High definition infrared chemical imaging of colorectal tissue using a Spero QCL microscope
B.
Bird
 * and
J.
Rowlette
* and
J.
Rowlette
Daylight Solutions Inc., 15378 Avenue of Science, Suite 200, San Diego, CA 92128, USA. E-mail: b.bird@daylightsolutions.com
First published on 11th January 2017
Abstract
Mid-infrared microscopy has become a key technique in the field of biomedical science and spectroscopy. This label-free, non-destructive technique permits the visualisation of a wide range of intrinsic biochemical markers in tissues, cells and biofluids by detection of the vibrational modes of the constituent molecules. Together, infrared microscopy and chemometrics is a widely accepted method that can distinguish healthy and diseased states with high accuracy. However, despite the exponential growth of the field and its research world-wide, several barriers currently exist for its full translation into the clinical sphere, namely sample throughput and data management. The advent and incorporation of quantum cascade lasers (QCLs) into infrared microscopes could help propel the field over these remaining hurdles. Such systems offer several advantages over their FT-IR counterparts, a simpler instrument architecture, improved photon flux, use of room temperature camera systems, and the flexibility of a tunable illumination source. In this current study we explore the use of a QCL infrared microscope to produce high definition, high throughput chemical images useful for the screening of biopsied colorectal tissue.
Introduction
The latest generation of Fourier transform infrared (FT-IR) spectrometers, that incorporate large liquid nitrogen cooled focal plane array (FPA) detectors within an infrared microscope system, have no doubt accelerated the development of the field of spectral histopathology. Significant improvements in data acquisition, processing, and classification times, in part due to the increased field of view (FOV) of these FPA based devices, have enabled spectroscopic investigations that now include clinically relevant patient populations.1–4 Nevertheless, despite these marked improvements when compared to linear detector array (LDA) or point detector based systems, data collection times from tissue micro-array (TMA) cores or whole tissue sections are still in the order of hours or days.6 All FT-IR based microscope systems are inevitably limited by a tradeoff between spatial resolution, signal-to-noise ratio, field of view and acquisition time. Traditionally with a conventional bench-top FT-IR based system, to achieve true diffraction-limited resolution, high magnification objectives with the largest possible numerical aperture (N.A.), or use of upstream optics to increase the overall system magnification are required that provide a much-reduced field of view (ca. 150 × 150 μm) and extended sample dwell times to achieve an acceptable signal-to-noise ratio for tissue classification purposes.7–9 More recently, however, the application of high resolution imaging for clinical diagnostics has shown clear advantages for the identification of small tissue structures that are essential for detecting early forms of disease.9–11 Identification of very small changes in the biochemical components of glandular, endothelial and myoepithelial cells, or intra-lobular stroma could be key for early pre-malignant changes to be identified and tracked.The recent development of the Spero® microscope (Daylight Solutions Inc., San Diego, USA), a laser-based mid-infrared microscope, provides the capability to perform diffraction-limited imaging across the molecular fingerprint region (900–1800 cm−1), at high N.A. (0.7) and with a much enhanced FOV (650 × 650 μm). This equates to a ∼20× enhancement in the FOV, making high resolution imaging of tissue samples more viable in practical timeframes.6,12 Currently the two major bottlenecks predominant in IR chemical imaging are long data collection times and spectral hypercube file size. The Spero QCL infrared microscope can address both issues by harnessing the power and flexibility of the tunable laser source. If diagnostic spectral features are located at a reduced number of frequencies across the full spectrum, which is often the case, the laser can be programmed to target these frequencies alone, reducing acquisition time and data file size proportionately.13,14
This contribution presents data acquired from biopsied colorectal tissue specimens as an example case study of this new capability. In addition to the acquisition of full band infrared spectra, sparse frequency datasets were recorded from 10 infrared bands previously identified as salient for tissue segmentation in the literature. This was conducted to create rapid chemical maps. Such images do not provide diagnostic contrast (i.e. the segmentation of cancerous cells), but could serve as methods for more rapid global screening of samples. As in traditional histology, identification of abnormal tissue architecture could potentially be visualized using these more rudimentary chemical images, prior to moving on to more rigorous analyses, including full-band infrared analysis, or by using new laser microdissection techniques for downstream proteomic and genomic testing of targeted regions.
