Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of a glial cell-derived neurotrophic factor-releasing artificial dura for neural tissue engineering applications
N. K.
Mohtaram
a,
J.
Ko
a,
A.
Agbay
b,
D.
Rattray
c,
P. O.
Neill
c,
A.
Rajwani
d,
R.
Vasandani
a,
H. L.
Thu
e,
M. B. G.
Jun
a and
S. M.
Willerth
*abf
aDepartment of Mechanical Engineering, University of Victoria, P.O. Box 1700, STN CSC, Victoria, BC V8W 2Y2, Canada. E-mail: willerth@uvic.ca
bDivision of Medical Sciences, University of Victoria, P.O. Box 1700, STN CSC, Victoria, BC V8W 2Y2, Canada
cDepartment of Biochemistry and Microbiology, University of Victoria, P.O. Box 1700, STN CSC, Victoria, BC V8W 2Y2, Canada
dDepartment of Biomedical Engineering, University of Victoria, P.O. Box 1700, STN CSC, Victoria, BC V8W 2Y2, Canada
eDepartment of Biomedical Engineering, International University-Vietnam National University, Vietnam
fInternational Collaboration on Repair Discoveries (ICORD), Vancouver, BC V5Z 1M9, Canada
First published on 3rd August 2015
Abstract
Encapsulated electrospun nanofibers can serve as an artificial dura mater, the membrane that surrounds the brain and spinal cord, due to their desirable drug delivery properties. Such nanofiber scaffolds can be used to deliver drugs such as glial cell-derived neurotrophic factor (GDNF). GDNF promotes the survival of both dopaminergic and motor neurons, making it an important target for treatment of central nervous system injuries and disorders. This work focuses on designing a novel class of encapsulated poly(ε-caprolactone) (PCL) nanofiber scaffolds with different topographies (random and aligned) that generate controlled release of GDNF to potentially serve as a suitable substitute for the dura mater during neurosurgical procedures. Random and aligned scaffolds fabricated using solution electrospinning were characterized for their physical properties and their ability to release GDNF over one month. GDNF bioactivity was confirmed using a PC12 cell assay with the highest concentrations of released GDNF (∼341 ng mL−1 GDNF) inducing the highest levels of neurite extension (∼556 μm). To test the cytocompatibility of aligned GDNF encapsulated PCL nanofibers, we successfully seeded neural progenitors derived from human induced pluripotent stem cells (hiPSCs) onto the scaffolds where they survived and differentiated into neurons. Overall, this research demonstrates the potential of such substrates to act as artificial dura while delivering bioactive GDNF in a controlled fashion. These scaffolds also support the culture and differentiation of hiPSC-derived neural progenitors, suggesting their biocompatibility with the cells of the central nervous system.
Introduction
The dura mater consists of a thick membrane that covers the brain and spinal cord, where it serves as a barrier to ensure that the cerebrospinal fluid remains in the central nervous system (CNS).1,2 This membrane can be damaged by trauma to the brain or spinal cord.3,4 Autografts are commonly used to repair such damaged regions of the dura during surgery since they do not initiate immunologic reactions.5 However, autografts require an accessible site and an additional operation for obtaining a healthy region of the dura. A promising alternative to traditional autografts is implanting a suitable substrate to act as an artificial dura to preserve the spinal cord after surgery. Many synthetic and natural materials are employed in neurosurgery to repair dural defects.2,5–16 Advances in biomaterials have led to the development of new artificial duras with some of these materials being translated from preclinical models to clinical trials. For instance, Esposito et al. have developed substitutes out of collagen and no toxicity was observed after implanting such substrates in 208 patients.2 Yamada et al. have used bioadsorbable poly(glycolic acid) (PGA) nonwoven polymer composite sheets for similar studies with no reported risk of inflammation after 35.5 months in 53 patients.6 In addition to the aforementioned options, commercially available artificial duras made from PGA and L-lactide and ε-caprolactone (P(LA/CL)) copolymer films exist as well.17,18Although major advances have been made in using synthetic polymers to fabricate artificial duras,6,9,12,15,16 these scaffolds do not accurately capture all of the biological functions of the dura.19 For example, there is no biocompatible substitute that delivers proteins, such as neurotrophic factors, to further enhance functionality. Neurotrophic factors regulate and promote the differentiation, growth, and survival of neurons.20 Expressed in the CNS, glial derived neurotrophic factor (GDNF) promotes the survival of motor and dopaminergic neurons, making it a promising therapeutic for the treatment of neurological disorders.21–30 In particular, we selected GDNF over other neurotrophic factors as it promotes the survival of multiple types of neurons, which increases its potential relevance for clinical applications.31,32 Therefore, the encapsulation of GDNF within artificial duras could be used to treat different CNS disorders where this membrane has been disrupted.
As discussed, polymeric substrates can be used to repair defects of the dura as demonstrated in pre-clinical and clinical trials.7–9 Polymeric nanofibers offer a number of advantages over other substrates such as synthetic films, including the ability to control the diameter of nanofibers, altering scaffold porosity, and the ability to tailor mechanical properties by changing topographical properties. Nanofiber scaffolds can be fabricated using solution electrospinning where an applied electric field stretches out fibers from a polymer solution.33,34 Such substrates can also be encapsulated with growth factors to act as biologically active materials.29 Previous studies focused on the encapsulation of different drugs into electrospun nanofibers and characterizing the resulting release rates from the nanofibers.35–42 For example, Valmikinathan et al. showed the sustained release of bovine serum albumin (BSA) and nerve growth factor (NGF) from poly(ε-caprolactone) (PCL) nanofibers after 28 days.43 Leong and co-workers reported that aligned poly(ε-caprolactone)-co-(ethyl ethylene phosphate) (PCLEEP) nanofibers could release GDNF for up to two months in vitro.44 Mohtaram et al. showed that PCL nanofibers could generate controlled release of retinoic acid (RA), a small molecule that regulates neurogenesis.45
In related work, radially aligned PCL nanofibers could be used as substrates for culturing dural fibroblast cells as a promising approach to developing a novel artificial dura as they could promote cellular migration and population compared to randomly aligned nanofibers.9 A different study showed that aligned electrospun scaffolds could direct neurite outgrowth from embryonic stem cell derived neurons, which may be desirable when promoting regeneration of damaged neural tissue.46 More recently, Mohtaram et al. have shown that RA encapsulated aligned PCL nanofibers are able to direct the neuronal differentiation of mouse induced pluripotent stem cells.47 Although it has been shown that fibrous structures could play a key role in the development of artificial dura, electrospun scaffolds with a porous structure can be further functionalized with specific proteins or encapsulated with neurotrophic drugs to introduce a better class of artificial dura substrates.
This study focuses on designing a novel biocompatible GDNF releasing artificial dura with aligned topography. In this study, we fabricated and characterized encapsulated PCL–BSA and PCL–BSA–GDNF nanofiber scaffolds with varied topographies (random and aligned) that could serve as an artificial dura. To further enhance the clinical relevance of our work, these scaffolds were fabricated with human GDNF protein produced through good manufacturing practices provided by MedGenesis Therapeutix. A release study was performed to determine the kinetics of protein release from these scaffolds over a 30 day time period and the released GDNF was confirmed to be bioactive using a PC12 cell differentiation assay. Finally, neural progenitors derived from human induced pluripotent stem cells (hiPSCs) were seeded upon the aligned PCL–BSA–GDNF nanofibers to determine if such scaffolds could support stem cell culture and neuronal differentiation. These scaffolds supported cell adhesion and neuronal differentiation of hiPSC-derived neural progenitors. This study represents an important step in developing such biomaterial scaffolds for use as an artificial dura.
