Open Access Article
Open Access ArticleThe origin of the band at around 730 cm−1 in the SERS spectra of bacteria: a stable isotope approach
Patrick
Kubryk
,
Reinhard
Niessner
and
Natalia P.
Ivleva
*
Technical University of Munich, Institute of Hydrochemistry, Chair of Analytical Chemistry, Marchioninistr. 17, 81377 Munich, Germany. E-mail: natalia.ivleva@ch.tum.de; Fax: +49 89 2180 78255; Tel: +49 89 2180 78238
First published on 20th April 2016
Abstract
Raman microspectroscopy is an emerging tool to analyze the molecular and isotopic composition of single microbial cells. It can be used to achieve an in situ understanding of metabolic processes. Due to the low sensitivity of the Raman effect, surface-enhanced Raman scattering (SERS) is utilized to enhance the Raman signal. The SERS spectra of bacteria are usually characterized by a pronounced band at around 730 cm−1, which is assigned to glycosidic ring vibrations or to adenine or even to CH2 deformation in different studies. In order to clarify the origin of this band, we employed a stable isotope approach and performed a SERS analysis of Escherichia coli bacteria using in situ prepared Ag nanoparticles. The cells were grown on unlabeled (12C, 14N) and labeled (13C, 15N) carbon and nitrogen sources in different combinations. The SERS band of the stable isotope labeled microorganisms showed a characteristic red-shift in the SERS spectra, which solely depends on the isotopic composition. It was therefore possible to confidently assign this band to adenine-related compounds. Furthermore, by utilizing the fingerprint area of single-cell SERS spectra as the input for the principal component analysis, one can clearly differentiate between E. coli bacteria incorporating different stable isotopes.
Introduction
Raman microspectroscopy (RM) offers fingerprint spectra with a spatial resolution down to 1 μm and with minimal or no water interference. This technique allows the analysis of single bacterial cells in situ, and can be applied to discriminate between microbial species or to monitor the physiological state of a cell. On the other hand, the sensitivity of Raman spectroscopy is quite limited. To overcome this shortcoming, surface-enhanced Raman scattering (SERS) is applied to enhance the Raman signal. When nanometer-sized metallic structures (Ag or Au) are brought into direct contact with target molecules, electromagnetic (“localized surface plasmon resonance”) and chemical (“charge transfer”) enhancement effects can increase the Raman signal by a factor of 103–1011.1 This improvement allows a rapid analysis and identification of single cells.2,3 However, the interpretation of the vibrational features of bacteria at the molecular level is a difficult task, since various vibrational bands typical of proteins, phospholipids, nucleic acids, and polysaccharides are anticipated to contribute to the SERS spectra.4–8 As different vibrational bands found in Raman spectra are often enhanced differently, while new bands may appear due to the chemical enhancement, the analysis of conventional Raman spectra can only partially help in explaining the SERS results.The main feature of the SERS spectra of bacteria is often the sharp band in the region of 720–735 cm−1.9 Previous SERS studies of bacteria proposed various assignments to this region. Usually reference compounds were compared with the SERS spectra and on this basis the conclusions and band assignments were made. The outer bacterial cell layer sensitivity of the SERS technique makes the assignment of this band to the components of the cell membrane possible. This band therefore may be assigned to the polysaccharide mode since the cell walls of bacteria are made up of peptidoglycan, which contains N-acetyl-D-glucosamine (NAG). It was therefore suggested that this band can be assigned to the vibrational glycosidic ring mode, since the SERS spectra of D-glucose and NAG