Materials and methods
Sample preparation
Colorectal tissue sections were cut from archival tissue banks of formalin fixed paraffin embedded (FFPE) tissue blocks (SDPathology, San Diego, USA). Two parallel sections were cut from each sample for analysis. The first section was cut at 10 μm thickness and floated onto a CaF2 disc (25 mm × 2 mm) for infrared microscopy. The second and adjacent section was cut at 5 μm thickness and floated onto a regular glass slide for brightfield visible microscopy. Both sections were subsequently de-paraffinised using standard protocols. The section mounted onto glass was further H&E stained and cover slipped before conventional brightfield imaging at 20× magnification using a whole slide imaging system (Hamamatsu, Japan). The section mounted onto CaF2 disc remained stain-free and stored in a desiccator prior to infrared microscopy.QCL-based infrared microscopy
QCL-based infrared microscopy6,12–16 was performed using a Spero microscope (Daylight Solutions Inc., San Diego, USA). Mosaic images were recorded from the tissue samples using the 12.5×, 0.7 N.A., compound refractive infrared objective of the microscope. This optic has a field of view 650 μm × 650 μm (480 × 480 pixels), and sample referenced pixel size of 1.35 μm.Mercury cadmium telluride (MCT) focal plane arrays (FPAs) have been the choice for FT-IR based infrared imaging and have been employed in some QCL microscope prototypes.16 These cameras can operate over 2–12 μm when cryogenically cooled with frame rates exceeding 1 kHz but suffer from poor linear dynamic range (ca. 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and long-term reliability.
1) and long-term reliability.
Room temperature operated microbolometer cameras based on VO2 or a-Si have intrinsic responsivity over the entire mid-IR band and can exhibit excellent linear dynamic ranges (>1000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). However, commercially available microbolometers are spectrally limited to 8–14 μm in order to suppress water vapor noise in thermal imaging applications. The Spero microscope uses a custom uncooled 480 × 480 microbolometer VO2 FPA having a 30 Hz frame rate and excellent responsivity over the 5–14 μm molecular fingerprint band.
1). However, commercially available microbolometers are spectrally limited to 8–14 μm in order to suppress water vapor noise in thermal imaging applications. The Spero microscope uses a custom uncooled 480 × 480 microbolometer VO2 FPA having a 30 Hz frame rate and excellent responsivity over the 5–14 μm molecular fingerprint band.
Currently, a full data cube containing 226 spectral images takes 5 minutes or about 9000 individual camera frames. Future improvements in instrumentation could allow spectral data collections rates to increase to about one spectral image per 1–2 camera frames collected.
As all molecules respond to the exciting infrared radiation and produce relatively complicated infrared spectra, the response observed for a single cell or an area of tissue is a complex superposition of all spectral features of all biomolecules in the sample. Although infrared spectroscopy is usually referred to as a “fingerprint” spectroscopic technique, which implies that every molecule known exhibits a distinct spectrum that identifies it, the superposition of such fingerprints leads to relatively broad spectral features that need to be decoded, or de-convolved, to enable an interpretation or diagnosis. Nevertheless, it is typical that small but reproducible changes in the spectral features can be identified between cell/tissue types. These changes can be identified by use of unsupervised methods of multivariate image analysis or mined and ranked using annotated spectral libraries and feature selection algorithms based on statistical measures such as the t-test. In general, a smaller number of salient spectral features can be used to train robust supervised algorithms that provide optimal classification.1,4
In this study, all infrared images were recorded in duplicate from the full fingerprint region between 900–1800 cm−1, and from a reduced set of 10 sparsely distributed salient frequencies within the same region, located at 940, 1044, 1180, 1300, 1336, 1360, 1480, 1544, 1656 and 1800 cm−1 respectively. These frequencies were selected after consultation of several rigorous FT-IR based studies performed by 3 independent research groups17–20 and are further summarized in Table 1. Each recorded spectral hypercube took 5 minutes for a full broadband spectrum and 0.9 minutes for a sparse frequency data collection. The data was recorded at 8 cm−1 spectral resolution with a 4 cm−1 data interval, and ratio'd against a background from a clean area of the substrate. Spectral hypercubes 480 × 480 × 256 (102 MB) or 480 × 480 × 10 (5 MB) were thus created.