Results and discussion
Fabrication and characterization of encapsulated nanofibers
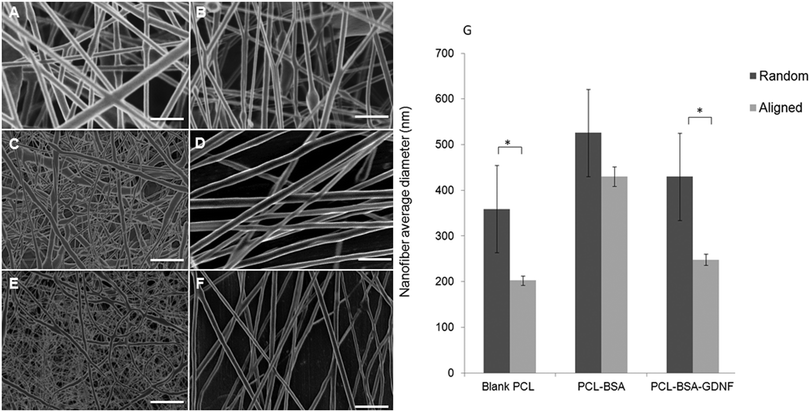
We fabricated blank PCL, PCL–BSA, and PCL–BSA–GDNF nanofibers scaffolds with random and aligned topographies used a previously published solution electrospinning set-up.47 The thickness of these scaffolds could vary between to 20 to 30 μm depending on the electrospinning process time and the type of topography. By tuning the solution electrospinning parameters such as collecting distance, applied voltage, and the drum speed, we could successfully fabricate nanofibers scaffolds with varied topographies with a very porous structure without any polymer beads or protein agglomeration. Parameters for fabricating nanofiber scaffolds with varied topographies are given in Table 1. We have previously shown that the degree of alignment of blank and RA-containing electrospun nanofibers can be improved significantly using a rotating drum.45 Random and aligned electrospun PCL–BSA nanofibers were fabricated at 15 kV and 12 kV applied voltage respectively and their structure has shown a very fine bead-free morphology at these aforementioned voltages. Scanning electron microscopy (SEM) images of both topographies for each type of scaffold are shown in Fig. 1. The morphology of random and aligned blank PCL nanofibers is shown in Fig. 1A and B respectively. For random blank PCL nanofibers, the average fiber diameter measured was 358.62 ± 95.42 nm (n = 100) and for aligned blank PCL nanofibers, the average diameter measured was 202.25 ± 145.87 nm (n = 100). Fig. 1C and D represent the morphology of random and aligned PCL–BSA nanofibers respectively. The average fiber diameter for random and aligned PCL–BSA nanofibers was 625.87 ± 240.48 nm (n = 100) and 429.57 ± 152.37 nm (n = 100). Fig. 1E and F show the topography of the random and aligned electrospun PCL–BSA–GDNF nanofibers fabricated at 5 and 2.5 mm collecting distance. For aligned scaffolds, the average fiber diameter measured was 247.78 ± 124.66 nm (n = 100) and for random scaffolds the average diameter was 525.42 ± 193.32 nm (n = 100).| Topography | Scaffold | Collecting distance (mm) | Voltage (kV) | Drum speed (rpm) |
|---|---|---|---|---|
| Random | PCL Blank | 5 | 15 | N/A |
| PCL–BSA | 5 | 15 | N/A | |
| PCL–BSA–GDNF | 5 | 15 | N/A | |
| Aligned | PCL Blank | 2.5 | 12 | 5000 |
| PCL–BSA | 2.5 | 12 | 5000 | |
| PCL–BSA–GDNF | 2.5 | 12 | 5000 |
Critical to the design of encapsulated electrospun nanofibers is the production of a bead free, smooth morphology. Our random and aligned blank PCL, PCL–BSA and PCL–BSA–GDNF nanofiber scaffolds had excellent bead-free structures. Due to a tensile force induced by the rotating drum, the aligned nanofibers tend to be stretched uniaxially along the flow direction which results in the aligned nanofibers being thinner when compared to the random nanofibers. Significant difference was observed in fiber diameter between random and aligned nanofibers for PCL–BSA–GDNF scaffolds. The same behaviour was seen for blank PCL nanofibers, however, no significant difference was observed between random and aligned PCL–BSA nanofibers. For aligned PCL–BSA–GDNF scaffolds, the average fiber diameter measured was 247.78 ± 124.66 nm (n = 100) and for random scaffolds the average diameter was 525.42 ± 193.32 nm (n = 100). For all nanofibers, the fiber diameter changed along each individual fiber so the fibers had non-uniform diameters. Controlling the uniform distribution of fiber size is very challenging. For instance, Yoshimoto et al. reported a broad fiber diameter distribution for PCL nanofibers (400 nm ± 200 nm).48 This variation in diameter may be due to the fast phase separation of PCL and the uncontrollable evaporation of volatile solvent during electrospinning. Controlling this variation will be even more difficult when proteins are encapsulated into nanofibers. For instance, it has been reported that encapsulation of NGF inside PCL nanofibers yielded nanofibers with a large fiber diameter distribution (548.89 ± 214.37 nm).43 Valmikinathan et al. have hypothesized that this variation in diameter happened due to the phase separation of PCL and NGF.43
The variation in fiber diameter can have a significant role in determining mechanical properties. These mechanical properties, including elastic modulus and strain at break, are highly dependent on the fiber diameter. For instance, Ko et al. have shown that the strain at break increased when the fiber diameter decreased.49 On the other hand, the elastic modulus increased with fiber diameter. In addition to fiber diameter, the porosity of electrospun scaffolds plays a key role in determining the mechanical properties. In the same study, our group showed that varying the porosity range from 75 to 92% led to significant changes in yield strength from 0.457 to 1.886 MPa.49 In addition to controlling the distribution of fiber size, the degree of alignment and fiber densities are interesting parameters to investigate particularly in cell-based studies since the physical three-dimensional environment has such a great effect on the growth of such cells. For instance, Park et al., have fabricated PCL nanofibers with controlled degree of alignment along fiber densities and where able to quantify the degree of alignment by using an elongation factor.50
The thickness of natural dura will depend on the sex and age of human being, and the natural dura consists of different parallel plates with varied thickness. Each layer ranges between 5–8 μm in thickness. Depending on type of the nanofibers produced (random/aligned and blank/encapsulated) the thickness of our scaffolds will vary between 20 to 30 μm. Additionally, we can spin different layers of nanofibers to make artificial duras with varied thicknesses. The approximate thickness of the naturally occurring dura is around 270 μm.51,52
Encapsulation efficiency and release study
We were able to encapsulate BSA and GDNF inside bead-free PCL–BSA and PCL–BSA–GDNF nanofibers with different scaffold topographies with varying efficiencies (Table 2). The encapsulation efficiency of BSA inside random nanofibers (54.3 ± 3.3%) was significantly higher than aligned PCL–BSA scaffolds (42.2 ± 4.5%). For random PCL–BSA–GDNF nanofibers the measured encapsulation efficiency of GDNF was 31.6 ± 2.4% and for aligned PCL–BSA–GDNF nanofibers the encapsulation efficiency was 27.5 ± 3.7%. No significant difference was observed in encapsulation efficiencies between random and aligned nanofibers. Chew et al. have shown that the encapsulation efficiency of NGF inside their PCLEEP nanofibers was very low which might be due to the immiscibility of NGF in the protein aqueous phase with PCLEEP solution phase.44| Topography | Scaffold | Encapsulation efficiency % |
|---|---|---|
| Random | PCL–BSA | 54.3 ± 3.3 |
| PCL–BSA–GDNF | 31.6 ± 2.4 | |
| Aligned | PCL–BSA | 42.2 ± 4.5 |
| PCL–BSA–GDNF | 27.5 ± 3.7 |
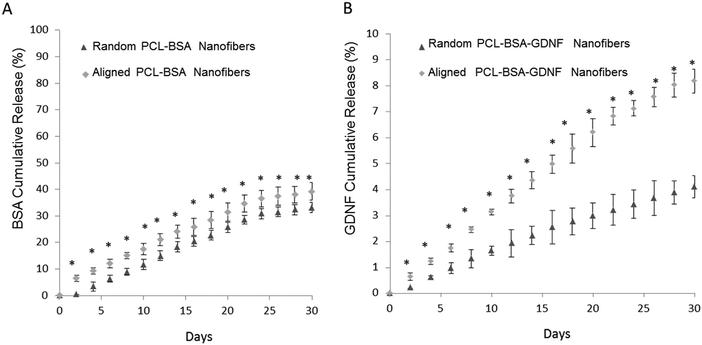
The percentage of BSA released over time is shown in Fig. 2A for both random and aligned PCL–BSA nanofiber scaffolds. At the end of a 30 day release study, random PCL–BSA nanofibers released around 30% of the total loaded BSA while aligned PCL–BSA nanofibers released around 40% of the total loaded BSA. Fig. 2B represents the release data for random and aligned nanofibers. Controlled release of GDNF from random and aligned PCL–BSA–GDNF nanofibers was characterized over 30 days as well (Fig. 2B). Within 2 days, 0.64 ± 0.14% of GDNF was released from the aligned PCL–BSA–GDNF nanofibers. This amount was greater than the amount released by random nanofibers (0.22 ± 0.01%). By the end of the study, random PCL–BSA–GDNF scaffolds released 4.1 ± 0.4% of its encapsulated GDNF and aligned PCL–BSA–GDNF scaffolds had a cumulative release of 8.17 ± 0.4% of its encapsulated GDNF. GDNF was released at an average rate around 0.28% and 0.15% every other day for the aligned and randomly oriented PCL–RA scaffolds respectively. There was a significant difference between the release data at each day for the random and aligned scaffold nanofibers (p < 0.05).
