Open Access Article
Open Access ArticleSynthesis of nickel–iron hydrogenase in Cupriavidus metallidurans is controlled by metal-dependent silencing and un-silencing of genomic islands†
Martin
Herzberg
,
Marcel
Schüttau
,
Matthias
Reimers
,
Cornelia
Große
,
Hans-Günther-Schlegel‡
and
Dietrich H.
Nies
*
Molecular Microbiology, Institute for Biology/Microbiology, Martin-Luther-University Halle-Wittenberg, Kurt-Mothes-Str. 3, 06120 Halle/Saale, Germany. E-mail: d.nies@mikrobiologie.uni-halle.de; Fax: +49-(0)-345-5527010; Tel: +49-(0)-345-5526352
First published on 19th February 2015
Abstract
Cupriavidus metallidurans CH34 is able to grow autotrophically as a hydrogen-oxidizing bacterium and produces nickel-dependent hydrogenases, even under heterotrophic conditions. Loss of its two native plasmids resulted in inability of the resulting strain AE104 to synthesize the hydrogenases and to grow autotrophically in phosphate-poor, Tris-buffered mineral salts medium (TMM). Three of eleven previously identified catabolic genomic islands (CMGIs; Van Houdt et al., 2009), two of which harbor the genes for the membrane-bound (CMGI-2) and the soluble hydrogenase (CMGI-3), were silenced in strain AE104 when cultivated in phosphate-poor TMM, explaining its inability to produce hydrogenases. Production of the soluble hydrogenase from the aut region 1 of CMGI-3, and concomitant autotrophic growth, was recovered when the gene for the zinc importer ZupT was deleted in strain AE104. The transcriptome of the ΔzupT mutant exhibited two up-regulated gene regions compared to its parent strain AE104. Expression of the genes in the aut region 1 increased independently of the presence of added zinc. A second gene region was expressed only under metal starvation conditions. This region encoded a TonB-dependent outer membrane protein, a putative metal chaperone plus paralogs of essential zinc-dependent proteins, indicating the presence of a zinc allocation pathway in C. metallidurans. Thus, expression of the genes for the soluble hydrogenase and the Calvin cycle enzymes on aut region 1 of CMGI-3 of C. metallidurans is under global control and needs efficient ZupT-dependent zinc allocation for a regulatory role, which might be discrimination of nickel.
Introduction
Currently, a picture is emerging as to how homeostasis of multiple transition metals in bacteria occurs.1,2 Requirements include transport reactions, more specifically import and efflux across the inner and outer membrane,3 a contribution from cellular thiols such as glutathione or bacillithiol,4–6 changes in the metallation of metal-binding sites as a response to oxidative stress,7 and the zinc repository, which has been recently identified in Cupriavidus metallidurans.8 We selected nickel as a starting point to investigate the link between nickel and zinc allocation pathways in C. metallidurans because bacteria usually contain only a few nickel-dependent proteins.9,10 Besides an urease, C. metallidurans possesses two [NiFe]-hydrogenases, a membrane-bound and a soluble, NAD+-reducing enzyme, both of which are synthesized under autotrophic as well as heterotrophic growth conditions in the wild type strain CH34.11 CH34 synthesizes neither a type 5 high-affinity hydrogenase12 nor a sensory hydrogenase,13 which is in alignment with the observation that the availability of molecular hydrogen is not required for synthesis of either hydrogenase.11C. metallidurans is a betaproteobacterium able to maintain its transition metal homeostasis over a wide range of metal concentrations.11,14,15 Metal import and export systems have been identified that interact in concert to establish a kinetic ‘flow’ equilibrium, which adjusts the concentration of the individual metal and the composition of all enzymes to meet the cellular requirements.15–19 The most important and sophisticated metal-resistance systems of strain CH34 are encoded by two indigenous plasmids, pMOL28 and pMOL30.11,14 These systems are absent in the plasmid-free strain AE104.11
Import of transition metal cations in C. metallidurans is achieved by a battery of highly redundant uptake systems, which have only minimal cation selectivity.15 In addition to its role as a metal importer with low substrate specificity, the zinc importer ZupT of the ZIP protein family20 is also essential for zinc supply under zinc starvation conditions, and even at higher zinc concentrations it is required for efficient allocation of the metal to zinc-dependent proteins21 despite the fact that the other zinc uptake systems are still available to transport the metal into the cytoplasm.15
In the case of zinc ions, the zinc ion repository seems to counterbalance the transport processes. C. metallidurans contains at least 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc-binding proteins per cell,8 sometimes with more than one binding site per protein, but only 70
000 zinc-binding proteins per cell,8 sometimes with more than one binding site per protein, but only 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms per cell when the metal is present at concentrations in the upper nM range. When μM concentrations (100–150 μM) of zinc are added, this number increases up to 120
000 zinc atoms per cell when the metal is present at concentrations in the upper nM range. When μM concentrations (100–150 μM) of zinc are added, this number increases up to 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms per cell, filling up the repository. Deletion of zinc efflux pumps (ΔzntA ΔcadA ΔdmeF ΔfieF) results in 250
000 atoms per cell, filling up the repository. Deletion of zinc efflux pumps (ΔzntA ΔcadA ΔdmeF ΔfieF) results in 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms per cell at 10 and 15 μM added zinc, and those cells are unable to grow at higher zinc concentrations. This indicates that efflux systems are needed to keep the number of zinc atoms at a level of 120
000 zinc atoms per cell at 10 and 15 μM added zinc, and those cells are unable to grow at higher zinc concentrations. This indicates that efflux systems are needed to keep the number of zinc atoms at a level of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per cell, and that there is an overflow of the zinc repository at 250
000 per cell, and that there is an overflow of the zinc repository at 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms per cell or above, which subsequently leads to zinc toxicity.8 Moreover, when C. metallidurans is treated with high concentrations of other transition metal cations, the resulting number of atoms per cell stayed in the range of 100
000 atoms per cell or above, which subsequently leads to zinc toxicity.8 Moreover, when C. metallidurans is treated with high concentrations of other transition metal cations, the resulting number of atoms per cell stayed in the range of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 200
000 to 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000,15,21 indicating that the repository might also be involved in storage and sorting of these other ions, e.g. Ni(II).
000,15,21 indicating that the repository might also be involved in storage and sorting of these other ions, e.g. Ni(II).
A ΔzupT mutant contains only 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms when grown in the presence of zinc at a concentration in the upper nM range. The size of the zinc repository is not different between the mutant and its parent but an efficient zinc allocation, e.g. to the RpoC subunit of the RNA polymerase, was impaired in the ΔzupT mutant, even when it contained 120
000 zinc atoms when grown in the presence of zinc at a concentration in the upper nM range. The size of the zinc repository is not different between the mutant and its parent but an efficient zinc allocation, e.g. to the RpoC subunit of the RNA polymerase, was impaired in the ΔzupT mutant, even when it contained 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms at medium μM zinc concentrations.8 Comparisons of the proteomes between mutant and parent did not yield any clues as to why the mutant had a problem with zinc allocation. Rather, the ΔzupT mutant up-regulated synthesis of a number of proteins important for chemolithoautotrophic growth, such as the soluble NAD-reducing hydrogenase and enzymes of the Calvin cycle. Synthesis of the hydrogenase is nickel-dependent, and is stimulated by carbon dioxide as electron sink during CO2-fixation.11 A defect in zinc allocation thus results in formation of a nickel-dependent protein, indicating an inter-connection between zinc and nickel homeostasis in C. metallidurans.
000 zinc atoms at medium μM zinc concentrations.8 Comparisons of the proteomes between mutant and parent did not yield any clues as to why the mutant had a problem with zinc allocation. Rather, the ΔzupT mutant up-regulated synthesis of a number of proteins important for chemolithoautotrophic growth, such as the soluble NAD-reducing hydrogenase and enzymes of the Calvin cycle. Synthesis of the hydrogenase is nickel-dependent, and is stimulated by carbon dioxide as electron sink during CO2-fixation.11 A defect in zinc allocation thus results in formation of a nickel-dependent protein, indicating an inter-connection between zinc and nickel homeostasis in C. metallidurans.
In this publication we demonstrate that the hydrogenases and Calvin cycle enzymes do indeed have the predicted functions and allow autotrophic growth of the ΔzupT mutant strain but not of its parent strain AE104 in phosphate-poor (642 μM phosphate) Tris-buffered mineral salts medium TMM. The genes encoding these proteins in CH34 are located on a genomic island, that was silenced in TMM-grown AE104 cells but which was ‘un-silenced’ again in the ΔzupT mutant. Unexpectedly, our approach also uncovered candidates for a zinc allocation pathway needed under conditions of severe zinc starvation. Together, these data indicate that C. metallidurans possesses ZupT-dependent zinc allocation pathways that “channel” the zinc repository to provide this metal efficiently to zinc-dependent proteins such as RpoC or maybe also HypA, which is crucial to discriminate nickel and to control further allocation of this metal to nickel-dependent proteins, such as hydrogenases.