also exhibit an intense peak at around 730 cm−1.5 Substances, such as amino acids and phospholipids, also show SERS bands in this region.5,10,11 Furthermore, in a normal Raman analysis of bacteria the band at 725 cm−1 was assigned to the CH2 rocking mode.12 Also the O–P–O vibrational mode of a phosphate group was suggested.3 In addition, the strong band at 735 cm−1 has been attributed to molecules possessing the adenine moiety in their structure, located on or in the cell wall structure, which is extremely sensitive to SERS enhancement.6,11,13 The strong 735 cm−1 vibration is especially characteristic of DNA and often serves as its marker.7,14–18 It is less probable that DNA in its native form is responsible for these adenine-based Raman bands, as bands derived from the other nucleic bases in addition to adenine would be expected.7 Therefore, the appearance of the strong band at 735 cm−1 can indicate DNA denaturation.15 Other common adenine derivatives or adenine-containing molecules include RNA, RF, FAD, NAD, NADH, AMP, ADP, or ATP, which are critical molecules for the biochemical processes in bacterial cells.6,14 Also the purine derivative hypoxanthine is suggested.19,20 A study by Guzelian et al. showed a comparison of molecules containing the adenine functionality with the SERS spectra of bacteria.11 In a study by Kahraman et al. different bacterial species, their isolated DNA and their isolated peptidoglycan layer were analyzed to obtain information about the interaction of the silver nanoparticles with the bacteria. They concluded that the SERS spectra of bacteria originate probably from adenine-containing molecules situated in the bacterial cell wall with possible contributions from molecular species detached from the cell surface.6 However, the presence of adenine containing compounds on the outermost surface of bacterial cells is not obviously explained.11 It should also be noted that all the proposed peak assignments at around 730 cm−1 on the SERS spectra of bacteria could be correct and signals originating from those various substances are overlapping.14
Nowadays this SERS marker band at around 730 cm−1 is usually assigned to adenine, although no clear evidence is available. Raman spectroscopy in combination with stable isotopes is an often used technique to assign Raman bands and to understand spectral features.21 Therefore, in this study we applied a stable isotope surface-enhanced Raman scattering approach to obtain more information about this band and to understand its origin. Polysaccharides, phospholipids and adenine-related substances have a vastly different molecular structure. Hence, the position of this band in the SERS spectra of stable isotope labeled microorganisms will help us to differentiate between the different compounds and determine the origin of this band. Since the feasibility of 15N labeling of E. coli cells was previously demonstrated for normal Raman studies,22,23 labeled nitrogen alone or in combination with labeled carbon will also be used here, for the first time, for the SERS analysis of E. coli. Furthermore, in this study we compare the red-shift of the SERS spectra of microorganisms for different isotopic compositions (12C–14N, 13C–14N, 12C–15N and 13C–15N). We demonstrate the possibilities of discriminating stable isotope labeled cells by means of SERS with the help of principal component analysis (PCA) of the fingerprint region of E. coli strain 1116 at the single cell level.
Experimental section
Cultivation of E. coli and in situ silver nanoparticle synthesis