Encapsulated electrospun nanofibers not only provide a membrane structure with controllable porosity, but can also improve the design of current artificial dura through controlled release of neurotrophic drugs. Biocompatible polymers can be used to fabricate drug delivery systems for providing controlled release of neurotrophic factors.29 Such nanofibers with high levels of encapsulation efficiencies have shown to provide the controlled release of small molecules and also various proteins while allowing tailored topographies.29,39,43,45,53
We have previously encapsulated the small molecule RA inside PCL nanofibers using solution electrospinning to fabricate random and aligned encapsulated nanofibers.45 Our previous data showed that 8.6%, and 18% of RA was released from aligned and random PCL nanofibers respectively over a month. In this paper, we could successfully encapsulate BSA and GDNF inside random and aligned electrospun nanofibers. Our data showed controlled release of BSA and GDNF over a month. Our PCL–BSA nanofiber scaffolds were able to provide around 40% and 30% release of BSA from aligned and random nanofibers respectively. Similarly, Valmikinathan et al. have reported almost the same amount of BSA release (∼50%) from their nanofibers.43 However, they have not studied the effect of topography on BSA release from such scaffolds. Our work has shown that the random PCL–BSA nanofibers released a maximum BSA amount of 0.5 mg on day 30 of release studies while aligned PCL–BSA nanofibers released a maximum of 0.6 mg BSA on day 30. Madduri et al. used electrospun nanofibers made from a natural biomaterial, silk fibroin, to encapsulate GDNF with varied topographies.37 To our knowledge, we are the first group to develop synthetic encapsulated PCL–BSA–GDNF nanofibers with varied topographies. In order to produce more uniform and higher encapsulated nanofibers, while preserving the bioactivity of GDNF, we co-encapsulated the growth factor with BSA inside the nanofibers. BSA has shown to act as a carrier protein to preserve bioactivity during the solution electrospinning process.43
Overall, the PCL–BSA aligned nanofibers showed 118% more release than the randomly oriented fibers and this amount was around ∼200% when comparing PCL–BSA–GDNF nanofibers to PCL–BSA–GDNF random nanofibers. This indicates that these scaffolds generate quicker release while having aligned topography for tissue engineering applications. One possible explanation for this observation is that the aligned nanofibers were thinner, creating a larger surface area for release compared to random nanofibers in our previous work, our group showed that the release of RA from random fibers was more rapid compared to aligned nanofibers. In this case, the randomly nanofibers were thinner than the aligned nanofibers, suggesting that nanofiber diameter plays an important role in controlling release.47 Additionally, the rate of protein delivery from aligned nanofibers was similar to the release rate of GDNF from PCL microspheres.45 In the future, using coaxial electrospinning may be an alternative strategy to increasing the encapsulation efficiency of protein loading into nanofiber scaffolds.54
Kinetics of neurite outgrowth of PC12 cells in response to released GDNF
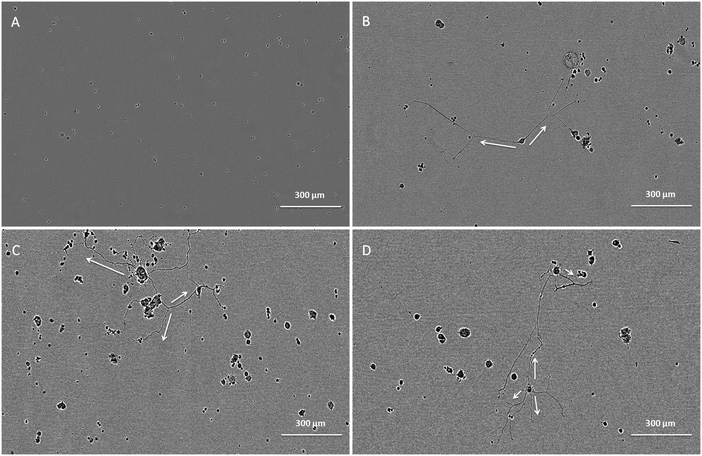
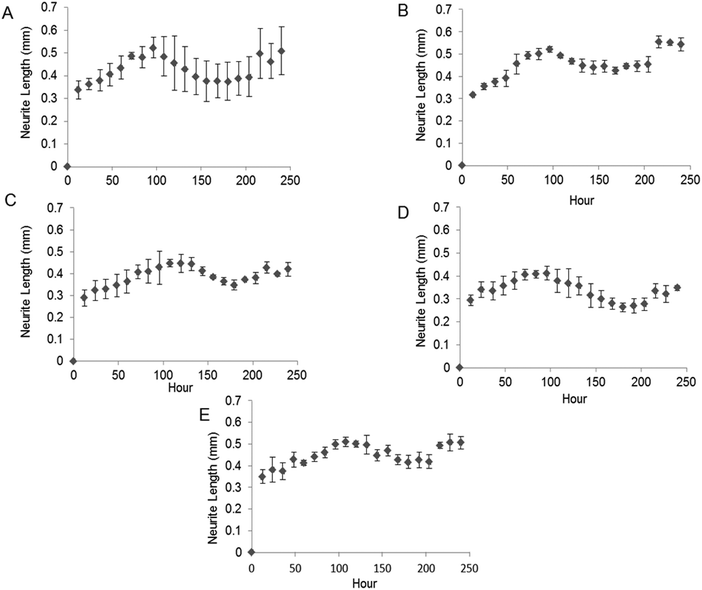
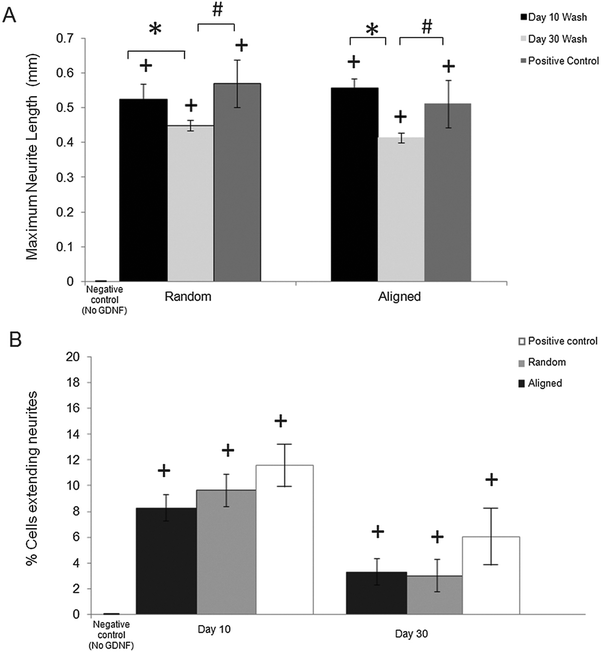
The bioactivity of released GDNF from random and aligned PCL–BSA–GDNF nanofibers was evaluated using PC12 cells, which differentiate and extend neurites in response to bioactive GDNF. Cells were exposed to GDNF washes from day 10 for random and aligned PCL–BSA–GDNF nanofibers, corresponding to 172 ng mL−1 (∼1.5%) and 341 ng mL−1 (∼3%) and also day 30 for random and aligned PCL–BSA–GDNF nanofibers, corresponding to 23 ng mL−1 (∼4%) and 14 ng mL−1 (∼8%). Fig. 3 shows representative images of neurite extensions observed at 10× magnification of cells exposed to the released GDNF from both random and aligned nanofibers. The GDNF released from random and aligned PCL–BSA–GDNF nanofibers at day 30 provided enhanced neurite extension from PC12 cells (Fig. 3C and D) compared to the negative control where no GDNF was present (Fig. 3A).Fig. 4 shows the kinetics of neurite differentiation from PC12 cells in the presence of GDNF for different dosages and topographies. The GDNF released for both day 10 (∼341 ng mL−1 GDNF) and day 30 (∼23 ng mL−1 GDNF) washes from the random and aligned PC–BSA–GDNF nanofibers initiated the neurite outgrowth of PC12 cells with an approximate length of 300 μm after the first 12 hours of seeding. Fig. 4A shows that the maximum neurite length (∼523 μm) for random PCL–BSA–GDNF nanofibers was observed after 96 hours for day 10 of wash (∼172 ng mL−1). Fig. 4B shows that the maximum neurite length (∼556 μm) was observed after 216 hours for day 10 of wash (∼341 ng mL−1 GDNF) for the aligned PCL–BSA–GDNF nanofibers. Overall, the released GDNF from aligned PCL–BSA–GDNF nanofibers induced the maximum neurite outgrowth from PC12, where this highest neurite outgrowth was induced from the highest concentration of GDNF (∼341 ng mL−1 GDNF) subsequently. Due to the lower concentration of GDNF washes at 30 (∼23 ng mL−1 for random and ∼14 ng mL−1 for aligned nanofibers), PC12 cells showed a shorter neurites compared to day 30 of wash for both set of nanofibers. Our data confirms that after 30 days, the released GDNF is still bioactive. Another study demonstrated even longer release of GDNF (∼56 days).55 The doses of GDNF being delivered are within the therapeutic range and can be safely tolerated by neural tissue.57