Results
Autotrophic growth
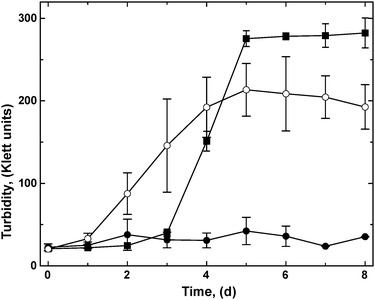
A bottom-up proteomic approach revealed that the ΔzupT mutant up-regulated synthesis of the NAD+-reducing hydrogenase and many Calvin-cycle enzymes when heterotrophically cultivated in phosphate-poor (642 μM phosphate) Tris-buffered mineral salts medium (TMM), while its parent strain AE104 did not,8 although strain AE104 was capable of synthesizing hydrogenases in phosphate-rich (36 mM) growth medium.11 To test if synthesis of these proteins had a physiological consequence, the C. metallidurans strains AE104, ΔzupT and, as a positive control the parental CH34 strain, were pre-cultivated heterotrophically to carbon exhaustion, diluted into fresh TMM with carbonate as sole carbon source, and cultivated with shaking at 30 °C under a “Knallgas” atmosphere.11 Strain CH34 needed a lag phase of about 2 days to adapt to these conditions, and subsequently grew to 275 Klett units (KE) in the following 3 days (Fig. 1). The plasmid-free strain AE104, however, failed to grow within 8 days of incubation in TMM without an organic carbon source. The ΔzupT mutant of strain AE104 grew even more rapidly than CH34 but to a lower overall turbidity and with higher deviations between the individual cultures. This demonstrated that the ΔzupT strain with its up-regulated Calvin cycle and soluble hydrogenase proteins was indeed able to switch rapidly to autotrophic growth as a hydrogen-oxidizing bacterium.Characterization of the key enzymes needed for autotrophic growth
To confirm the identity and biochemical function of the key enzymes needed for autotrophic growth, crude extracts, soluble supernatant and membrane fractions from cells of the three strains CH34, AE104 and ΔzupT were separated on a non-denaturing polyacrylamide gel. Subsequently, an activity stain for hydrogenases was performed (Fig. 2). After autotrophic growth, extracts from strain CH34 exhibited activity of the membrane-bound hydrogenase while the corresponding activity band from the ΔzupT cells was significantly weaker. Strain AE104 did not grow autotrophically in phosphate-poor TMM (but heterotrophically as published11). After heterotrophic cultivation in TMM, only CH34 wild type showed activity of a membrane-bound hydrogenase (Fig. 2 bottom).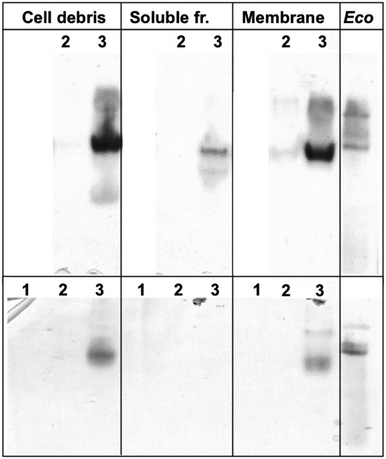 | ||
| Fig. 2 Activity of membrane-bound hydrogenases. C. metallidurans strains were cultivated autotrophically (top) or heterotrophically (bottom) in phosphate-poor TMM, E. coli W3110 cells as control were grown heterotrophically in LB. Crude extracts of ultrasonicated cells, soluble and membrane fraction separated by ultracentrifugation were separated on a non-denaturing polyacrylamide gel in MOPS buffer pH 7 plus 4% (w/v) Triton X100, and an activity stain was performed as published.22E. coli crude extract (200 μg protein per lane) served as positive control. C. metallidurans strains were AE104 (1, no growth under autotrophic conditions in TMM), ΔzupT (2) or CH34 (3), each loaded at 50 μg protein per lane. | ||
The specific activity of the soluble, NAD+-reducing hydrogenase was determined as hydrogen-dependent NAD+-reduction in crude extracts and soluble cell fractions (Table 1). After autotrophic growth in phosphate-poor TMM, hydrogenase activity above background was determined for strain CH34 and the ΔzupT mutant of strain AE104. After heterotrophic growth, significant hydrogenase activity could be measured for the ΔzupT mutant.
| Strain | Cultivation | NAD+-reduction rate, U g−1 protein | |
|---|---|---|---|
| Crude extract | Soluble fraction | ||
| CH34 | Heterotrophically | 1.1 ± 0.1 | 1.3 ± 0.6 |
| AE104 | Heterotrophically | 1.2 ± 0.0 | 2.4 ± 2.6 |
| ΔzupT | Heterotrophically | 23.5 ± 19.2 | 58.8 ± 36.1 |
| CH34 | Autotrophically | 16.0 ± 0.0 | 60.0 ± 0.0 |
| AE104 | Autotrophically | No growth | No growth |
| ΔzupT | Autotrophically | 20.5 ± 0.0 | 10.9 ± 3.8 |
The four key enzymes for autotrophic growth, soluble and membrane-bound hydrogenase, ribulose-bisphosphate carboxylase and phosphoribulo-kinase, were purified from crude extract of C. metallidurans CH34 wild type cells. The specific activities and molecular masses confirmed that these proteins indeed performed the predicted functions (Table S1, ESI†), although the molecular masses of the hydrogenases were over- and that of the ribulose-bisphosphate carboxylase was under-estimated. The membrane-bound hydrogenase exhibited a Km value of 38 μM for the artificial electron acceptor methylene blue in a 100% hydrogen atmosphere The procion dye HERD used for the last purification step competitively inhibited methylene blue reduction with a Ki of 4.1 μM (data not shown).
The kinetic mechanism of the NAD+-reducing hydrogenase was analyzed in more detail using Hanes–Woolf plots as described by Segel.23 The enzyme exhibited a typical ping-pong reaction mechanism with an apparent Km for molecular hydrogen of 38 μM, NAD+ of 242 μM and NADH (reverse reaction) of 60 μM (data not shown). In analogy to the protein from Ralstonia eutropha (synonym Cupriavidus eutrophus),24,25 the enzyme from C. metallidurans was oxygen-tolerant and could be re-activated after oxygen treatment with NADH and molecular hydrogen (data not shown).
This demonstrated that deletion of the zupT gene for the zinc importer led to synthesis of the NAD+-reducing hydrogenase and the Calvin cycle enzymes, which were fully functional, had the predicted biochemical functions, and enabled autotrophic growth.
Isolation of autotrophic mutants
It was unexpected that the plasmid-free strain AE104 failed to grow autotrophically in phosphate-poor (642 μM) TMM because it was able to grow in a phosphate-rich (36 mM) Schlegel–Gottschalk–Kaltwasser (SGK) medium.11 Strain AE104 was treated with mutagenic agents (nitrite, EMS, NMG, mitomycin C) and mutants unable to grow autotrophically even in phosphate-rich SGK medium were isolated. A total of 16 mutants from 6400 colonies analyzed in two independent mutagenesis experiments were unable to grow autotrophically in SGK (7 from experiment 1, 9 from experiment 2; 3 nitrite-generated mutants, 5 EMS, 5 NMG, 3 mitomycin C, data not shown). A D-cycloserine selection procedure did not influence the number of successfully isolated mutant strains, and all mutagenic agents used generated mutants despite their different modes of action. Most of the mutant strains had a stable Aut− phenotype and did not generate revertants. Exceptions were one EMS mutant (spontaneous reversion rate 10−6, reversion rate after a second EMS treatment 10−1), and all mitomycin C mutants that reverted back to autotrophic growth with a spontaneous reversion rate of 10−2 (data not shown). In a control experiment with untreated cells, no mutants could be identified among 3200 tested colonies indicating that the mutants did not arise spontaneously.Of the 16 mutants, 13 were unable to produce the soluble hydrogenase and the Calvin cycle enzymes, and 3 were additionally unable to synthesize the membrane-bound hydrogenase. Thus, absence of the native metal-resistance plasmids pMOL28 and pMOL30 in strain AE104 prevented autotrophic growth or synthesis of hydrogenases under heterotrophic conditions in phosphate-poor TMM but allowed both processes in phosphate-rich SGK medium. Treatment of strain AE104 with mutagenic compounds efficiently yielded pleiotropic mutants no longer able to synthesize the soluble hydrogenase and the Calvin cycle enzymes, even in phosphate-rich SGK-medium. A second or additional mutagenic event also prevented the synthesis of the membrane-bound hydrogenase. A metal and phosphate-dependent regulatory process is thus in control of hydrogenase gene expression in C. metallidurans.
Comparison of AE104 with CH34
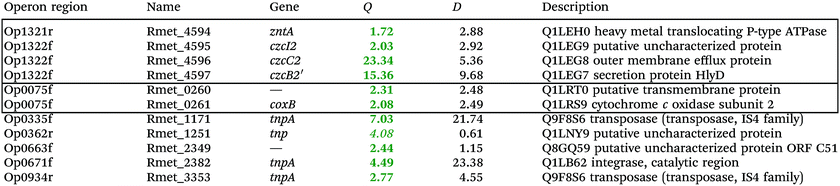
To investigate the consequences of the plasmid loss on TMM-grown C. metallidurans cells, the transcriptome of the plasmid-free derivative AE104 was compared to that of parental strain CH34 (Table S2, ESI†). The cells were heterotrophically cultivated in phosphate-poor TMM without additions. To facilitate analysis of the data, a group of adjacent genes in the same direction of transcription that was not interrupted by other genes transcribed in the opposite orientation was designated as “operon region”, and all operon regions in the C. metallidurans genome were numbered from Op0001f on chromosome 1 starting with Rmet_0001 to Op1929r on plasmid pMOL28, with “f” indicating the forward and “r” the reverse direction of transcription compared to the genome annotation14 (data not shown). The Q values (ratios) AE104/CH34, sorted according to their protein products that were up- or down-regulated, are presented in Table S2 (ESI†). Only Q values were considered that were ≥2 or ≤0.5 with D-values (difference of the two data points divided by the sum of the standard deviations) >1 (Table S2, ESI†).Only 11 genes were up-regulated (Table 2), four of which were in the divergently organized region Op1321r–Op1322f. These genes coded for the zinc-exporting P-type ATPase ZntA (Op1321r) and the 5′ end of a region czcI2C2B2′ related to the czcICBAD resistance determinant on plasmid pMOL30; however, this region was interrupted by an inversion of a large part of chromosome 2 in the middle of the czcB2 gene. Two genes in Op0335f encoded putative cytochrome c oxidase subunits of the respiratory chain, the others transposon-related proteins. Up-regulation of the Op1321r–Op1322f divergently oriented region indicated an impaired zinc homeostasis of strain AE104 when cultivated in TMM.