| Spectral descriptor type | Peak 1 band position (cm−1) | Vibrational mode/assignment | Peak 2 band position (cm−1) | Molecular assignment | Baseline positions (cm−1) |
|---|---|---|---|---|---|
| Protein | 1656 | Combination of: | 1544 | Combination of: | 1480, 1800 |
| Peak height ratio | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (mainly) O stretching (mainly) |
N–H bending | |||
| N–H bending | C–N stretching vibration | ||||
| C–N stretching | |||||
| Amide I of proteins | Amide II of proteins | ||||
| 1656 cm−1 position is correlated with α-helix structures | |||||
| Ratio highlights gross protein compositional changes i.e. changes in secondary structure/amino acids | |||||
| Collagen | 1336 | CH2wagging | 1300, 1360 | ||
| Peak height | Amino acid proline side chains | ||||
| Abundant in connective tissues | |||||
| Mucin | 1044 | COH deformation | 940, 1180 | ||
| Peak height | Glycosylated protein mucin | ||||
| Abundant in mucus membranes |
Data pre-processing
Standard pre-processing techniques for mid-infrared spectroscopy were used. These have been reported at great length previously.5,21,22 Raw data sets were imported into the multisensor imaging software package ImageLab.23 The data processing included:• Noise reduction via the maximum noise fraction transform.24
• Spectral quality test to remove pixel spectra from regions not occupied by tissue (pixels describing an amide I intensity at 1656 cm−1 <0.05 au were rejected).
• Full band spectra were converted to second derivatives (Savitzy–Golay algorithm, 9-point smooth) to provide better resolved peaks and eliminate background slopes.
• All spectral vectors were scaled to a zero mean and a standard deviation of 1.0 (standard normal variate normalisation) to reduce the influence of intensity changes caused by differences in cellular density and tissue thickness.
Data analysis
Sparse frequency absorbance datasets were used to create chemical images as follows:• Protein map: peak height ratio of the amide I (1656 cm−1)/amide II (1544 cm−1) bands. The ratio was linear baseline corrected using intensity values recorded at 1480 and 1800 cm−1. The ratio image produced was mapped against a white–blue (min–max) colour palette.
• Collagen map: peak height of collagen CH2 side chain vibrations (1336 cm−1). The measurement was linear baseline corrected using intensity values recorded at 1300 and 1360 cm−1. The intensity image produced was mapped against a white–red (min–max) colour palette.
• Mucin map: peak height of the strongest mucin glycosylation band (1044 cm−1). The measurement was linear baseline corrected using intensity values recorded at 1180 and 940 cm−1. The intensity image produced was mapped against a white–green (min–max) colour palette.
• RGB image: each chemical map mentioned above was merged into a single red, green, blue colour image.
These chemical maps or stains were generated in an effort to describe the potential of sparse frequency collection protocols to provide meaningful and easily interpretable chemical images for rapid inspection/screening of samples.
Full band (900–1800 cm−1) second derivative datasets were analysed using the unsupervised algorithm, k-means clustering, a non-hierarchical iterative method that obtains “hard” class membership for each spectrum. A total of 6 clusters was chosen for analyses. False colour images were constructed, where a pixel's cluster membership is defined by a corresponding colour in the image. Pixels rejected by the quality test are provided a black colour. These datasets were collected to provide further evidence that QCL-based infrared spectral imaging data can produce high fidelity images and spectra sufficient for rigorous multivariate analyses and allay concerns from early reports of potentially problematic coherence effects.25
Results and discussion
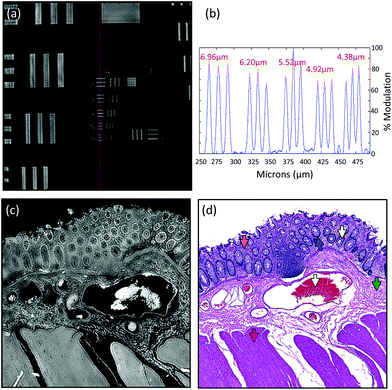
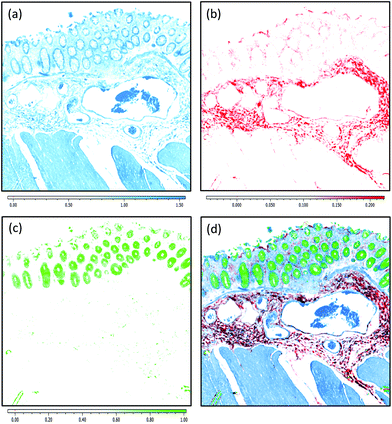