Shock frozen E. coli DSM 1116 strains were purchased from DSM Nutritional Products GmbH (Grenzach, Germany). The cells were transferred into 250 mL shake flasks containing 25 mL M9 minimal medium without addition of trace metals.24,25 For 15N labeling NH4Cl was replaced with 15NH4Cl (3.42 mM; Sigma-Aldrich, Steinheim, Germany). The flasks were incubated overnight at 100 rpm and 37 °C with 0.4% (w/v) 12C- or 13C-glucose (Sigma-Aldrich, Steinheim, Germany) as the single carbon source. The technique of coating cells with silver colloid follows the work of Efrima and Zeiri, which consists of adding the different precursor substances (silver nitrate and reduction agents, e.g. sodium borohydride or hydroxylammonium chloride) successively to the bacterial sample.26–28 The so called in situ Ag nanoparticles were prepared according to a literature procedure.29 Briefly, 1 mL of washed bacteria was pelleted and resuspended in 180 μL of hydroxylammonium chloride solution (1.67 mM; Merck, Hohenbrunn, Germany) containing NaOH (3.3 mM; Carl Roth GmbH & Co. KG, Karlsruhe, Germany). After addition of 20 μL of silver nitrate solution (10 mM; Merck, Hohenbrunn, Germany) and incubation for 1 hour at 4 °C, 5 μL of the bacterial sample were transferred onto a glass slide (26 mm × 76 mm × 1 mm; Carl Roth GmbH & Co. KG, Karlsruhe, Germany) and dried for 1 hour at room temperature.Surface-enhanced Raman microspectroscopic analysis
A Lab-RAM HR Raman microscope (Horiba Scientific, Japan) was used for Raman analysis. The system was equipped with an integrated Olympus BXFM microscope, a He–Ne-Laser (633 nm, 4 mW at the sample) and a Peltier-cooled CCD detector. A 100× objective (Olympus MPlan N, NA = 0.9) was used to focus the laser beam onto the sample and to collect the scattered light. The light then passes a diffraction grating (600 lines per mm) and a confocal pinhole (100 μm). A laser power of 0.4 mW at the sample and an acquisition time of 1 s were applied for the SERS analysis of E. coli cells. The spectral range of the recorded Raman spectra was set to 50 cm−1–3750 cm−1. A silicon wafer with its first-order phonon band at 520.7 cm−1 was used for wavelength calibration.Results and discussion
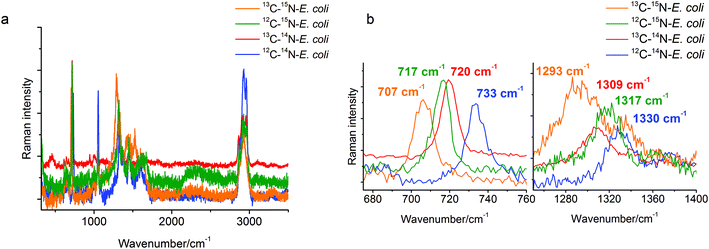
A stable isotope surface-enhanced Raman scattering approach was applied here to clarify the origin of the SERS marker band at around 730 cm−1. Four samples of E. coli, each grown on unlabeled (12C, 14N) and labeled (13C, 15N) carbon and nitrogen sources in different combinations, were cultivated and analyzed. The spectral changes for different labeled E. coli cells can be seen in Fig. 1a. Particularly the SERS band at 733 cm−1 for 12C–14N-E. coli shows a red-shift to 720 cm−1 for 13C–14N-E. coli, to 717 cm−1 for 12C–15N-E. coli and even to 707 cm−1 for 13C–15N-E. coli in the baseline-corrected SERS spectra (see Fig. 1b). Another relatively strong band at 1330 cm−1 of 12C–14N-E. coli is red-shifted to 1317 cm−1 for 12C–15N-E. coli, to 1309 cm−1 for 13C–14N-E. coli, and to 1293 cm−1 for 13C–15N-E. coli.The characteristic band at 733 cm−1 is sharp and shows a complete red-shift without any residual intensity at the original position for all of the different stable isotope labels. Also no peak broadening or peak splitting, which would indicate the presence of different substances or isotopologues, could be detected. For example, a broadening of this specific peak was reported for 50% 13C labeled E. coli cells in an earlier study.29 In that study it was also stated that the red-shift of these SERS bands is caused only by the incorporation of the stable isotope. This band can therefore be attributed to only one vibrational mode. In addition, due to the red-shift when using 15N labeling, only vibrational modes containing nitrogen are possible. Therefore, compounds without nitrogen cannot contribute to this band. Hence, C–H, C–C and O–P–O modes and many polysaccharides (glycosidic ring breathing) and carbohydrates can be excluded. The high red-shift of 16 cm−1 also requires a vibrational mode where nitrogen is directly involved. The nitrogen in N-acetyl-D-glucosamine is not located in the ring structure and therefore the influence of the 15N labeling and thus the red-shift of the glycosidic ring breathing mode would