| a Only the up-regulated genes are shown, while the complete gene-set is in the Table S2 (ESI). Genes in the same or adjacent operon regions are boxed. Provided are the Q-ratios AE104/CH34, bold-faced letters indicate significant differences, letters in italics not significant differences. The D-value is the distance of both mean values divided by the sum of both deviations. (If D > 1 the deviation bars do not touch or overlap) the data points are different with a probability in the t-test of >95%. |
|---|
|
A total of 250 genes were down-regulated in strain AE104, most of them located in 6 gene regions (Fig. 3). Region 1 (aut region 1) from Op0421f to Op0431f contained 55 genes for the soluble, NAD+-reducing hydrogenase and the Calvin cycle enzymes, region 3 had a total of 63 genes, including those for the membrane-bound hydrogenase (aut region 2) and degradation of aromatic carbohydrates.
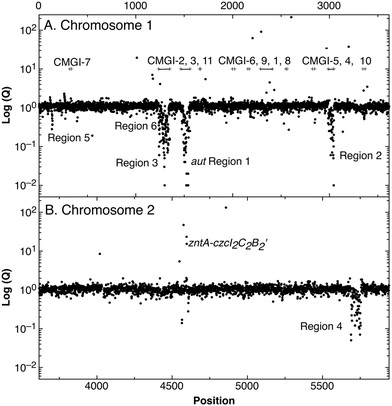 | ||
| Fig. 3 Difference in gene expression between C. metallidurans parental strain CH34 and its plasmid-free derivative AE104. After cultivation of both strain in TMM without added metals, the cells were harvested, a gene array experiment performed and the ratios (Q values, closed circles) AE104/CH34 plotted against the position (Rmet number) on chromosome 1 (panel A) or chromosome 2 (panel B). Please note that the significance of the Q values is not indicated. The position of the zntA-czcI2C2B2′ divergon and six large gene regions that were down-regulated in strain AE104 are indicated. The bars in panel A indicate proposed genomic islands on chromosome 1.27 | ||
Region 2 showed similarity to a catabolic genomic island.26 Eleven catabolic genomic islands, CMGI-1 to CMGI-11, had been identified on chromosome 1 of C. metallidurans.27 The regions 1 to 3, which were strongly down-regulated in strain AE104, were indeed identical with the predicted catabolic genomic islands CMGI-2 (= region 3 containing aut region 2), CMGI-3 (= aut region 1) and CMGI-4 (= region 2), respectively (Fig. 3). No CMGI had been identified on chromosome 2, so region 4 could not be matched in a similar way. With the exception of region 5 at the beginning of chromosome 1 and region 4 on chromosome 2, most of the genes down-regulated in AE104 compared to CH34 were located in CMGIs 1, 2, 3, 4. Expression of the pitA gene for the metal:phosphate importer and pst for the phosphate-specific ABC importer were not changed (data not shown).
When cultivated in phosphate-poor TMM, the plasmid-free strain AE104 exhibited silencing of some CMGIs including the aut region 1 as central part of CMGI-3 and aut region 2 on CMGI-2, activated recombination activity of four CMGIs, and increased the expression of the gene for the PIB1-type zinc-exporting ATPase ZntA plus an adjacent inactivated paralogous region of the czcICBA metal resistance determinant on plasmid pMOL30. When grown instead in phosphate-rich SGK medium, gene products of this region could be found in AE104 cells but mutants could be readily isolated with these aut regions silenced even in phosphate-rich SGK medium. The metal and phosphate-dependent regulatory process in control of hydrogenase gene expression in C. metallidurans functioned by silencing the responsible aut regions located within CMGIs. The mutants indicated that both regions were silenced independently. Silencing required a increased cellular zinc availability, as demonstrated by the up-regulation of zntA-czcI2C2B2′. Silencing occurred as a consequence of plasmid loss but only in the phosphate-poor TMM medium and not in the phosphate-rich, metal-complexing SGK medium.
Comparison of the proteome with the transcriptome
To analyze the connection between transcriptomic and proteomic results, the cellular content of a single protein in TMM-grown AE104 cells was compared with its gene expression signal measured by the gene array experiment. The luminescence of the single spots was normalized to a number of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 gene-specific and intergenic RNAs. The overall data field resulting from this comparison resembled a triangle in the semi-logarithmical plot with the elongation factor TufA (Rmet_3324) on top of the triangle (Fig. S1, ESI†). Other proteins nearby were subunits of the ribosome (RpiL/Rmet_3335, RpsI/Rmet_0411, RplM/Rmet_0410, RpsU1/Rmet_2455, RpmL/Rmet_1162), the chaperone GroEL (Rmet_0616), alkyl hydroperoxidase Rmet_1950, and the extracytoplasmic solute-binding protein Rmet_0521, which is highly abundant in proteobacteria.28
000 gene-specific and intergenic RNAs. The overall data field resulting from this comparison resembled a triangle in the semi-logarithmical plot with the elongation factor TufA (Rmet_3324) on top of the triangle (Fig. S1, ESI†). Other proteins nearby were subunits of the ribosome (RpiL/Rmet_3335, RpsI/Rmet_0411, RplM/Rmet_0410, RpsU1/Rmet_2455, RpmL/Rmet_1162), the chaperone GroEL (Rmet_0616), alkyl hydroperoxidase Rmet_1950, and the extracytoplasmic solute-binding protein Rmet_0521, which is highly abundant in proteobacteria.28
The protein abundance was not a discrete function of the mRNA abundance measured as spot intensity in a gene array experiment. Nevertheless, the envelope curve that describes the increasing side of the triangle (Fig. S1, ESI†) suggested that a high cellular abundance of a given protein requires also a certain transcript level, as can be expected. On the other hand, a strong signal in the gene array experiment did not necessarily relate to an high protein abundance. This suggested a strong influence of translation control and protein stability on the protein copy-number, and that a protein copy number cannot be easily derived from transcriptomic data. A general lack of an mRNA–protein correlation has already been shown on the single-cell level29 and was demonstrated here also on the population level. Nevertheless, absence of a signal in the gene array experiment should lead to a corresponding absence of the respective protein in the proteome, so that silencing of CMGIs was indeed the reason for the absence of hydrogenases and Calvin cycle proteins in AE104 cells grown in TMM.
Operon regions up-regulated at 100 μM Zn(II) were involved in iron–sulfur cluster biosynthesis, the phosphate starvation response, cadmium/zinc efflux by the P-type ATPases CadA and ZntA, and response to unfolded proteins in the periplasm (Table S3, ESI†). The interrupted zntA-czcI2C2B2′ region was again up-regulated, indicating that the increased environmental zinc availability resulted in increased cellular zinc availability. Up-regulated zinc-containing proteins were fructose-bisphosphate aldolase, alcohol dehydrogenases and alkaline phosphatases. Down-regulated genes included those encoding TonB-dependent outer membrane proteins, several ABC transporters, periplasmic substrate-binding proteins, and di- and tricarboxylic acid transporters that might be involved in metal import. Two zinc-dependent but B12-independent methionine synthases were also strongly down-regulated (0.06-fold, 0.17-fold; Table S3, ESI†). This all indicated an expected effect of a moderate zinc shock on AE104 cells: decreased import and increased export, repair of zinc-mediated damage to proteins and iron-dependent processes, and a shift in zinc-dependent proteins. But there was no influence on expression of the hydrogenase genes located in CMGI-2 and 3.
Eight genes were specifically up-regulated by zinc starvation in the ΔzupT mutant strain, Q(EDTA) ≥ 2 and Q(Zn) ≤ 0.5, seven of them significantly (D > 1). Six were all located in the same operon region Op0317f (Table 3). The region Op0318r adjacent to Op0317f contained the remaining gene (Rmet_1104, Table 3). The direction of transcription of the operon regions Op0317f and Op0318r was pointing towards each other and they were not divergently oriented. Regulation of expression of the genes in these regions was not due to a common promoter region but probably by the same regulator(s). The first of the 6 genes in Op0317f encoded CobW1, a putative metal chaperone, followed by 5 genes for paralogs of important zinc-dependent proteins. The single gene in Op0318r might encode a TonB-dependent outer membrane receptor. None of these proteins was found in the proteome of AE104 or ΔzupT cells grown without additions in mineral salts medium.8 This region (cobW1 cluster) might encode an outer membrane uptake system for zinc or zinc complexes, and the possible zinc chaperone CobW1, which might deliver Zn(II) specifically to the other five proteins under zinc starvation conditions.