Due to the longer wavelengths of mid-infrared light in the biological fingerprint region (λ = 5.5–11 μm) the lateral resolution of IR based microscopes is reduced compared to visible light microscopes by an order of magnitude. However, this inherent drawback is offset by the rich chemical information that can be visualised by this non-destructive, label free technique. The performance of the Spero microscope's 12.5× optic is demonstrated in Fig. 1(a)–(d), whereby the contrast of a USAF 1951 resolution test pattern was used to quantify spatial resolution in reflection geometry. Fig. 1(a) displays a reflectance image of groups 6 and 7 of the target recorded at 1555 cm−1 (6.4 μm). Fig. 1(b) displays a line profile plot across group 6 elements that has patterns of spatial frequencies down to 4.38 μm (widths of each group of elements are displayed in red). Spatial resolution is governed by the diffraction limit. A microscope can be considered diffraction limited if able to distinguish objects separated by 0.61λ/N.A. The line plot clearly indicates that features of 5 microns can be resolved, meeting the Rayleigh criterion of 26% image contrast. Fig. 1(c) shows an image captured by the microscope from a microtome tissue section. The greyscale IR chemical image was acquired from a healthy colorectal tissue section (3 × 3 image tile mosaic) using the amide 1 (1656 cm−1) vibration of proteins. Fig. 1(d) displays a brightfield image captured from the parallel H&E stained section. By direct comparison of the images it is clear the small diagnostic features of the colorectal tissue can be adequately resolved with this optic, including microstructures such as blood vessel walls and the colonocytes that surround the goblet cells of the crypts (a reference is provided by the coloured arrows in the stained image).Fig. 2(a)–(c) display chemical images from the same tissue region shown in Fig. 1. These were calculated using the sparse frequency absorbance datasets that comprise 10 salient IR frequencies (45MB in size, ca. 20× smaller than a full band dataset of a mosaic the same size). Fig. 2(a) shows a chemical image based on the gross protein composition of the tissue and was calculated by ratio of the two most prominent bands of proteins, the amide I and amide II vibrations. After normalisation of the spectral vectors, a ratio of the protein bands yields greater contrast between tissue structures than using a single peak height alone. The colonocytes and blood cells are provided the greatest contrast in this chemical map. Fig. 2(b) shows a chemical image based on the presence of collagen and was calculated using the CH2 side chain vibrations of collagen. The submucosa, being comprised mostly of collagen, has the highest contrast when using this chemical map. Fig. 2(c) shows a chemical image based on the presence of mucin and was calculated using a characteristic glycosylation absorbance band in the low wavenumber part of the IR spectrum. The greatest contrast for the goblet cells and lumen of the crypts was achieved using this chemical map, which again makes biochemical sense since they both comprise high concentrations of muco- and glyco-proteins. The final image in Fig. 2(d) is an RGB fused image of the chemical maps shown in Fig. 2(a)–(c) respectively. By direct comparison to the brightfield image captured from the parallel H&E stained section in Fig. 1(d), the RGB image provides clear contrast of the major colorectal tissue structures, including the mucosa, submucosa and muscularis externa. Such chemical based images could help identify discrete regions within large areas of tissue where the mucosa no longer provides normal architecture, i.e. the appearance of clusters of enlarged crypts (aberrant crypts) and other structural hallmarks of epithelial dysplasia and neoplasia. The combination of these RGB type chemical images with morphometric descriptors used in digital pathology analysis may be worth consideration. In the example presented, colour channels were selected with colon tissue in mind, however these can be adjusted to suit the application. For example, lipid associated diseases (e.g. glioblastoma multiforme) could be visualised using the lipid ester band at 1740 cm−1, whereas disease associated changes in cellular metabolism (e.g. hypoxia driven changes) could be tracked using one of the characteristic glycogen peaks at 1030, 1080 or 1152 cm−1 respectively. Such simplified chemical representations of the tissue could serve both pre-clinical and clinical needs in routine screening applications. More recently, it has been pondered whether all the frequencies that are routinely collected in a broadband FT-IR measurement are required for infrared spectral staining/diagnostics.1,4,26,27 Subsequent proof of concept studies performed using QCL microscopy technology have shown great promise.13–15 However, more exhaustive investigations into clinically relevant patient populations is still required to ascertain whether frequency targeting protocols can be sufficiently robust.