only be very small. Hence, the band cannot be assigned to the glycosidic ring breathing mode of NAG. Phospholipids and amino acids on the other hand would only show a C–N stretching vibration in this area. This mode should follow the simple rule that the exact value of the red-shift is dependent on the relative mass change of the involved atoms. The relative mass change when substituting 12C with 13C is slightly higher than substituting 14N with 15N. Therefore, the position of the 13C–14N band in the spectra should be at a lower wavenumber than the band of 12C–14N. But actually, the band of 13C–14N-E. coli can be found at 720 cm−1, whereas the band of 12C–15N-E. coli can be found at 717 cm−1. This leads to the conclusion that a simple C–N stretching vibration cannot be attributed to this obviously more complex spectral feature in the spectra. In the SERS spectra of adenine or adenine related substances like adenosine or ATP a very strong band in the area of 730–735 cm−1 and a second relatively strong band at around 1330 cm−1 can be found.7,8 The exact match of these modes to the bands in the SERS spectra of E. coli is remarkable. The band around 730 cm−1 is assigned to the in-plane pyrimidine and imidazole ring breathing modes.30,31 It is known that ring breathing modes are preferentially highly enhanced in the SERS analysis of organic compounds. Furthermore, the molecular structure possesses four nitrogen atoms situated in the ring structure and one nitrogen atom in the side chain, which perfectly explains the high red-shift from 733 cm−1 for 12C–14N-E. coli to 717 cm−1 for 12C–15N-E. coli in the SERS spectra of 15N labeled E. coli. Additionally, the band at around 1330 cm−1 shows a red-shift to 1293 cm−1 in 15N labeled E. coli cells, therefore it is safe to assume that nitrogen is also involved in this vibration mode. It can therefore be concluded that the band at around 730 cm−1 can be assigned to adenine and more precisely to the in-plane ring breathing mode of adenine. It should be noted that a range of adenine-containing compounds (FAD, NAD, etc.) as well as different products of the purine degradation pathway (e.g. hypoxanthine19) contain a purine moiety and may contribute to this band. Therefore, a further detailed analysis of unlabeled and stable isotope labeled reference compounds with a purine moiety is required for the unambiguous band assignment. This will be the subject of our subsequent study.
In order to confirm the observed band shifts and to demonstrate the reproducibility of the single cell signatures of the different labeled E. coli samples, the obtained spectra were subjected to a principal component analysis (PCA) using MATLAB R2013b. The spectra were denoised with the integrated denoising method of LabSpec 6.3 to improve the spectral quality for PCA. The first derivatives were generated to eliminate the contribution of background signals and were then utilized as the input for PCA.
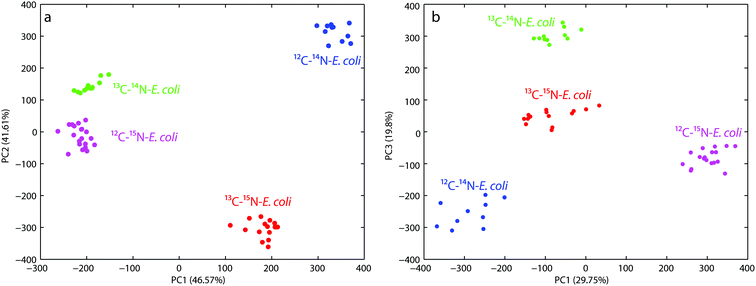
For the first PCA the wavenumber range from 600–800 cm−1 was used to evaluate the observed red-shift of the band at 733 cm−1. Fig. 2a shows that both the principal components PC1 and PC2 have a nearly similar total explained variance (TEV) with 46.57% and 41.61%, respectively. The first four principal components explained 94.3% of the total data variance. PC1 reflects the shift of the 12C–14N and 13C–15N samples from a combined 13C–14N and 12C–15N cluster. This clustering perfectly separates the non-(12C–14N), partly-(13C–14N, 12C–15N) and fully-labeled (13C–15N) samples, but fails to separate the partly labeled samples from each other. Whereas the clustering based on PC2 reflects directly the red-shift of the band of different samples and therefore succeeds in separating each sample from each other. Both the principal components in combination therefore show a proper clustering of the different labeled E. coli cells.