| Name | Q(Zn) | Q(EDTA) | Description |
|---|---|---|---|
| a None of the corresponding gene products was found in the proteome. The ratio of ΔzupT/AE104 in cells cultivated in the presence of 10 μM Zn, Q(Zn), or 50 μM EDTA, Q(EDTA) is shown. Values in bold-faced letters indicate significant changes (≥2 fold, D-value ≥1). The genes are located in the operon region Op317f–Op318r and a similar regulatory pattern indicates that they might be present in operons. | |||
| Op0317f | |||
| Rmet_1098, CobW1 |
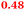
|
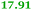
|
Cobalamin synthesis protein |
| Rmet_1099 |
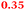
|
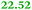
|
GTP cyclohydrolase |
| Rmet_1100 |
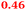
|
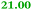
|
Cysteinyl-tRNA synthetase |
| Rmet_1101 |
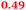
|
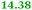
|
6-Pyruvoyl-tetrahydropterin synthase-like protein |
| Rmet_1102 |
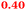
|
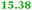
|
Carbonic anhydrases |
| Rmet_1103 |
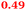
|
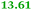
|
Q1LPD7 Dihydroorotase |
| Rmet_1104 |
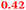
|
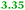
|
TonB-dependent receptor |
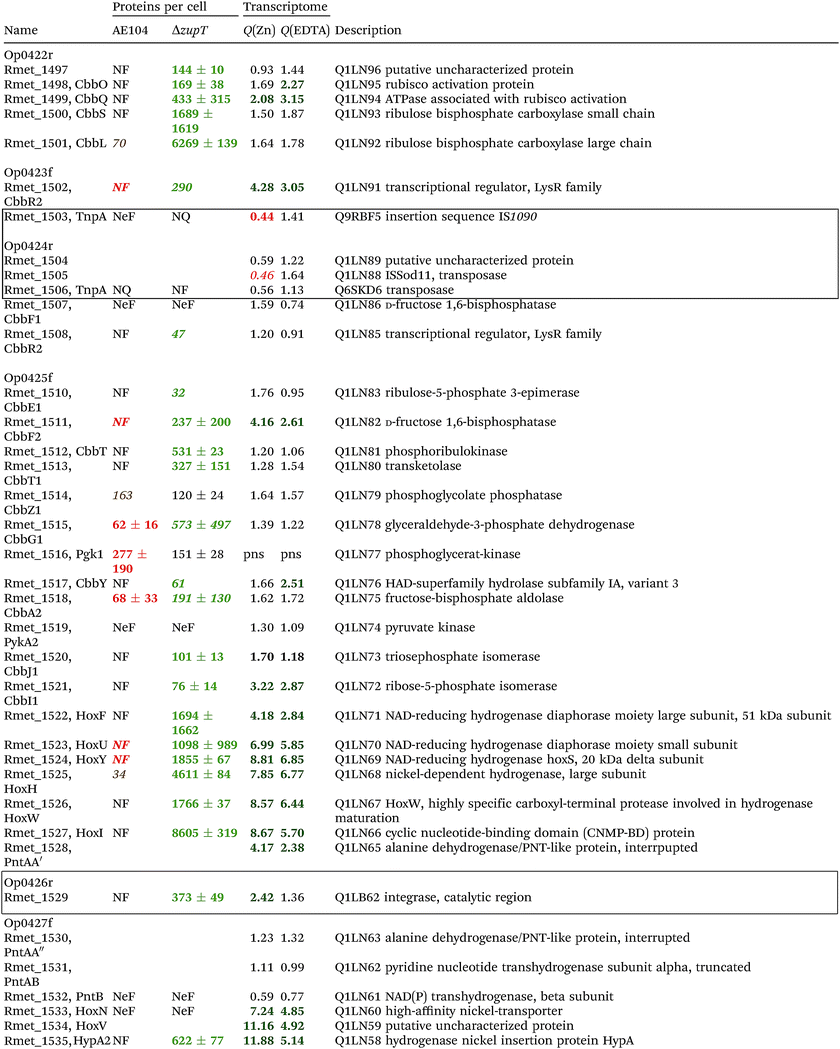
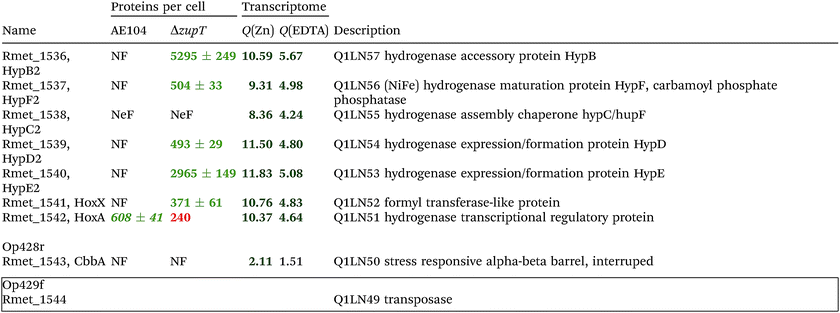
The condition Q(EDTA) ≥ 2 and Q(Zn) ≥ 2, specifically up-regulated in the ΔzupT strain but independent of the two growth conditions, identified 21 genes in the operon regions Op0422r to Op0427f, which encode the enzymes of the Calvin cycle and those involved in formation of the soluble hydrogenase (Table 4) as aut region 1 of CMGI-2, confirming the proteomic, biochemical and physiological results. Two genes on Op1926f, which were transposon-associated (Table S4, ESI†) are also up-regulated.
| a The protein numbers were taken from ref. 8. The transcriptome results were from Table S4 (ESI), and ratios ΔzupT/AE104 in zinc- or EDTA-treated cells are indicated. The proteins and genes were sorted according to the operon region (genes in the same orientation of transcription that are not interrupted by other genes in another orientation), from Op422r (r for reverse) to Op429f (f for forward). The region in chromosome 1 that contains these operon regions possesses three transposons or insertion elements in this region, which are marked by boxes. NF, not found in these two strains; NeF, never found in the proteome analysis done so far; NQ found but could not be quantified. Numbers in red indicate down-regulation, in green up-regulation, with bold numbers indicating significant differences. Up- and down-regulation in the proteome experiment was compared to strain CH34 (AE104 only) or to AE104 (ΔzupT) only. |
|---|
|
|
A total of 654 genes were specifically down-regulated in the ΔzupT strain, Q(EDTA) ≤ 0.5 (Table S4, ESI†) and 345 genes had Q(EDTA) ≤ 0.5 and Q(Zn) ≤ 0.5. Among these were those for the sigma factor FliA and other proteins involved in cell motility, and as an artifact the ΔzupT gene, probably resulting from some transcripts coming from the deletion scar. Down-regulation of the genes for cell motility was also observed in the proteomic approach.8 A total of 308 genes had Q(EDTA) ≤ 0.5 but Q(Zn) ratios that were not significantly changed. Among these were the gene for the zinc-exporting P-type ATPase ZntA and the metal-phosphate importer PitA, which is in agreement with the proteomic approach8 (Table S4, ESI†).
If two additional aut region 1 genes (Rmet_1498, Rmet_1517) that were up-regulated only 1.7-fold by zinc are included, then the cobW1 and aut region 1 genes were the only ones specifically up-regulated in the ΔzupT mutant strain compared to its parent strain. The cobW1 cluster was repressed by zinc and might encode a specific zinc allocation pipeline to zinc-dependent proteins. The genes in aut cluster 1, in contrast, were not repressed by zinc.
The ΔzupT deletion in strain AE104 also influenced gene expression of catabolic metabolic islands CMGI-1, 10 and a few genes in CMGIs-2, 5, 7 (Table S4, ESI†), while loss of the plasmids resulted in down-regulation of CMGI-2, 3, 4. The expression pattern of the aut region 1 genes in CMGI-3 in the ΔzupT mutant compared to parent AE104 was unique: the genes at both ends of CMGI-3 were down-regulated similar to CMGI-1 and 10, while aut region 1 in the middle was strongly up-regulated in the ΔzupT mutant strain (Table 4 and Table S4, ESI†). This indicated that silencing of CMGI-3 as the result of plasmid loss was a process different from the regulatory event that specifically ‘un-silenced’ the aut region 1 on CMGI-3 but not aut region 2.
The number of proteins per cell, which were encoded by the genes in the CMGIs, was compared between the ΔzupT mutant cell and its parent AE104 (Table 5). For most CMGIs, gene expression was decreased in the ΔzupT mutant and consequently the number of proteins per cell was lower in the mutant. This was true even for CMGI-6, 8, 9, and 11 although no decreased gene expression had been observed here. This finding confirmed the transcriptomic data, because CMGI-3 encoded 11-times more protein in ΔzupT than in AE104 (Tables 4 and 5) due to increased synthesis of the aut region 1 gene products.