Neurite length analysis
We quantitatively assessed the bioactivity of released GDNF using PC12 cells. After 10 days of culture following exposure to day 10 and 30 washes, we measured the maximum neurite length and also the percentage of cells extending neurites for both random and aligned nanofibers. Fig. 5A shows the maximum neurite length in total cells for washes on days 10 and 30 for both random and aligned PCL–GDNF nanofibers. There was a significant difference between washes on day 10 and day 30 for random GDNF nanofibers. There was no significant difference between the percentage of cells extending neurites for random and aligned PCL–BSA–GDNF nanofibers for day 10 and 30 washes.Despite the advantages of using solution electrospinning to fabricate nanoscale drug delivery systems, one of the main biological concerns is using toxic solvents such as hexafluoroisopropanol (HFIP). In order to prove that the encapsulated GDNF has not been denatured in the presence of HFIP, we evaluated the bioactivity of released GDNF random and aligned electrospun nanofibers by quantitative and qualitative assays. We studied the neurite kinetics over 10 days for different washes of GDNF when exposed to PC12 cells to further understand the effect of dosage and time on neurite outgrowth length from such cells. GDNF washes (day 10 and 30) from the both random and aligned PCL–BSA–GDNF nanofibers were observed to initiate PC12 neurite outgrowth after 12 hours of seeding. Day 10 of GDNF wash (∼341 ng mL−1) samples from random nanofibers yielded longer neurite extensions compared to day 30 of GDNF wash (∼23 ng mL−1), and this amount was not significantly different compared to the positive controls, indicating the potential of such nanofibers to serve as a possible device for GDNF delivery. The same behaviour has been demonstrated for the aligned nanofibers as well showing the effect of concentration on the neurite length (Fig. 5A). For all cases, the experimental groups and the positive controls where significantly different from when no GDNF was present. Moreover, these GDNF concentrations are therapeutically effective in enhancing axonal regeneration.55,56 The percentage of cells extending neurites compared to no GDNF (negative control) was not statistically different between random and aligned nanofibers for day 10 and day 30 of washes (Fig. 5B). Although the released GDNF has been shown to be bioactive, we believe that there is still a possibility that a fraction of the GDNF bioactivity may have been be lost due to the exposure to a strong organic solvent. Day 30 wash (∼23 ng mL−1) yielded neurite extension comparable to 25 ng mL−1 of soluble GDNF, however, the higher concentration (∼173 ng mL−1) wash did not produce a greater neurite extension as expected. Instead, the resulting extension was also comparable to the positive control suggesting some degree of loss in bioactivity. Therefore, possible alternative strategies to provide better bioactivity should be investigated.
Evaluating the compatibility of aligned GDNF nanofibers with hiPSCs culture and differentiation
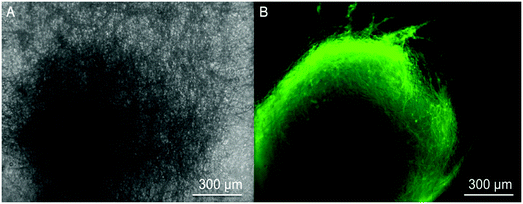
These encapsulated nanofiber scaffolds are designed to serve as a novel class of artificial duras, which will interact with neural cells in an in vivo setting. In order to investigate the compatibility of such scaffolds with neural cells, we cultured neural progenitors derived from hiPSCs onto scaffolds and aimed to differentiate them into neurons. HiPSCs-derived neural progenitors attached to these nanofibers and neuronal differentiation was observed after 12 days of culture. Fig. 6 shows the differentiation of neural progenitors into neurons when seeded upon such scaffolds. Fig. 6A shows a bright field image of an individual neural progenitor. Fig. 6B shows the neuronal differentiation of such cells. These images verify that such scaffolds could support the neuronal differentiation of hiPSCs-derived neural progenitors. Our work shows that GDNF encapsulated nanofibers supported the attachment and differentiation of hiPSCs into neurons.In order to preserve the bioactivity of GDNF, we co-encapsulated GDNF with BSA which is an animal-derived protein. Future studies should use human-derived proteins as a carrier alongside GDNF to replace BSA for future studies. Another approach could be to use a core-shell electrospinning technique in which human GDNF can be safely dissolved in water-soluble polymers without the use of toxic solvents where there may be a potential risk of protein denaturation.