The second PCA plot (see Fig. 2b) targeted the fingerprint region from 600–1500 cm−1 to demonstrate the overall discrimination of the different labeled E. coli samples. The first four principal components explained 80% of the total data variance. The score plot for PC1 versus PC3 yields the best discrimination for all the different labeled E. coli samples. PC1 mostly reflects the changes in the spectra due to 15N incorporation and therefore separates the 12C–14N/15N labeled samples, but is not able to discriminate the samples labeled with 13C regardless of the nitrogen label. In contrast PC3 perfectly separates the 13C labeled and unlabeled samples, but only when no 15N is incorporated. The combination of PC1 and PC3 therefore shows proper clustering of the different labeled E. coli cells directly correlated with the incorporation of the different isotopes.
Conclusions
This study demonstrates the unique ability of the stable isotope labeling approach to understand certain spectral features of the SERS spectra of microorganisms. With the help of 15N labeling the pronounced SERS band at around 730 cm−1 could successfully be assigned to adenine-related compounds. Furthermore, the discrimination of E. coli bacteria with different isotopic compositions of 12C/13C/14N/15N based on single-cell SERS spectra could be shown for the first time. The results were confirmed by a principal component analysis of the fingerprint area of the SERS-spectra, which clearly proves that the SERS fingerprint spectra of stable isotope labeled bacteria can help in identifying and characterizing specific bacteria at the single cell level. To our knowledge these are the first reproducible SERS data of 15N and 15N–13C stable isotope labeled microorganisms. Although this study mainly focused on the band at around 730 cm−1 in the SERS spectra of E. coli, this technique should also be usable to study more complex spectral features in other microorganisms and thus can be used as a microbiological tool to ultimately understand metabolic pathways and track the carbon and nitrogen flows in microbial communities.Acknowledgements
The authors gratefully acknowledge the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG, project IV 110/2-1) and the Helmholtz Wasserzentrum München (HWZM) within the Helmholtz Water Network for financial support.References
- E. C. Le Ru and P. G. Etchegoin, Annu. Rev. Phys. Chem., 2012, 63, 65–87 CrossRef CAS PubMed.
- R. M. Jarvis and R. Goodacre, Chem. Soc. Rev., 2008, 37, 931–936 RSC.
- Y. Liu, Y.-R. Chen, X. Nou and K. Chao, Appl. Spectrosc., 2007, 61, 824–831 CrossRef CAS PubMed.
- W. R. Premasiri, D. T. Moir, M. S. Klempner, N. Krieger, G. Jones II and L. D. Ziegler, J. Phys. Chem. B, 2005, 109, 312–320 CrossRef CAS PubMed.
- R. M. Jarvis and R. Goodacre, Anal. Chem., 2004, 76, 40–47 CrossRef CAS PubMed.
- M. Kahraman, K. Keseroglu and M. Culha, Appl. Spectrosc., 2011, 65, 500–506 CrossRef CAS PubMed.
- L. Zeiri, B. V. Bronk, Y. Shabtai, J. Eichler and S. Efrima, Appl. Spectrosc., 2004, 58, 33–40 CrossRef CAS PubMed.
- A. Walter, A. Marz, W. Schumacher, P. Rosch and J. Popp, Lab Chip, 2011, 11, 1013–1021 RSC.
- W. R. Premasiri, Y. Gebregziabher and L. D. Ziegler, Appl. Spectrosc., 2011, 65, 493–499 CrossRef CAS PubMed.
- K. J. Rothschild, J. R. Andrew, W. J. De Grip and H. E. Stanley, Science, 1976, 191, 1176–1178 CAS.
- A. A. Guzelian, J. M. Sylvia, J. A. Janni, S. L. Clauson and K. M. Spencer, Proc. SPIE-Int. Soc. Opt. Eng., 2002, 4577, 182–192 CrossRef CAS.