| Region | CMGI | AE104 | ΔzupT | % |
|---|---|---|---|---|
| a Protein numbers taken from ref. 8, CMGI regions from ref. 27. | ||||
| Rmet_0317–0333 | CMGI-7 | 139 ± 10 | 235 ± 0 | 169 |
| Rmet_1236–1351 | CMGI-2 | 6961 ± 1310 | 3698 ± 713 | 53 |
| Rmet_1465–1560 | CMGI-3 | 3956 ± 1087 | 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 396 ± 7366 396 ± 7366 |
1122 |
| Rmet_1660–1668 | CMGI-11 | 1693 ± 0 | 0 ± 0 | 0 |
| Rmet_1997–2020 | CMGI-6 | 644 ± 47 | 164 ± 33 | 26 |
| Rmet_2156–2172 | CMGI-9 | 1306 ± 237 | 573 ± 118 | 44 |
| Rmet_2287–2408 | CMGI-1 | 3593 ± 874 | 2138 ± 339 | 60 |
| Rmet_2549–2561 | CMGI-8 | 178 ± 0 | 0 ± 0 | 0 |
| Rmet_2824–2847 | CMGI-5 | 1497 ± 296 | 1165 ± 143 | 78 |
| Rmet_2987–3045 | CMGI-4 | 1984 ± 143 | 1828 ± 135 | 92 |
| Rmet_3347–3368 | CMGI-10 | 1237 ± 711 | 534 ± 255 | 43 |
Importance of CobW1
To investigate the cobW1 cluster in more detail, the cobW1 gene for Rmet_1098/CobW1 was interrupted using a vector that inserted a lacZ gene with its own ribosome-binding site directly downstream of the interrupted gene. This mutation was introduced in the parent strain AE104 and its isogenic ΔzupT mutant. Metal resistance of the strains AE104, AE104(ΔzupT), AE104(ΔcobW1::lacZ) and AE104(ΔzupT ΔcobW1::lacZ) was compared in liquid culture (Fig. 4). The ΔzupT-strain showed a 3-fold decrease in cobalt and cadmium resistance and a slight decrease in zinc and EDTA resistance in liquid culture, as previously published.15 Introduction of the ΔcobW1::lacZ mutation into strain AE104 lowered resistance to these substances significantly but not completely to the level of the ΔzupT mutant. Resistance of the ΔzupT ΔcobW1::lacZ double deletion strain, however, was very similar to that of the ΔzupT single deletion strain (Fig. 4). This effect argued for a similar function of the uptake system ZupT and the putative zinc chaperone CobW1 in managing zinc starvation conditions and homeostasis of multiple metal ions.As judged by the β-galactosidase fusion, expression of the cobW1 gene was strongly up-regulated by metal starvation and down-regulated by metals in the ΔzupT background, leading to a 37-fold increase when comparing EDTA- and zinc-treated cells (Table 6). In the parent strain, expression levels of the cobW1::lacZ fusion were much lower, addition of zinc had no effect, and β-galactosidase activity in EDTA-treated cells was 20% of the value of the corresponding ΔzupT strain. These values were in full agreement with the gene array data (Table 3) and explained why CobW1 protein was not found in the proteome of (non-starving) ΔzupT mutant or AE104 parent strains.8 The CobW1 system was only needed under conditions of severe metal starvation.
| Additions | β-Galactosidase activity (U mg−1 d.w.) | |
|---|---|---|
| AE104(ΔcobW1::lacZ) | ΔzupT (ΔcobW1::lacZ) | |
| a Derivatives of strains AE104 and ΔzupT, each containing a ΔcobW1:lacZ fusion, were cultivated in TMM to the late exponential phase of growth, diluted into fresh TMM to a turbidity of 30 Klett units and incubated with shaking at 30 °C until the turbidity has reached 60 KE. Subsequently, they were distributed into 96-well plates, the indicated compounds were added, incubation was continued for 3 h or for 16 h, and the specific β-galactosidase activity was determined. | ||
| None | 1.20 ± 0.16 | 4.43 ± 1.05 |
| 10 μM Zn(II) | 0.98 ± 0.15 | 3.12 ± 1.61 |
| 50 μM EDTA | 24.3 ± 3.1 | 115 ± 20 |
The lacZ reporter constructs all contained an integration of lacZ into the respective target gene, thereby interrupting it, so that the reporter strains were also all insertion mutants. To circumvent possible biases resulting from this situation and to exclude feedback effects, a gfp reporter plasmid was constructed and used to clone in front of gfp the 150 nucleotides upstream of zupT (zupTp) or cobW1 (cobW1p). The resulting constructs were transferred by conjugation into various C. metallidurans strains, which were subsequently cultivated in TMM with or without added EDTA or zinc chloride (Table S5, ESI†). The basal level of the GFP-mediated fluorescence coming from those promoters was the specific fluorescence activity of TMM-grown cells divided by that from the negative control, the promoter-less gfp fusion. Subsequently, the specific fluorescence activities from the cultures with added EDTA or zinc were compared to these basal levels.
All regulatory events strictly depended on the presence of Zur/FurC, so that this protein was indeed required for regulation of cobW1 expression in a similar fashion as previously shown for zupT. In the Δzur mutant strain, zupTp and cobW1p expressed gfp at an increased high level of 7-fold and 5-fold compared to the negative control, respectively (Table S5, ESI†). Activity of the promoters was not different between strains CH34 and AE104 although the values coming from CH34 were slightly lower than those from AE104. Plasmid-borne genes were not required for these Zur-dependent regulatory processes but seem to ameliorate metal starvation mediated by EDTA to some extent. Deletion of the most important zinc efflux systems (strain Δe4 = ΔzntA ΔcadA ΔdmeF ΔfieF) did not influence the expression pattern but the additional deletion of zupT in Δe4 increased the basal level of expression while it decreased EDTA-dependent up-regulation. In AE104, activity of zupTp was at a high basal level (10-fold higher than the negative control), but it could be up-regulated 2-fold under extreme metal starvation conditions (10 mM EDTA). However, it was down-regulated 50% by addition of zinc. In contrast, the expression of the cobW1p promoter was at the level of the negative control in AE104, although it was also up-regulated by EDTA, and there was no difference between AE104 and its ΔzupT mutant. The cobW1p promoter was up-regulated by higher EDTA concentrations than the zupTp promoter (Table S5, ESI†).
The cobW1 region was the only one specifically up-regulated in the ΔzupT strain under conditions of metal starvation and it was regulated mainly by zinc availability in a pattern reminiscent of the regulation of expression in ΔzupT; however, it was expressed more prominently during starvation. Moreover, Zur/FurC was essential for regulation of cobW1p, as it was observed for zupTp, Zur did not require ZupT-dependent zinc uptake and allocation for its function, and there seemed to be some feed-back regulatory pathway that specifically up-regulates zupT expression in a ΔzupT mutant. Together, these data indicated that a specific zinc allocation pathway exists in C. metallidurans under conditions of severe zinc starvation. This pathway is composed of a TonB-dependent outer membrane protein, ZupT, and CobW1 that may “channel” the zinc repository to allocate zinc efficiently to paralogs of important zinc-dependent proteins, which are also encoded by genes of the cobW1 cluster.
Discussion
Global silencing of horizontally acquired genes
Three facts are important to explain the up-regulation of the aut region 1, which encodes the soluble hydrogenase and the Calvin cycle enzymes, in the ΔzupT mutant of the plasmid-free C. metallidurans derivative strain AE104 when cultivated in phosphate-poor Tris-buffered mineral salts medium TMM. First, aut region 1 is located in the middle of the catabolic genomic island CMGI-3.27 Genomic islands are emerging as important factors for niche differentiation in bacteria30–33 in a similar way to transposons and plasmids. Second, loss of the native plasmids pMOL28 and pMOL30 of the parental strain CH34 resulted in a silencing (or repression) of the CMGIs 2, 3, and 4, whereby CMGI-2 harbors the aut region 2 genes including those encoding the membrane-bound hydrogenase of C. metallidurans, while CMGI-4 harbors genes for catabolic enzymes not required for autotrophic growth. Third, in a unique fashion, expression of only the genes in aut region 1 were up-regulated, activated or ‘un-silenced’ (de-repressed) in the ΔzupT mutant of AE104, indicating a mechanism specific for aut region 1.Silencing or repression of large gene regions can be accomplished on the DNA level by ‘wrapping’ these regions such that access by RNA polymerase is hindered, or by influencing transcription by other means. Chromosome 1 of C. metallidurans contains 11 CMGIs, which could be the product of horizontal gene transfer and/or recombination events.27 In Escherichia coli, the proteins HU and HN-S are important factors in genome organization, silence horizontally acquired genetic elements, and are both global transcription regulators. HU, present as homodimers HupA2 and HupB2 and as heterodimer HupAB depending on the growth conditions, re-organizes the transcriptome, binds non-coding RNAs, is involved in stress resistance, and is required for production of the stationary phase sigma factor RpoS.34–38 Moreover, presence of polyphosphate stimulates degradation of HupA, which is linked to the exponential phase of growth and nucleoid organization,39 linking growth phases and the phosphate status of the cell. Seven putative HupAB-related proteins are encoded by the C. metallidurans genome, three on the plasmids (Rmet_6397, 6191, 6090) and four on the chromosomes (Rmet_3538, 4749, 4742 and 5558). These 4 proteins could be found in the proteome of C. metallidurans with copy-numbers of about 6000, 6000, 8500 and 1700 per cell, respectively, which is maintained between CH34, AE104 and ΔzupT.8 These numbers were 10-fold lower than those determined for E. coli,40 perhaps due to the different methods used.
H-NS silences foreign DNA41 by wrapping the DNA.42 The proteins with the highest similarity to E. coli H-NS are Rmet_3677 with about 3800 copies per cell and the more distantly related Rmet_5562 with about 450 copies per cell. Both did not change their numbers between CH34, AE104 and ΔzupT8 but the gene for Rmet_3677 was up-regulated 2.5-fold in AE104 cells treated with 100 μM Zn(II) (Table S3, ESI†), clearly indicating the influence of zinc on H-NS in C. metallidurans.