Overall, this work demonstrates the promising potential of GDNF aligned electrospun nanofiber scaffolds fabricated via solution electrospinning for neural drug delivery and tissue engineering applications. Such scaffolds were successfully presented the controlled release of bioactive GDNF over 30 days. In the future, the effects of aligned scaffolds and controlled delivery of GDNF from such scaffolds on enhancing neural differentiation of pluripotent stem cells will be studied.
Experimental
Materials
Poly(ε-caprolactone) (PCL, Mn ∼ 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) (PCL), bovine serum albumin (BSA) (Mw 66 kDa, lyophilized powder) was purchased from Sigma-Aldrich (St. Louis, MO, USA). 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP), 99+% was purchased from Alfa Aesar (Ward Hill, MA, USA). Phosphate-buffered saline (PBS) was purchased from Life Technologies (Burlington, ON, Canada) (pH 7.4 from Gibco®). Dichloromethane (DCM) (reagent/ACS grade) was purchased from VWR International (Edmonton, AB, Canada). Polyclonal goat IgG human glial derived neurotrophic factor (GDNF) biotinylated antibody, monoclonal rat IgG anti-GDNF antibody, and wash buffer were purchased from R&D Systems (Minneapolis, MN, USA). ImmunoPure® streptavidin horseradish peroxidase conjugate and fetal bovine serum (FBS) were purchased from Thermo Scientific (Rockford, IL, USA). Good manufacturing practice (GMP) liatermin (r-metHuGDNF), referred to as GDNF in this paper, was provided by MedGenesis Therapeutix Inc. (Victoria, BC, Canada). Milk protein solution (0.1% Milk, 0.01% Tween 20 in 1× PBS) was purchased from Mandel Scientific (Guelph, ON, Canada). PC12 cells (ATCC) (Rockville, MD, USA) were provided by the Burke lab (Department of Biochemistry and Microbiology, University of Victoria). Dulbecco's modified eagle medium (DMEM; 1×, 4.5 g L−1D-glucose, L-glutamine) was purchased from Invitrogen. Laminin derived from the basement membrane of Engelbreth–Holm–Swarm murine sarcoma cells and 0.01% poly-L-ornithine (PLO) solution was purchased from Sigma-Aldrich (St. Louis, MO, USA). Human induced pluripotent stem cells (hiPSCs) were purchased from WiCell (the 1-MCB-01 line from WiCell, Madison, Wisconsin, USA). STEMdiff™ Neural Induction Medium was purchased from STEMCELL Technologies (NIM, Vancouver, BC, Canada).
000) (PCL), bovine serum albumin (BSA) (Mw 66 kDa, lyophilized powder) was purchased from Sigma-Aldrich (St. Louis, MO, USA). 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP), 99+% was purchased from Alfa Aesar (Ward Hill, MA, USA). Phosphate-buffered saline (PBS) was purchased from Life Technologies (Burlington, ON, Canada) (pH 7.4 from Gibco®). Dichloromethane (DCM) (reagent/ACS grade) was purchased from VWR International (Edmonton, AB, Canada). Polyclonal goat IgG human glial derived neurotrophic factor (GDNF) biotinylated antibody, monoclonal rat IgG anti-GDNF antibody, and wash buffer were purchased from R&D Systems (Minneapolis, MN, USA). ImmunoPure® streptavidin horseradish peroxidase conjugate and fetal bovine serum (FBS) were purchased from Thermo Scientific (Rockford, IL, USA). Good manufacturing practice (GMP) liatermin (r-metHuGDNF), referred to as GDNF in this paper, was provided by MedGenesis Therapeutix Inc. (Victoria, BC, Canada). Milk protein solution (0.1% Milk, 0.01% Tween 20 in 1× PBS) was purchased from Mandel Scientific (Guelph, ON, Canada). PC12 cells (ATCC) (Rockville, MD, USA) were provided by the Burke lab (Department of Biochemistry and Microbiology, University of Victoria). Dulbecco's modified eagle medium (DMEM; 1×, 4.5 g L−1D-glucose, L-glutamine) was purchased from Invitrogen. Laminin derived from the basement membrane of Engelbreth–Holm–Swarm murine sarcoma cells and 0.01% poly-L-ornithine (PLO) solution was purchased from Sigma-Aldrich (St. Louis, MO, USA). Human induced pluripotent stem cells (hiPSCs) were purchased from WiCell (the 1-MCB-01 line from WiCell, Madison, Wisconsin, USA). STEMdiff™ Neural Induction Medium was purchased from STEMCELL Technologies (NIM, Vancouver, BC, Canada).
Fabrication of encapsulated nanofibers
400 mg of PCL was added to 5 mL of HFIP to make 8% (w/v) solution which was mixed overnight. Then 50 mg of BSA was added for day 2 of mixing to make the final PCL–BSA (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) solution. 107 μL of the GDNF stock solution (9.3 mg mL−1) was added to make a homogenous solution resulting in a theoretical loading level of 0.22 wt/wt% GDNF in the polymer solution. The solution electrospinning setup used for the fabrication of encapsulated nanofibers was previously reported.45 The PCL, PCL–BSA, and PCL–BSA–GDNF solutions were pumped at the constant flow rate of 2 mL h−1 into electrospinning syringe in order to fabricate random and aligned scaffolds respectively. Random nanofibers were collected on top of an aluminum foil and aligned fibers were spun on a rotating drum as previously described.45 Each scaffold (∼10 mg total weight) was spun for 5 minutes and dried overnight.
20) solution. 107 μL of the GDNF stock solution (9.3 mg mL−1) was added to make a homogenous solution resulting in a theoretical loading level of 0.22 wt/wt% GDNF in the polymer solution. The solution electrospinning setup used for the fabrication of encapsulated nanofibers was previously reported.45 The PCL, PCL–BSA, and PCL–BSA–GDNF solutions were pumped at the constant flow rate of 2 mL h−1 into electrospinning syringe in order to fabricate random and aligned scaffolds respectively. Random nanofibers were collected on top of an aluminum foil and aligned fibers were spun on a rotating drum as previously described.45 Each scaffold (∼10 mg total weight) was spun for 5 minutes and dried overnight.
Morphological characterization
The nanofibers were characterized using scanning electron microscopy (SEM) (Hitachi S-4800 SEM). A standard SEM procedure was followed for obtaining high magnification images (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×), including carbon coating the samples prior to imaging as previously reported.45 The average fiber diameter was determined using Quartz-PCI Image Management Systems® software tools with x fibers being measured per sample.
000×), including carbon coating the samples prior to imaging as previously reported.45 The average fiber diameter was determined using Quartz-PCI Image Management Systems® software tools with x fibers being measured per sample.
In vitro release studies
Release studies were conducted over 30 days to measure the release rate of BSA from the nanofibers. Initially, nanofibers were immersed in 1 mL of PBS and stored at 37 °C. Every 24 hours the nanofibers were washed by removing the PBS via a needle and syringe. The collected washes were then frozen (−20 °C) for future analysis and 1 mL of fresh PBS was added to each scaffold. The concentration of protein in the extracted solution was measured using the Bio-Rad®Protein Assay. The BSA standard curve covered concentrations ranging from 8 μg mL−1 to 80 μg mL−1. This assay measures the total amount of protein in the sample. The absorbance was measured using spectroscopy (Infinite M200 Pro Softmax Pro version 5.2) at 595 nm. To determine the concentration of protein present, the absorbance of the samples was compared to a standard curve generated from known concentrations of BSA.
Bioactivity assay
In order to determine the bioactivity of released GDNF, undifferentiated PC12 cells were exposed to GDNF samples retrieved from release points at day 0, 10 and 30 as previously described.45 Undifferentiated PC12 cells were grown in suspension using DMEM supplemented with 15% FBS at 37 °C and 5% CO2. Cells were passaged with a 22-gauge needle to separate cell clumps.45 24-well tissue culture plates (Grenier) were first coated with PLO and then with laminin.45 The plates were seeded with PC12 cells at a concentration of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–60
000–60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and exposed to the released GDNF at concentrations of 0.01 ng mL−1 and 1.0 × 10−5 μg mL−1 corresponding to day 10 and day 30 of the release study. The plates were incubated inside of an IncuCyte® ZOOM Essen BioScience® microscope (Ann Arbor, Michigan, USA) and scanned at 12 hour intervals for neurite outgrowth tracking.