- K. Maquelin, L.-P. Choo-Smith, T. Van Vreeswijk, H. P. Endtz, B. Smith, R. Bennett, H. A. Bruining and G. J. Puppels, Anal. Chem., 2000, 72, 12–19 CrossRef CAS PubMed.
- S. Farquharson, W. W. Smith, V. Y. Lee, S. Elliott and J. F. Sperry, Proc. SPIE-Int. Soc. Opt. Eng., 2002, 4575, 62–72 CrossRef CAS.
- M. Kahraman, M. M. Yazici, F. Sahin and M. Culha, Langmuir, 2008, 24, 894–901 CrossRef CAS PubMed.
- K. Kneipp, H. Kneipp, I. Itzkan, R. R. Dasari and M. S. Feld, J. Phys.: Condens. Matter, 2002, 14, R597–R624 CrossRef CAS.
- K. Kneipp, A. S. Haka, H. Kneipp, K. Badizadegan, N. Yoshizawa, C. Boone, K. E. Shafer-Peltier, J. T. Motz, R. R. Dasari and M. S. Feld, Appl. Spectrosc., 2002, 56, 150–154 CrossRef CAS.
- L. A. Gearheart, H. J. Ploehn and C. J. Murphy, J. Phys. Chem. B, 2001, 105, 12609–12615 CrossRef CAS.
- G. D. Chumanov and T. M. Cotton, Proc. SPIE-Int. Soc. Opt. Eng., 1999, 3608, 204–210 CrossRef CAS.
- W. Ranjith Premasiri, P. Lemler, Y. Chen, Y. Gebregziabher and L. D. Ziegler, in Frontiers of Surface-Enhanced Raman Scattering, John Wiley & Sons, Ltd, 2014, ch. 12, pp. 257–283, DOI:10.1002/9781118703601.
- L. Cui, P. Chen, S. Chen, Z. Yuan, C. Yu, B. Ren and K. Zhang, Anal. Chem., 2013, 85, 5436–5443 CrossRef CAS PubMed.
- D. L. Perry, Applications of Analytical Techniques to the Characterization of Materials, Springer, 1991 Search PubMed.
- H. Muhamadali, M. Chisanga, A. Subaihi and R. Goodacre, Anal. Chem., 2015, 87, 4578–4586 CrossRef CAS PubMed.
- Y. Wang, Y. Ji, E. S. Wharfe, R. S. Meadows, P. March, R. Goodacre, J. Xu and W. E. Huang, Anal. Chem., 2013, 85, 10697–10701 CrossRef CAS PubMed.
- S. Sabri, L. K. Nielsen and C. E. Vickers, Appl. Environ. Microbiol., 2013, 79, 478–487 CrossRef CAS PubMed.
- M. R. Green and J. Sambrook, Molecular Cloning : a Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y, 2012 Search PubMed.
- L. Zeiri, B. V. Bronk, Y. Shabtai, J. Czege and S. Efrima, Colloids Surf., A, 2002, 208, 357–362 CrossRef CAS.
- S. Efrima and L. Zeiri, J. Raman Spectrosc., 2009, 40, 277–288 CrossRef CAS.
- H. Zhou, D. Yang, N. P. Ivleva, N. E. Mircescu, R. Niessner and C. Haisch, Anal. Chem., 2014, 86, 1525–1533 CrossRef CAS PubMed.
- P. Kubryk, J. S. Kölschbach, S. Marozava, T. Lueders, R. U. Meckenstock, R. Niessner and N. P. Ivleva, Anal. Chem., 2015, 87, 6622–6630 CrossRef CAS PubMed.
- A. Toyama, N. Hanada, Y. Abe, H. Takeuchi and I. Harada, J. Raman Spectrosc., 1994, 25, 623–630 CrossRef CAS.
- B. Giese and D. McNaughton, J. Phys. Chem. B, 2002, 106, 101–112 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2016 |