Sigma factors and metal homeostasis
The activity of the RNA polymerase in the global control of transcription initiation can be influenced by sigma factors or activators while repressors usually act upon one or a few operons only. C. metallidurans contains 11 sigma factors of the extracytoplasmic function (ECF) family,43–46 arranged in four groups of related proteins. C. metallidurans often contains between two and four paralogs of these proteins, which substitute each other in deletion strains, e.g. the zinc-exporting P-type ATPases ZntA, CadA, PbrA and CzcP.17,47 To characterize the contribution of these ECF sigma factors to metal resistance we therefore produced double or triple deletion mutants of C. metallidurans CH34 in which all genes encoding related ECF sigma factors were deleted. These mutants were tested for the impact of these deletions on the cellular metal content, the transcriptome, and the ability of the respective strain to handle a metal mixture composed of 30 μM each of CoCl2, NiCl2, CuCl2, ZnCl2 and CdCl2 (C. Große and D. H. Nies, unpublished). C. metallidurans CH34 is able to deal with all of these metals in parallel,15 however, treatment with this metal mixture resulted in down-regulation of aut region 2 on CMGI-2 and aut region 1 on CMGI-3 but not of CMGI-4, indicating that the CMGIs 2 and 3 are indeed silenced under conditions of metal stress. Deletion of groups of sigma factors had no effect except in the triple deletion strain CH34 (ΔrpoEP cnrH::pLO2-lacZ). This strain contained a significantly higher nickel content and expressed the aut region 1 similar to AE104ΔzupT (C. Große and D. H. Nies, unpublished). CnrH is the sigma factor required for expression of the cnrCBA nickel resistance determinant on plasmid pMOL28,48–50 which explains the higher nickel content of the cells. So, when the metal content of C. metallidurans was increased, CMGIs were silenced, but when specifically the nickel content was increased even more, aut region 1 was un-silenced again. Un-silencing of this region is prevented by an efficient zinc allocation but is facilitated by nickel.These results demonstrated that a perturbed metal ion homeostasis was probably responsible for silencing of CMGI-2, -3, and -5 in strain AE104, CMGI-1 and -11 in the isogenic ΔzupT mutant, and CMGI-2 and -3 in metal mixture-treated CH34 cells. Interestingly, such a link between metal homeostasis and hydrogenase synthesis has also been shown for E. coli.51 Up-regulation of the zntA-czcI2C2B2′ region on chromosome 2 is also in agreement with a disturbed metal homeostasis or increased zinc availability in TMM-grown AE104 cells. Moreover, strain AE104 readily produced hydrogenases and was able to grow autotrophically in phosphate-rich SGK mineral salts medium;11 however, this ability was easily lost again in AE104 mutants.
Zinc speciation in the growth medium and zinc uptake
The phosphate-poor TMM11 contains 642 μM phosphate, 1 mM magnesium chloride, 200 μM calcium chloride, 5 μM iron ammonium citrate and nM concentrations of the other essential transition metal cations, e.g. 200 nM zinc.8,21 Calculated from the solubility constants of the respective metal phosphate compounds, which are in the in the region of 10−30 to 10−40 for the transition metal cations and Ca(II), and 10−25 for Mg(II),52 most of the transition metal cations and also Ca(II) should reside in disperse metal phosphate precipitates, with magnesium filling up the remaining phosphate ions. The “free” Zn(II) concentrations under these conditions is calculated from the solubility constant to be 16.3 pM. The other anions, such as hydroxide, carbonate, sulfate, and the Tris buffer possess much lower solubility constant than phosphate, are thus not able to compete with phosphate for these cations, and do not need to be considered to understand the speciation of divalent metal cations in the growth media used.In phosphate-rich SGK,53 the phosphate concentration of 36 mM is much higher than that of the metal cations, so that only 3.3% of the phosphate pool in the growth medium should harbor a metal cation. The remaining 96.7% of the phosphate should be HPO42− and H2PO4− at neutral pH values. In this medium, the “free” Zn(II) concentration is calculated to be only 1.12 pM.
Uptake of Zn(II) and the other transition metal cations (iron not considered here) is by ZupT and PitA. Since a ΔzupT ΔpitA double mutant is still able to import zinc, at least one more metal cation importer is involved, e.g. one of the three CorAs.15 The ability of ZupT to fetch zinc ions at low concentrations can be judged by the decreased EDTA resistance of the ΔzupT mutant on solid TMM medium. The MIC value is 300 μM compared to 1.5 mM of strain AE104.15 Using the stability constant of zinc EDTA complexes,54 the “free” zinc concentration at 300 μM EDTA and 200 nM Zn(II) is 53 aM. So, ZupT is needed to gather Zn(II) at “free” concentrations of 50 aM and below, which is sufficient to access the “free” zinc concentration in the phosphate-containing TMM and SGK media.
The PitA protein imports metal:phosphate complexes in C. metallidurans15 and other prokaryotes.55,56 As judged by the activity of the lacZ reporter gene fusion15 PitA is the most strongly expressed metal uptake system in C. metallidurans. In SGK with most of the phosphate anions not bound to metal cations, PitA should mainly import just phosphate and only very few metal:phosphate complexes, resulting in a low PitA-mediated influx rate of metals. In contrast, in phosphate-poor TMM, with all phosphate ions bound to metal cations, PitA should import much more metal:phosphate complexes into the cell. In C. metallidurans CH34 wild type, the Czc system encoded on plasmid pMOL30 interferes with a too extensive zinc accumulation by CzcP- and CzcD-mediated export from the cytoplasm to the periplasm, and from here by the CzcCBA transenvelope efflux complex further on to the outside.17 CzcP and CzcD export rapidly loosely bound zinc while the P-type APTases ZntA and CadA more tightly bound zinc, albeit with a lower export rate.17 Assuming that phosphate-bound zinc could represent loosely bound zinc and zinc bound within the zinc repository or to thiols more tightly bound zinc, the products of the Czc system would be the antagonists of PitA in phosphate-poor environments and remove surplus transition metals from phosphate complexes in the cytoplasm and periplasm.
Consequently, the plasmid-free and Czc-free strain AE104 is not able to counterbalance the increased influx of zinc by the PitA system in TMM, leading to the observed mild zinc stress in these cells as indicated by the up-regulation of zntA and czcI2C2B2′. In contrast, PitA imports not so many metal:phosphate complexes when strain AE104 grows in SGK, and there is consequently no need for czc-mediated zinc efflux. The resulting mild zinc stress is responsible for the specific silencing of the CMGIs specifically in TMM-grown AE104 cells, and also in metal-shocked CH34 cells as outlined above.
Expression of pitA is up-regulated with increasing phosphate content of the growth medium (up to 4 mM phosphate) but is down-regulated again at higher phosphate concentrations, except in the ΔzupT deletion strain.15 This indicates that expression of pitA is influenced by zinc availability rather than by phosphate. Indeed, addition of zinc led to down-regulation of the pitA-lacZ reporter but this process did not require the Zur (FurC) zinc uptake regulator in C. metallidurans,57 indicating the presence of additional zinc-dependent regulatory pathways in this bacterium. This pathway is also responsible for zinc-dependent down-regulation of other metal uptake systems such as CorA1, CorA2, CorA3 (ref. 57) and may have a global regulatory function, which might also include silencing of CMGIs.
The Zur regulon
Zur (previously FurC) is required for control of expression of zupT.57 Besides expression of the genes of the aut region 1 on CMGI-3, which were up-regulated independent of zinc availability (Table 4), the only genes up-regulated in the ΔzupT mutant under metal starvation conditions were those in the adjacent operon regions Op0371f and Op0318r (Table 3). Op0318r contains the gene for a TonB-dependent outer membrane protein Rmet_1104, which could catalyze the transport of zinc compounds across the outer membrane as in the cyanobacterium Anabaena sp.58 The preceding operon region Op0371f comprises 6 genes in the same direction of transcription, which might be a hexacistronic operon. Rmet_1103 is a dihydroorotase, Rmet_1102 a carbonic anhydrase, Rmet_1101 a 6-pyrovoyl-tetrahydropterin synthase, Rmet_1100 a cysteinyl-tRNA synthetase and Rmet_1099 a GTP cyclohydrolase (Table 3), all are zinc-dependent proteins59–62 with paralogs encoded in other parts of the genome. In the case of the cysteinyl-tRNA synthetase, the zinc ion is essential to discriminate between the related amino acids cysteine and serine63 and the metal has a similar importance for the other proteins.The first gene of the putative operon in region Op0317f encodes Rmet_1098 or CobW1, a homolog of YeiR from E. coli,64 which is a zinc-binding metal chaperone and GTPase. Deletion of the respective gene from E. coli leads to sensitivity to EDTA or cadmium caused by zinc depletion.64 When cobW1 was interrupted by a lacZ insertion, the resulting strain displayed a similar EDTA and metal resistance as the ΔzupT strain, and the phenotype of the double mutant was similar to that of the single mutants, and to that of ΔyeiR in E. coli. As judged by the reporter activity, cobW1 was up-regulated by zinc starvation in strain AE104 but more strongly so in the ΔzupT strain. CobW1 and YeiR belong to the COG0523 subfamily of G3E-GTPases, and the other three families of the G3E family of GTPases are represented by HypB, UreG and MeaB, which insert nickel or cobalt ions into hydrogenases, ureases or methylmalonyl-CoA mutases, respectively.64 CobW1 might insert zinc into the proteins encoded downstream of cobW1, and the similar phenotype of the cobW1 insertion mutant and the ΔzupT deletion mutant indicates that the Zn(II) ion might originate from ZupT. CobW1 might therefore “channel” the cytoplasmic zinc repository in C. metallidurans to provide the metal rapidly to zinc-requiring proteins under zinc starvation conditions.