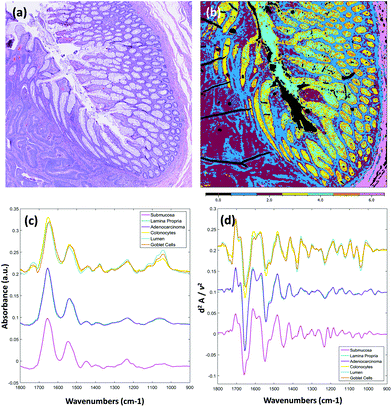
Fig. 3 displays results obtained from the multivariate analysis of a full band second derivative dataset acquired from a diseased colorectal tissue section (3 × 3 mosaic image, 918 MB in size, ca. 20× larger than a sparse 10 frequency dataset of the same mosaic size). After pre-processing and quality check the mosaic composed 1.65 million pixel spectra. Fig. 3(a) displays the brightfield image captured from the parallel H&E stained section. Fig. 3(b) displays the k-means clustering image (6 clusters) calculated from the derivative infrared dataset. Both the mean absorbance and second derivative spectra were calculated for the cluster analysis and are displayed in Fig. 3(c) and (d) respectively. The absorbance data was baseline corrected using an asymmetric least squares approach (Eilers algorithm, 5 iterations),28 and normalised for clarity before calculation of mean cluster spectra. This is the first report, to the authors knowledge, of rigorous analysis performed on full fingerprint region second derivative spectra recorded using a QCL infrared microscope. It has been reported that scattering effects, amplified by the coherent light source, can cause unwanted artefacts.25 Unsupervised methods of multivariate analyses are very sensitive to these types of structures in the spectral and spatial domain but were not observed as problematic in these tissue studies. By observation of the mean cluster spectra, it appears segmentation of the different structures within the colonic crypts are predominantly caused by intensity differences at several characteristic glycosylation bands located at 1044, 1076, 1120 and 1374 cm−1 associated with mucin. A strong lipid ester band at 1740 cm−1 is also observed for cell types of the crypts. The submucosa, in contrast, provides a number of strong bands that can be directly attributed to the structural protein collagen with bands located at 1204, 1236, 1280, 1336 (amide III), and 1452 cm−1 respectively. Spectroscopic differences between the lamina propria and adenocarcinoma are far more subtle, and are located at nucleic acid-related vibrations at ca. 964, 1062, 1090 and 1236 cm−1 respectively, with again very small differences in the amide I and amide II band profiles.
Conclusions
The results presented in this contribution lend further evidence to the great promise of QCL infrared microscopy. The technology can provide high resolution, high throughput IR chemical images useful for infrared based spectral pathology. The efficiency of frequency targeting protocols will be subject to continued debate within the spectroscopic community in the coming years, especially when considering confounding band lineshape distortions that can be caused by scattering, which may necessitate collection of full band spectra for robust correction. However, such concerns shall likely dissipate as QCL imaging technology achieves even faster data acquisition speeds. With a myriad of possible diagnostic applications, it is reasonable to expect that for some cases, sparse frequency protocols can realise higher throughput without sacrificing accuracy. Proof of concept studies performed on breast tissue microarrays13 and blood sera14 have shown great promise. However, further work needs to be performed on significantly larger patient numbers to fully understand and realise the potential of the technology.Acknowledgements
The authors would like to acknowledge Dr Tim Robbins and Dr Shirley Reidy at SDPathology for sourcing of archival tissue blocks and pathology consult. We would in addition like to thank Dr Hans Lohninger and Elisabeth Renner at Epina for many fruitful conversations and support. We would also like to acknowledge Dr Edeline Fotheringham for building and calibrating the Spero microscope used in this work.Notes and references
- B. Bird, M. Miljkovic, S. Remiszewski, A. Akalin, M. Kon and M. Diem, Lab. Invest., 2012, 92, 1358–1373 CrossRef PubMed.
- N. Bergner, B. F. M. Romeike, R. Reichart, R. Kalff, C. Krafft and J. Popp, Analyst, 2013, 138, 3983–3990 RSC.
- A. Akalin, B. Bird, M. Miljković, X. Mu, M. A. Kon, A. Ergin, S. H. Remiszewski, C. M. Thompson, D. J. Raz and M. Diem, Lab. Invest., 2015, 95, 406–421 CrossRef PubMed.