000 cells per mL and exposed to the released GDNF at concentrations of 0.01 ng mL−1 and 1.0 × 10−5 μg mL−1 corresponding to day 10 and day 30 of the release study. The plates were incubated inside of an IncuCyte® ZOOM Essen BioScience® microscope (Ann Arbor, Michigan, USA) and scanned at 12 hour intervals for neurite outgrowth tracking.
Quantitative analysis of neurite outgrowth
An IncuCyte® ZOOM Essen BioScience® microscope was used to analyze bright field images of seeded PC12 exposed to GDNF washes with a 10× objective. 24-well plates were placed inside the Incucyte and each well was scanned every 12 hours for a period of 10 days to study the kinetics of neurite growth. At the end of scanning schedule, images were analyzed using the IncuCyte® ZOOM NeuroTrack SoftwareTM. Neurite length was calculated for each day of the 10 day time course and the maximum neurite length after 10 days was calculated.58Stem cell culture and neuronal differentiation
HiPSCs were cultured on Vitronectin XF™ coated surfaces in the presence of TeSR™-E8™ media in 6 well plates to maintain pluripotency as previously described.58–61 Undifferentiated hiPSCs were subsequently removed and cultured into a single well of an Aggrewell™ 800 plate in the presence of 2 mL of STEMdiff™ NIM.58 During this process, the media was changed with 1.5 mL of fresh NIM every day. The resulting aggregate consists entirely of neural progenitors (after 5 days). Aligned scaffolds spun on coverslips were sterilized in using a Trans-UV 302 cleaner; GelDoc XR Bio-Rad®. Under aseptic conditions, the scaffolds were adhered to the bottom of each well in a 6-well polystyrene tissue culture plate. Individual neural aggregates were removed from suspension and seeded onto scaffolds. HiPSCs-derived neural progenitors were cultured on aligned PCL–BSA–GDNF nanofiber scaffolds for 12 days before image analysis. Neuronal differentiation of seeded cells was qualitatively assessed after 12 days by immunocytochemistry targeting the neuron-specific protein Tuj1 as previously published.47,58 Images were captured for green fluorescence and fluorescent images were acquired on a LEICA 3000B inverted microscope using an X-cite series 120Q fluorescent light source (Lumen Dynamics) coupled to a Retiga 2000R fast cooled mono 12-bit camera (Q-imaging).Statistical analysis
Data are presented as mean values ± standard deviation of the mean. Statistical analysis was performed using STATISTICA 9 by applying a standard t-test to compare differences between experimental groups. Significance was considered at the p < 0.05 level.Conclusions
In this study, the ability of random and aligned PCL electrospun nanofibers scaffold to release bioactive GDNF was investigated. We were able to fabricate bead-free nanofibers containing clinical grade human GDNF while controlling scaffold topography. We achieved reasonable encapsulation efficiency of both BSA and GDNF into our nanofiber scaffolds (∼30–50%). Controlled GDNF release from random and aligned PCL–BSA–GDNF nanofibers was observed over 30 days. PC12 cell neurite outgrowth confirmed the bioactivity of released GDNF from such nanofibers. The released GDNF from nanofibers initiated the neurite outgrowth as indicated by tracking extension over time. Finally, aligned scaffolds were able to support the adhesion and neuronal differentiation of hiPSCs-derived neural progenitors, making them a useful tool for tissue engineering applications. Such drug releasing scaffolds could serve as a novel approach for engineering artificial dura mater.Acknowledgements
The authors would like to acknowledge the funding support from the Natural Sciences and Engineering Research Council's Discovery Grant program, Canada Research Chair program and the Rick Hansen Foundation. The authors would also like to acknowledge the Advanced Microscopy Facility at the University of Victoria and Mr Nathan Bodie one of our lab volunteers who helped us with neurite outgrowth data analysis. The corresponding author acknowledges a previously held an Engage Grant with MedGenesis Therapeutix, who supported this project through donation of GDNF.References
- B. Dubrovsky, Fundamental neuroscience and the classification of psychiatric disorders, Neurosci. Biobehav. Rev., 1995, 19(3), 511–518 CrossRef CAS.
- F. Esposito, P. Cappabianca, M. Fusco, L. M. Cavallo, G. G. Bani, F. Biroli, A. Sparano, O. de Divitiis and A. Signorelli, Collagen-only biomatrix as a novel dural substitute Examination of the efficacy, safety and outcome: Clinical experience on a series of 208 patients, Clin. Neurol. Neurosurg., 2008, 110(4), 343–351 CrossRef PubMed.
- P. Liu, X. Zeng, F. Liu, J. Zhao, M. Liu and W. Fan, Surgical management of dural injuries and postoperative cerebrospinal fluid fistulas in spinal surgeries, Zhongguo Xiufu Chongjian Waike Zazhi, 2008, 22(6), 715–718 Search PubMed.
- M. H. Khan, J. Rihn, G. Steele, R. Davis, W. F. Donaldson, J. D. Kang 3rd and J. Y. Lee, Postoperative management protocol for incidental dural tears during degenerative lumbar spine surgery: a review of 3,183 consecutive degenerative lumbar cases, Spine, 2006, 31(22), 2609–2613 CrossRef PubMed.
- T. D. McCall, D. W. Fults and R. H. Schmidt, Use of resorbable collagen dural substitutes in the presence of cranial and spinal infections – report of 3 cases, Surg. Neurol., 2008, 70(1), 92–97 CrossRef PubMed.
- K. Yamada, S. Miyamoto, M. Takayama, I. Nagata, N. Hashimoto, Y. Ikada and H. Kikuchi, Clinical application of a new bioabsorbable artificial dura mater, J. Neurosurg., 2002, 96(4), 731–735 CrossRef PubMed.
- O. J. Lee, H. W. Ju, J. H. Kim, J. M. Lee, C. S. Ki, J. H. Kim, B. M. Moon, H. J. Park, F. A. Sheikh and C. H. Park, Development of Artificial Dermis Using 3D Electrospun Silk Fibroin Nanofiber Matrix, J. Biomed. Nanotechnol, 2014, 10(7), 1294–1303 CrossRef CAS PubMed.
- X. F. Wang, B. Ding and B. Y. Li, Biomimetic electrospun nanofibrous structures for tissue engineering, Mater. Today, 2013, 16(6), 229–241 CrossRef CAS PubMed.
- J. W. Xie, M. R. MacEwan, W. Z. Ray, W. Y. Liu, D. Y. Siewe and Y. N. Xia, Radially Aligned, Electrospun Nanofibers as Dural Substitutes for Wound Closure and Tissue Regeneration Applications, ACS Nano, 2010, 4(9), 5027–5036 CrossRef CAS PubMed.
- R. Gazzeri, M. Neroni, A. Alfieri, M. Galarza, A. Faiola, S. Esposito and M. Giordano, Transparent equine collagen biomatrix as dural repair. A prospective clinical study, Acta Neurochir., 2009, 151(5), 537–543 CrossRef PubMed.
- H. E. Bernd, C. Kunze, T. Freier, K. Sternberg, S. Kramer, D. Behrend, F. Prall, M. Donat and B. Kramp, Poly(3-hydroxybutyrate) (PHB) patches for covering anterior skull base defects – an animal study with minipigs, Acta Oto-Laryngol., 2009, 129(9), 1010–1017 CrossRef PubMed.