Similar operon regions, all containing the gene for a CobW-like protein and paralogs of zinc-requiring proteins, exist in other bacteria of the Burkholderiales.65 In C. metallidurans, the strong influence of metal starvation (Table 3) and a predicted Zur-binding site upstream of the operon region65 indicates that Op0317f is likely a Zur-dependent operon, although binding of Zur to this site remains to be demonstrated. Zur bound to the predicted site upstream of zupT57 and another Zur-binding site is located upstream of the zur gene. The zur gene itself (Rmet_0128, furC) is the first gene of operon region Op0032r and has a predicted Zur-binding site upstream in the regulatory region. The zur gene is followed by two genes for additional members of the COG0523 protein family, cobW2 (Rmet_0127) and cobW3 (Rmet_0125), which contains another Zur-box upstream, an uncharacterized protein (Rmet_0124), a TonB-dependent outer membrane protein (Rmet_0123), and, between the cobW genes, a dksA gene (Rmet_0126). While the TonB-dependent outer membrane and the uncharacterized protein were not found in the proteome of C. metallidurans, CobW2 was present in about 2000 copies per cell, CobW3 in 300, the DksA protein in 3000 and Zur in 100 copies per cell. These numbers did not change when zupT was deleted.8 These are all no longer putative proteins and are all clearly required for growth under mild zinc starvation conditions in TMM.
DksA
DksA is a global regulator in E. coli that interacts with ppGpp as cofactor, binds through a zinc-finger motif to the RNA polymerase, altering the kinetics of transcription initiation.66C. metallidurans has three genes for DksA paralogs in addition to Rmet_0126, Rmet_4453, 4470, and 4602 on chromosome 2. Although the Rmet_0126 DksA is present in a high copy number in the cell, the other three paralogs were not found in the proteome.8 Nevertheless, expression of Rmet_4602 was down-regulated in ΔzupT compared to AE104 when cultivated in the presence of EDTA. The three putative DksA paralogs Rmet_4453, 4470, and 4602 contained two pairs of conserved cysteine residues that are involved in formation of the zinc-finger motif (CcXCh–X16–CX2C).67,68 In contrast, the Rmet_0126 DksA has three of these cysteine residues changed to threonine, serine or alanine, respectively (CX2T–X16–SX2A). As the copy number of the Rmet_0126 DksA approaches the number of RNA polymerase molecules in the cell, this protein might link the status of the overall transition metal homeostasis with the upper levels of the cellular hierarchical control mechanisms.Experimental
Bacterial strains and growth conditions
Strains used for experiments were derivatives of the strain AE104, a megaplasmid-free derivative of C. metallidurans CH3411 and its ΔzupT deletion mutant.15 Tris-buffered mineral salts medium11 containing 2 g sodium gluconate per L (TMM) was used to cultivate these strains aerobically with shaking at 30 °C. Analytical grade salts of heavy metal chlorides were used to prepare 1 M stock solutions, which were sterilized by filtration. Solid Tris-buffered media contained 20 g agar per L.Autotrophic growth in TMM was performed in serum bottles with a “Knallgas” atmosphere of 80% H2, 10% CO2, and 10% O2 in 90% of the total volume of the bottle. The remaining 10% was filled with the growth medium. The pre-culture was cultivated heterotrophically in TMM on gluconate to carbon exhaustion and subsequently diluted 100-fold with TMM without gluconate for the autotrophic culture.
For autotrophic growth in phosphate-buffered mineral salts medium (SGK),53 the gas atmosphere also contained a mixture of H2, O2, and CO2 (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, vol/vol/vol). Growth rates were determined by optical density measurements of suspensions growing in 300 mL Erlenmeyer flasks, which were shaken in a water bath at 30 °C. For the heterotrophic de-repression of the four key enzymes of the autotrophic metabolism IND medium was used. This medium contained per 1 L: 9.0 g Na2HPO4·12H2O; 1.5 g KH2PO4; 3.0 g NH4CI; 0.2 g MgSO4·7H2O; 0.02 g CaCl2·2H2O; 36 mg iron-ammonium-citrate; 0.1 mL SL6 according to Pfennig 10-fold concentrated;69 18 μg Na2MoO4·2H2O; 42 μg MnCl2·4H2O; 140 μg ZnSO4·7H2O; 3 g glycerol and 9 g sodium gluconate. The pH value was adjusted to 6.9 ± 0.1.
1, vol/vol/vol). Growth rates were determined by optical density measurements of suspensions growing in 300 mL Erlenmeyer flasks, which were shaken in a water bath at 30 °C. For the heterotrophic de-repression of the four key enzymes of the autotrophic metabolism IND medium was used. This medium contained per 1 L: 9.0 g Na2HPO4·12H2O; 1.5 g KH2PO4; 3.0 g NH4CI; 0.2 g MgSO4·7H2O; 0.02 g CaCl2·2H2O; 36 mg iron-ammonium-citrate; 0.1 mL SL6 according to Pfennig 10-fold concentrated;69 18 μg Na2MoO4·2H2O; 42 μg MnCl2·4H2O; 140 μg ZnSO4·7H2O; 3 g glycerol and 9 g sodium gluconate. The pH value was adjusted to 6.9 ± 0.1.
For enzyme purification cells were cultivated in a 10 L fermentor (Biostat, Braun, Melsungen, Germany) at 30 °C, 600 rpm and a gas flow of 500 mL min−1. Cultures grown in IND medium were sparged with air, and cultures of autotrophically growing cells with a mixture of 80% H2, 10% O2 and 10% CO2. Both media additionally contained 2 mL per 8 L polypropylenglycol P200, 20% in 96% ethanol to prevent foam formation. The cells were harvested at the end of the exponential growth phase, washed with KP50 (50 mM potassium phosphate buffer, pH 7.0, containing 10 mM MgCl2) and stored at −20 °C.
Dose–response growth curves in 96-well plates
Growth curves for C. metallidurans were conducted in TMM. A pre-culture was incubated at 30 °C, 250 rpm up to early stationary phase, then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in fresh medium and incubated for 24 h at 30 °C and 250 rpm. Overnight cultures were used to inoculate (at a ratio of 1
20 in fresh medium and incubated for 24 h at 30 °C and 250 rpm. Overnight cultures were used to inoculate (at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) parallel cultures with increasing metal or EDTA concentrations in 96-well plates (Greiner). Cells were cultivated for 20 h at 30 °C and 1300 rpm in a neoLab Shaker DTS-2 (neoLab, Heidelberg, Germany) and the optical density was determined at 600 nm as indicated in a TECAN infinite 200 PRO reader (TECAN, Männersdorf, Switzerland). To calculate the IC50 value (metal concentration that led to turbidity reduction by half) and the corresponding b-value (measure of the slope of the sigmoidal dose–response curve), the data were adapted to the formula OD(c) = OD0/{1 + exp((c − IC50)/b)}, which is a simplified version of a Hill-type equation as introduced by Pace and Scholtz70 as published.71 OD(c) is the turbidity at a given metal concentration, OD0 is the turbidity with no added metal and c the metal concentration.
10) parallel cultures with increasing metal or EDTA concentrations in 96-well plates (Greiner). Cells were cultivated for 20 h at 30 °C and 1300 rpm in a neoLab Shaker DTS-2 (neoLab, Heidelberg, Germany) and the optical density was determined at 600 nm as indicated in a TECAN infinite 200 PRO reader (TECAN, Männersdorf, Switzerland). To calculate the IC50 value (metal concentration that led to turbidity reduction by half) and the corresponding b-value (measure of the slope of the sigmoidal dose–response curve), the data were adapted to the formula OD(c) = OD0/{1 + exp((c − IC50)/b)}, which is a simplified version of a Hill-type equation as introduced by Pace and Scholtz70 as published.71 OD(c) is the turbidity at a given metal concentration, OD0 is the turbidity with no added metal and c the metal concentration.
β-Galactosidase assay and lacZ-reporter constructions
C. metallidurans cells with a lacZ reporter gene fusion were cultivated as a pre-culture in TMM containing 1.5 mg L−1 kanamycin at 30 °C, 250 rpm for 30 h, then diluted into fresh medium to a density of 30 Klett units and incubated at 30 °C. At a cell density of 60 to 70 Klett units, metal salts were added in different concentrations and cells were incubated with shaking for a further 3 h and harvested by centrifugation (30 min at 5000 rpm). The specific β-galactosidase activity was determined in permeabilized cells as previously published with 1 U defined as the activity forming 1 nmol of o-nitrophenol min−1 at 30 °C.72 The lacZ reporter gene was inserted into the target gene (Rmet_1098) to construct reporter operon fusions. This was done by single cross-over recombination in C. metallidurans strains. A 300–400 bp PCR-product of the central region of the genes (Rmet_1098/cobW1) were amplified from total DNA of strain CH34 (primer: Rmet_1098 PstI Dis – AAACTGCAGGGCCGCAGTCTCAATGAGG & Rmet_1098 XbaI Dis – AAATCTAGAGGGCGCTTTCGATGCTTCC) and the resulting fragments were cloned into plasmid pECD794 (pLO2-lacZ).17 The respective operon fusion cassettes were inserted into the open-reading frame of the target gene by conjugation and single cross-over recombination.Fluorescence assay and gfp-reporter plasmid constructions
C. metallidurans cells with a gfp reporter gene promoter fusion plasmid were cultivated as a pre-culture in TMM containing 1.5 mg L−1 kanamycin at 30 °C. Cultures were shaken at 250 rpm for 30 h, then diluted into fresh medium to a density of 30 Klett units and incubated further at 30 °C. At a cell density of 60 to 70 Klett units, metal salts or EDTA were added to different final concentrations and cells were incubated with shaking for 18 h at 30 °C in a neoLab Shaker DTS-2 (neoLab, Heidelberg, Germany).The specific GFP fluorescence was calculated as nmol μg−1 (dry weight) by using an extinction coefficient (EGFP) at 488 nm of ε = 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 l mol−1 cm−1, with a layer thickness of the well and coefficient of culture volume to the volume of the reaction mixture. Measurement of the emission wavelength at 518 nm was performed after excitation at 488 nm and the optical density at 600 nm was also measured using a TECAN infinite 200 PRO reader (TECAN, Männersdorf, Switzerland). The dry weight per volume was calculated from the turbidity measurements, using a calibration curve.