- D. C. Fernandez, R. Bhargava, S. M. Hewitt and I. W. Levin, Nat. Biotechnol., 2005, 23, 469–474 CrossRef CAS PubMed.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sule-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protoc., 2014, 9(8), 1771–1791 CrossRef CAS PubMed.
- P. Bassan, M. J. Weida, J. Rowlette and P. Gardner, Analyst, 2014, 139, 3856–3859 RSC.
- L. S. Leslie, A. Kadjacsy-Balla and R. Bhargava, Medical Imaging: Digital Pathology, 2015, p. 9420 Search PubMed.
- V. Varma, A. Kajdacsy-Balla, S. K. Akkina, S. Setty and M. J. walsh, Kidney Int., 2016, 85(5), 1153–1159 CrossRef PubMed.
- H. Sreedhar, V. K. Varma, P. L. Nguyen, B. Davidson, S. Akkina, G. Guzman, S. Setty, A. Kajdacsy-Balla and M. Walsh, JoVE, 2015, 95, e52332 Search PubMed.
- M. J. Walsh, S. E. Holton, A. Kajdacsy-Balla and R. Bhargava, Vib. Spectrosc., 2012, 60, 23–28 CrossRef CAS PubMed.
- M. Walsh, D. Mayerich, A. Kajdacsy-Balla and R. Bhargava, Proc. SPIE, 2012, 8219 Search PubMed.
- H. Shreedhar, V. K. Varma, F. V. Gambacorta, G. Guzman and M. J. Walsh, Biomed. Opt. Express, 2016, 7(6), 2419–2424 CrossRef PubMed.
- M. J. Pilling, A. Henderson, B. Bird, M. D. Brown, N. W. Clarke and P. Gardner, Faraday Discuss., 2016, 187, 135–154 RSC.
- C. Hughes, G. Clemens, B. Bird, T. Dawson, K. M. Aston, M. D. Jenkinson, A. Brodbelt, M. Weida, E. Fotheringham, M. Barre, J. Rowlette and M. J. Baker, Sci. Rep., 2016, 6, 20173 CrossRef CAS PubMed.
- N. Kroger-Lui, N. Getz, K. Hasse, B. Kranzlin, S. Neudecker, A. Pucci, A. Regenscheit, A. Schonhals and W. Petrich, Analyst, 2015, 140, 2086 RSC.
- K. Yeh, S. Kenkel, J. Lui and R. Bhargava, Anal. Chem., 2015, 87(1), 485–493 CrossRef CAS PubMed.
- P. Lasch, W. Haensch, D. Naumann and M. Diem, Biochim. Biophys. Acta, 2004, 1688, 176–186 CrossRef CAS PubMed.
- P. Lasch and D. Naumann, Biochim. Biophys. Acta, 2006, 1758, 814–829 CrossRef CAS PubMed.
- A. Kallenback-Thieltges, F. Groβerϋschkamp, A. Mosig, M. Diem, A. Tannapfel and K. Gerwert, Biophotonics, 2013, 6(1), 88–100 CrossRef PubMed.
- C. Krafft, D. Codrich, G. Pelizzo and V. Serfo, Biophotonics, 2008, 1(2), 154–169 CrossRef CAS PubMed.
- M. Miljkovic, B. Bird, K. Lenau, A. I. Mazur and M. Diem, Analyst, 2013, 138, 3975 RSC.
- P. Lasch, Chemom. Intell. Lab. Syst., 2013, 117, 100–114 CrossRef.
- H. Lohninger and E. Renner, IMAGELAB, Epinasoftware, Austria, 2016 Search PubMed.
- A. A. Green, M. Berman, P. Switzer and M. D. Craig, IEEE Trans. Geosci. Electron., 1988, 26, 65–74 CrossRef.
- M. R. Kole, R. K. Reddy, M. V. Schulmerich, M. K. Gelber and R. Bhargava, Anal. Chem., 2012, 84(23), 10366–10372 CrossRef CAS PubMed.
- P. Bassan, J. Mellor, J. Shapiro, K. J. Williams, M. P. Lisanti and P. Gardner, Anal. Chem., 2014, 86, 1648–1653 CrossRef CAS PubMed.
- R. Bhargava, Anal. Bioanal. Chem., 2007, 389, 1155–1169 CrossRef CAS PubMed.
- P. H. C. Eilers and H. F. M. Boelens, Technical report, Leiden University Medical Center, 2005.
| This journal is © The Royal Society of Chemistry 2017 |