. Technol., 2011, 45(21), 9175–9179 CrossRef CAS PubMed.
- R. Dris, J. Gasperi, M. Saad, C. Mirande and B. Tassin, Mar. Pollut. Bull., 2016, 104, 290–293 CrossRef CAS PubMed.
- L. C. Woodall, C. Gwinnett, M. Packera, R. C. Thompson, L. F. Robinson and G. L. J. Paterson, Mar. Pollut. Bull., 2015, 95(1), 40–46 CrossRef CAS PubMed.
- L. C. de Sá, L. G. Luís and L. Guilhermino, Environ. Pollut., 2015, 196, 359–362 CrossRef PubMed.
- C. M. Rochman, E. Hoh, T. Kurobe and S. J. Teh, Sci. Rep., 2013, 3, 03263 Search PubMed.
- C. M. Rochman, T. Kurobe, I. Flores and S. J. Teh, Sci. Total Environ., 2014, 493, 656–661 CrossRef CAS PubMed.
- E. Bergami, E. Bocci, M. L. Vannuccini, M. Monopoli, A. Salvati, K. A. Dawson and I. Corsi, Ecotoxicol. Environ. Saf., 2016, 123, 18–25 CrossRef CAS PubMed.
- R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, 2013, Vienna, Austria, http://www.R-project.org/ Search PubMed.
- EFSA CONTAM Panel EFSA Panel on Contaminants in the food chain, EFSA Journal, 2016, 14(6), e04501, DOI:10.2903/j.efsa.2016.4501.
- M. A. Urbina, A. J. R. Watts and E. E. Reardon, Nature, 2015, 528, 479, DOI:10.1038/528479c.
- K. Mattsson, M. T. Ekvall, L. A. Hansson, S. Linse, A. Malmendal and T. Cedervall, Environ. Sci. Technol., 2015, 49(1), 553–561 CrossRef CAS PubMed.
- C. D. Rummel, M. Adolfsson-Erici, A. Jahnke and M. MacLeod, Environ. Sci.: Processes Impacts, 2016, 18, 788–795 CAS.
- N. A. Welden and P. Cowie, Environ. Pollut., 2016, 218, 895–900 CrossRef CAS PubMed.
- E. Davarpanah and L. Guilhermino, Estuarine, Coastal Shelf Sci., 2015, 167, 269–275 CrossRef CAS.
- D. Mazurais, B. Ernande, P. Quazuguel, A. Severe, C. Huelvan, L. Madec, O. Mouchel, P. Soudant, J. Robbens, A. Huvet and J. Zambonino-Infante, Mar. Environ. Res., 2015, 112, 78–85 CrossRef CAS PubMed.
- L. G. Luís, P. Ferreira, E. Fonte, M. Oliveira and L. Guilhermino, Aquat. Toxicol., 2015, 164, 163–174 CrossRef PubMed.
- M. Oliveria, A. Riberio, K. Hylland and L. Guilhermino, Ecol. Indic., 2013, 34, 641–647 CrossRef.
- J. R. Clarke, M. Cole, P. K. Lindeque, E. Fileman, J. Blackford, C. Lewis, T. M. Lenton and T. S. Galloway, Front. Ecol. Environ., 2016, 14, 317–324 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ay02415g |
| This journal is © The Royal Society of Chemistry 2017 |