- Y. Shimada, M. Hongo, N. Miyakoshi, T. Sugawara, Y. Kasukawa, S. Ando, Y. Ishikawa and E. Itoi, Dural substitute with polyglycolic acid mesh and fibrin glue for dural repair: technical note and preliminary results, J. Orthop. Sci., 2006, 11(5), 454–458 CrossRef CAS PubMed.
- V. A. Zerris, K. S. James, J. B. Roberts, E. Bell and C. B. Heilman, Repair of the dura mater with processed collagen devices, J. Biomed. Mater. Res., Part B, 2007, 83(2), 580–588 CrossRef PubMed.
- N. Ohbayashi, T. Inagawa, Y. Katoh, K. Kumano, R. Nagasako and H. Hada, Complication of Silastic Dural Substitute 20 Years after Dural Plasty, Surg. Neurol., 1994, 41(4), 338–341 CrossRef CAS.
- T. Mukai, N. Shirahama, B. Tominaga, K. Ohno, Y. Koyama and K. Takakuda, Development of watertight and bioabsorbable synthetic dural substitutes, Artif. Organs, 2008, 32(6), 473–483 CrossRef PubMed.
- K. Yamada, S. Miyamoto, I. Nagata, H. Kikuchi, Y. Ikada, H. Iwata and K. Yamamoto, Development of a dural substitute from synthetic bioabsorbable polymers, J. Neurosurg., 1997, 86(6), 1012–1017 CrossRef CAS PubMed.
- http://www.gunze.co.jp/e/medical/product/pdf/seamdura_catalog.pdf, SEAMDURA online, accessed March 2015.
- http://www.goremedical.com/resources/dam/assets/AE0346EN4.PDX.TRIFOLD.BROCHURE.FNL.mr.pages.pdf, GORE online, accessed March 2015.
- J. A. Spector, J. A. Greenwald, S. M. Warren, P. J. Bouletreau, R. C. Detch, P. J. Fagenholz, F. E. Crisera and M. T. Longaker, Dura mater biology: autocrine and paracrine effects of fibroblast growth factor 2, Plast. Reconstr. Surg., 2002, 109(2), 645–654 CrossRef.
- M. Barbacid, Neurotrophic Factors and Their Receptors, Curr. Opin. Cell Biol., 1995, 7(2), 148–155 CrossRef CAS.
- C. Apostolides, E. Sanford, M. Hong and I. Mendez, Glial cell line-derived neurotrophic factor improves intrastriatal graft survival of stored dopaminergic cells, Neuroscience, 1998, 83(2), 363–372 CrossRef CAS.
- B. J. Hoffer, A. Hoffman, K. Bowenkamp, P. Huettl, J. Hudson, D. Martin, L.-F. H. Lin and A. Gerhardt, Glial cell line-derived neurotrophic factor reverses toxin-induced injury to midbrain dopaminergic neurons in vivo, Neurosci. Lett., 1994, 182, 107–111 CrossRef CAS.
- A. Buj-Bello, V. L. Buchman, A. Horton, A. Rosenthal and A. M. Davies, GDNF is an age-specific survival factor for sensory and autonomic neurons, Neuron, 1995, 15(4), 821–828 CrossRef CAS.
- M. Chermenina, P. Schouten, N. Nevalainen, F. Johansson, G. Orädd and I. Strömberg, GDNF is important for striatal organization and maintenance of dopamine neurons grown in the presence of the striatum, Neuroscience, 2014, 270, 1–11 CrossRef CAS PubMed.
- A. Heller, S. Price and L. Won, Glial-derived neurotrophic factor (GDNF) induced morphological differentiation of an immortalized monoclonal hybrid dopaminergic cell line of mesencephalic neuronal origin, Brain Res., 1996, 725(1), 132–136 CrossRef CAS.
- C. Henderson, H. Phillips, R. Pollock, A. Davies, C. Lemeulle, M. Armanini, L. Simmons, B. Moffet, R. Vandlen and L. C. Simpson, et al., GDNF: a potent survival factor for motoneurons present in peripheral nerve and muscle, Science, 1994, 266(5187), 1062–1064 CAS.
- D. Martin, G. Miller, N. Fischer, D. Dix, T. Cullen and D. Russell, Glial Cell Line-derived Neurotrophic Factor: the Lateral Cerebral Ventricle as a Site of Administration for Stimulation of the Substantia Nigra Dopamine System in Rats, Eur. J. Neurosci., 1996, 8(6), 1249–1255 CrossRef CAS PubMed.
- M. S. Ramer, J. V. Priestley and S. B. McMahon, Functional regeneration of sensory axons into the adult spinal cord, Nature, 2000, 403, 312–316 CrossRef CAS PubMed.
- N. K. Mohtaram, A. Montgomery and S. M. Willerth, Biomaterial-based drug delivery systems for the controlled release of neurotrophic factors, Biomed. Mater., 2013, 8(2), 022001 CrossRef PubMed.
- L. E. Tomac A, L. F. Lin, S. O. Ogren, D. Young, B. J. Hoffer and L. Olson, Protection and repair of the nigrostriatal dopaminergic system by GDNF in vivo, Nature, 1995, 373(6512), 335–339 CrossRef PubMed.
- D. P. Meka, A. K. Muller-Rischart, P. Nidadavolu, B. Mohammadi, E. Motori, S. K. Ponna, H. Aboutalebi, M. Bassal, A. Annamneedi, B. Finckh, M. Miesbauer, N. Rotermund, C. Lohr, J. Tatzelt, K. F. Winklhofer and E. R. Kramer, Parkin cooperates with GDNF/RET signaling to prevent dopaminergic neuron degeneration, J. Clin. Invest., 2015, 125(5), 1873–1885 Search PubMed.
- N. J. Lamas, B. Johnson-Kerner, L. Roybon, Y. A. Kim, A. Garcia-Diaz and H. Wichterle, Neurotrophic Requirements of Human Motor Neurons Defined Using Amplified and Purified Stem Cell-Derived Cultures (vol. 9, e110324, 2014), PLoS One, 2015, 10(3) DOI:10.1371/journal.pone.0119195.
- T. J. Sill and H. A. von Recum, Electrospinning: applications in drug delivery and tissue engineering, Biomaterials, 2008, 29(13), 1989–2006 CrossRef CAS PubMed.
- J. Xie, X. Li and Y. Xia, Putting Electrospun Nanofibers to Work for Biomedical Research, Macromol. Rapid Commun., 2008, 29(22), 1775–1792 CrossRef CAS PubMed.
- L. Binan, C. Tendey, G. De Crescenzo, R. El Ayoubi, A. Ajji and M. Jolicoeur, Differentiation of neuronal stem cells into motor neurons using electrospun poly-L-lactic acid/gelatin scaffold, Biomaterials, 2014, 35(2), 664–674 CrossRef CAS PubMed.
- J. Du, E. Tan, H. J. Kim, A. Zhang, R. Bhattacharya and K. J. Yarema, Comparative evaluation of chitosan, cellulose acetate, and polyethersulfone nanofiber scaffolds for neural differentiation, Carbohydr. Polym., 2014, 99, 483–490 CrossRef CAS PubMed.
- S. Madduri, M. Papaloizos and B. Gander, Trophically and topographically functionalized silk fibroin nerve conduits for guided peripheral nerve regeneration, Biomaterials, 2010, 31(8), 2323–2334 CrossRef CAS PubMed.
- E. Masaeli, M. Morshed, M. H. Nasr-Esfahani, S. Sadri, J. Hilderink, A. van Apeldoorn, C. A. van Blitterswijk and L. Moroni, Fabrication, characterization and cellular compatibility of poly(hydroxy alkanoate) composite nanofibrous scaffolds for nerve tissue engineering, PLoS One, 2013, 8(2), e57157 CAS.
- S. K. Min, S. H. Kim, C. R. Kim, S.-M. Paik, S.-M. Jung and H. S. Shin, Effect of topography of an electrospun nanofiber on modulation of activity of primary rat astrocytes, Neurosci. Lett., 2013, 534, 80–84 CrossRef CAS PubMed.
- T.-Y. Wang, J. S. Forsythe, D. R. Nisbet and C. L. Parish, Promoting engraftment of transplanted neural stem cells/progenitors using biofunctionalised electrospun scaffolds, Biomaterials, 2012, 33(36), 9188–9197 CrossRef CAS PubMed.
- A. Weightman, S. Jenkins, M. Pickard, D. Chari and Y. Yang, Alignment of multiple glial cell populations in 3D nanofiber scaffolds: Toward the development of multicellular implantable scaffolds for repair of neural injury, Nanomedicine, 2014, 10(2), 291–295 CrossRef CAS PubMed.
- J. M. Zuidema, M. C. Hyzinski-García, K. Van Vlasselaer, N. W. Zaccor, G. E. Plopper, A. A. Mongin and R. J. Gilbert, Enhanced GLT-1 mediated glutamate uptake and migration of primary astrocytes directed by fibronectin-coated electrospun poly-L-lactic acid fibers, Biomaterials, 2014, 35(5), 1439–1449 CrossRef CAS PubMed.
- C. M. Valmikinathan, S. Defroda and X. Yu, Polycaprolactone and Bovine Serum Albumin Based Nanofibers for Controlled Release of Nerve Growth Factor, Biomacromolecules, 2009, 10(5), 1084–1089 CrossRef CAS PubMed.
- S. Y. Chew, R. F. Mi, A. Hoke and K. W. Leong, Aligned protein-polymer composite fibers enhance nerve regeneration: A potential tissue-engineering platform, Adv. Funct. Mater., 2007, 17(8), 1288–1296 CrossRef CAS PubMed.
- M. N. Agbay A and S. M. Willerth, Controlled release of glial cell line-derived neurotrophic factor from poly(ε-caprolactone) microspheres, Drug Delivery Transl. Res., 2014, 4(2), 159–170 CrossRef PubMed.
- J. W. Xie, S. M. Willerth, X. R. Li, M. R. Macewan, A. Rader, S. E. Sakiyama-Elbert and Y. N. Xia, The differentiation of embryonic stem cells seeded on electrospun nanofibers into neural lineages, Biomaterials, 2009, 30(3), 354–362 CrossRef CAS PubMed.
- N. Mohtaram, J. Ko, A. Montgomery, M. Carlson, L. Sun, A. Wong, M. Robinson, M. Jun and S. M. Willerth, Multifunctional Electrospun Scaffolds for Promoting Neuronal Differentiation of Induced Pluripotent Stem Cells, J. Biomater. Tissue Eng., 2014, 906–914 CrossRef PubMed.
- H. Yoshimoto, Y. M. Shin, H. Terai and J. P. Vacanti, A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering, Biomaterials, 2003, 24(12), 2077–2082 CrossRef CAS.
- J. Ko, N. K. Mohtaram, F. Ahmed, A. Montgomery, M. Carlson, P. C. Lee, S. M. Willerth and M. B. Jun, Fabrication of poly(-caprolactone) microfiber scaffolds with varying topography and mechanical properties for stem cell-based tissue engineering applications, J. Biomater. Sci., Polym. Ed., 2013, 25(1), 1–17 CrossRef PubMed.
- S. H. Park, J. W. Hong, J. H. Shin, D. Y. Yang and Y. Yang, Quantitatively Controlled Fabrication of Uniaxially Aligned Nanofibrous Scaffold for Cell Adhesion, J. Nanomater., 2011, 1–17, DOI:10.1080/09205063.2013.830913.
- M. A. Reina, A. Lopez, V. Badorrey, J. A. De Andres and S. Martin, Dura-arachnoid lesions produced by 22 gauge Quincke spinal needles during a lumbar puncture, J. Neurol., Neurosurg. Psychiatry, 2004, 75(6), 893–897 CrossRef CAS.
- M. A. Reina, A. Lopez-Garcia, M. Dittmann and J. A. de Andres, Structural analysis of the thickness of human dura mater with scanning electron microscopy, Rev. Esp. Anestesiol. Reanim., 1996, 43(4), 135–137 CAS.
- X. Jiang, H. Q. Cao, L. Y. Shi, S. Y. Ng, L. W. Stanton and S. Y. Chew, Nanofiber topography and sustained biochemical signaling enhance human mesenchymal stem cell neural commitment, Acta Biomater., 2012, 8(3), 1290–1302 CrossRef CAS PubMed.
- I. C. Liao, S. Y. Chew and K. W. Leong, Aligned core-shell nanofibers delivering bioactive proteins, Nanomedicine, 2006, 1(4), 465–471 CrossRef CAS PubMed.
- A. Aubert-Pouessel, M. C. Venier-Julienne, A. Clavreul, M. Sergent, C. Jollivet, C. N. Montero-Menei, E. Garcion, D. C. Bibby, P. Menei and J. P. Benoit, In vitro study of GDNF release from biodegradable PLGA microspheres, J. Controlled Release, 2004, 95(3), 463–475 CrossRef CAS PubMed.
- L. Q. Zhang, Z. W. Ma, G. M. Smith, X. J. Wen, Y. Pressman, P. M. Wood and X. M. Xu, GDNF-Enhanced Axonal Regeneration and Myelination Following Spinal Cord Injury is Mediated by Primary Effects on Neurons, Glia, 2009, 57(11), 1178–1191 CrossRef PubMed.
- H. Taylor, N. Barua, A. Bienemann, M. Wyatt, E. Castrique, R. Foster, M. Luz, C. Fibiger, E. Mohr and S. Gill, Clearance and Toxicity of Recombinant Methionyl Human Glial Cell Line-Derived Neurotrophic Factor (r-metHu GDNF) Following Acute Convection-Enhanced Delivery into the Striatum, PLoS One, 2013, 8(3), e56186, DOI:10.1371/journal.pone.0056186.
- N. K. Mohtaram, J. Ko, C. King, L. Sun, N. Muller, M. B. Jun and S. M. Willerth, Electrospun biomaterial scaffolds with varied topographies for neuronal differentiation of human-induced pluripotent stem cells, J. Biomed. Mater. Res., Part A, 2014, 2591–2601, DOI:10.1002/jbm.a.35392.
- S. R. Braam, L. Zeinstra, S. Litjens, D. Ward-van Oostwaard, S. van den Brink, L. van Laake, F. Lebrin, P. Kats, R. Hochstenbach, R. Passier, A. Sonnenberg and C. L. Mummery, Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alpha V beta 5 integrin, Stem Cells, 2008, 26(9), 2257–2265 CrossRef CAS PubMed.
- G. K. Chen, D. R. Gulbranson, Z. G. Hou, J. M. Bolin, V. Ruotti, M. D. Probasco, K. Smuga-Otto, S. E. Howden, N. R. Diol, N. E. Propson, R. Wagner, G. O. Lee, J. Antosiewicz-Bourget, J. M. C. Teng and J. A. Thomson, Chemically defined conditions for human iPSC derivation and culture, Nat. Methods, 2011, 8(5), 424–429, DOI:10.1038/nmeth.1593.
- M. Robinson, S. Yau, L. Sun, N. Gabers, E. Bibault, B. R. Christie and S. M. Willerth, Optimizing differentiation protocols for producing dopaminergic neurons from human induced pluripotent stem cells for tissue engineering applications, Biomarker Insights, 2015 DOI:10.4137/BMI.S20064.
| This journal is © The Royal Society of Chemistry 2015 |