000 l mol−1 cm−1, with a layer thickness of the well and coefficient of culture volume to the volume of the reaction mixture. Measurement of the emission wavelength at 518 nm was performed after excitation at 488 nm and the optical density at 600 nm was also measured using a TECAN infinite 200 PRO reader (TECAN, Männersdorf, Switzerland). The dry weight per volume was calculated from the turbidity measurements, using a calibration curve.
The gfp reporter gene was amplified (primer: GFP13 KpnI – 5′ AAAGGTACC-ATACATATGGCTAGCAAAG 3′ and GFP13 NsiI 5′ TTAATGCAT-AGTGCTCGAATTCATTATTT 3′) from source Plasmid pMUTIN-GFP+73 and inserted into pBBR1-MSC274 after digestion (NsiI/KpnI) to construct the reporter plasmid. A 100–150 bp PCR-product of the promotor region of the genes (Rmet_2621 zupT & Rmet_1098/cobW1) was amplified from total DNA of strain CH34 or AE104 (primer: zupTp HindIII – 5′ AAAAAGCTT-CTGCGCTGGCCGCTTCTTC 3′ and zupTp KpnI – 5′ AAAGGTACC-CGATTAACGCAACAATGTTGC 3′/cobW1p KpnI – 5′ AAAGGTACC-GGATTTGGTTTGCCCGCAAG 3′ and cobW1p SpeI – 5′ AAAACTAGT-TTCAGGCGACGCAGATAGAC 3′) and the resulting fragments were cloned after digestion (KpnI & SpeI) into plasmid pBBR1-MSC2Φgfp+. The zupT promoter region had to be subcloned into the pGEM T-easy (Promega) prior to subsequent subcloning.
Activity stain for membrane-bound hydrogenases
Hydrogen-dependent activities of hydrogenases were visualized by chromogenic detection in non-denaturing polyacrylamide gel electrophoresis (PAGE) using 7.5% (w/v) polyacrylamide, pH 8.5 included 0.1% (w/v) Triton X-100 in the gels. Cell debris, soluble extracts and/or membrane fractions were dissolved in 50 mM Tris-HCl pH 8.0; 150 mM NaCl buffer. Samples (50 μg of protein) were incubated with 4% (w/v) Triton X-100 prior to application to the gels. Hydrogenase activity staining was done as described in Ballantine and Boxer22 except that the buffer used was 50 mM MOPS pH 7.0, 0.5 mM BV (benzyl viologen) and 1 mM TTC (2,3,5-triphenyltetrazolium chloride) and under anaerobic conditions at RT.Determination of total hydrogenase enzyme activity
Hydrogenase enzyme activity (H2-dependent reduction of benzyl viologen) determines the activities of hydrogenases and was measured according to Ballantine and Boxer22 except that the buffer used was 50 mM MOPS, pH 7.0. No detergent was added to extracts measured by this method. In order to measure hydrogenase activity in solution a wavelength of 578 nm was used, and a molar extinction coefficient value of 8600 M−1 cm−1 was assumed for reduced benzyl viologen (BV). One unit of activity corresponded to the reduction of 1 μmol of hydrogen per min. Experiments were performed minimally two times and each time in duplicate. Data are presented as standard deviation of the mean.75Enzyme assays with purified proteins
NAD+ reduction catalyzed by the soluble hydrogenase was measured photometrically as outlined.76 The method of Schink and Schlegel (1979) was used for the photometric determination of methylene blue reduction catalyzed by the membrane-bound hydrogenase,77 but glucose, glucose-oxidase, and catalase were omitted. Anaerobic conditions were established by flushing the cuvette, sealed with a rubber stopper, for 10 min with oxygen-free hydrogen. The activity of ribulosebisphosphate carboxylase was assayed radiometrically,78 the activity of the phosphoribulokinase either radiometrically or photometrically.79 Protein was determined by the method of Bradford.80Electrophoretic methods
Polyacrylamide gel electrophoresis was performed in a vertical slab gel apparatus (Pantaphor, Fa. Müller, Hann. Münden, Germany) with gels of various concentration and TRIS borate buffer (pH 8.9 or 7.9) as described.81 To stain gels for hydrogenase activity,82 the gels were incubated between 1 and 3 h at 30 °C in the dark under a hydrogen atmosphere in 200 mL of 50 mM potassium phosphate buffer, pH 8.0, containing 12 mg 4-nitrobluetetrazolium chloride, 9 mg phenazine methosulfate and, in the case of staining the soluble hydrogenase, 200 μmol of NAD+. Protein was stained with Coomassie brilliant blue.83 Sodium dodecylsulfate polyacrylamidgel electrophoresis (SDS-PAGE) was performed in the same apparatus using various polyacrylamide concentrations as described.84 Electrophoresis was carried out in 100 mM sodium phosphate buffer, pH 7.2, containing 2% SDS. Samples were prepared as follows: solutions of 1 mg protein per mL in 10 mM sodium phosphate buffer, pH 7.2, were incubated with 2% (w/v) SDS and 5% (v/v) 2-mercaptoethanol for 3 min at 100 °C. Cytochrome c, chymotrypsinogen, ovalbumin, bovine serum albumin, aldolase, catalase and ferritin were used as molecular mass standards.Determination of the molecular mass
The relative molecular masses of the enzymes were determined by gel filtration on a Sephacryl S300 column.85 The same proteins as mentioned above were used as standard proteins. The relative molecular masses were also determined by sucrose gradient centrifugation.86 A linear sucrose density gradient (5 to 30% sucrose in 50 mM potassium phosphate buffer, pH 7.0) of 11 mL volume was used. The experiment was performed at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 28 h in a swing-out rotor and an OmegalI-ultracentrifuge (Christ, Osterode, Germany). Catalase and lactate-dehydrogenase were used as standard proteins.
000 rpm and 4 °C for 28 h in a swing-out rotor and an OmegalI-ultracentrifuge (Christ, Osterode, Germany). Catalase and lactate-dehydrogenase were used as standard proteins.
Purification procedures
One aim of this work was to purify all of the four key enzymes of the autotrophic metabolism from one cell suspension. Therefore, the purification procedures had steps in common. All purifications involved affinity chromatography using Procion-dye sepharose. The Procion dyes were coupled to Sepharose 4B following the procedure of Atkinson et al.87 They were a gift from the German ICI (Frankfurt).To prepare the crude extract, the cells were suspended in buffer A (20 mM imidazole-HCI buffer containing 10 mM MgCl2 and 500 μM dithioerythrol) to a final volume of 3 mL per g wet weight. 1 mg DNAse per mL of cell suspension was added, then the suspension was passaged twice through a French press (American Instruments Inc., Silver Springs, Maryland, USA) at 1560 kPa cm−2 and the mixture was subsequently centrifuged (10 min 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, Zeta 20, Christ) at 4 °C to remove unbroken cells and cell debris. The resulting cell-free extract was referred to as crude extract for the soluble enzymes, and was centrifuged for 1 h at 35
000 rpm, Zeta 20, Christ) at 4 °C to remove unbroken cells and cell debris. The resulting cell-free extract was referred to as crude extract for the soluble enzymes, and was centrifuged for 1 h at 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm in an Omicron ultracentrifuge (Christ). The sediment was used for the preparation of the membrane-bound hydrogenase, the supernatant for the three soluble enzymes, the soluble hydrogenase, the ribulose bisphosphate carboxylase and the phosphoribulokinase.
000 rpm in an Omicron ultracentrifuge (Christ). The sediment was used for the preparation of the membrane-bound hydrogenase, the supernatant for the three soluble enzymes, the soluble hydrogenase, the ribulose bisphosphate carboxylase and the phosphoribulokinase.
To purify the membrane-bound hydrogenase, the sediment from the ultracentrifugation step was re-suspended in 20 mM potassium phosphate buffer (KP20), pH 7.0, containing 0.086 g sucrose per mL and 0.031 mL sodium chloride solution (5 M) per mL suspension, stirred for 15 min at 23 °C and sedimented in the ultracentrifuge (conditions as above). The second sediment also was re-suspended in KP20, pH 7.0, to a final volume of 1 mL per 200 mg membranes. Per mL, 0.1 g sucrose and 0.05 mL solubilization mixture77 (100 mL L−1 Triton X-100, 20 g L−1 sodium deoxycholate, 200 mM EDTA) were added. The solution was carefully stirred at room temperature for 30 min and centrifuged (10 min at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, Zeta 20). This solution was applied onto a column of Procion-Blue HERD-sepharose matrix equilibrated with KP20, pH 7.0, and washed with the same buffer. The hydrogenase was eluted with 1 M KCI in KP20, pH 7.0. Fractions with high hydrogenase activity were combined, concentrated by ultrafiltration in a diaflow chamber (Amicon Corp., USA) using a PM10 filter and layered on the top of a Sephacryl S300 column. KP20, pH 8.0, was used for pre-equilibration and for elution. The most active fractions were combined, concentrated by ultrafiltration as above and applied onto a column of DEAE-Sephacel. This latter column was washed with KP20, pH 8.0, and eluted with a linear gradient of 0 to 1 M KCl in the same buffer.
000 rpm, Zeta 20). This solution was applied onto a column of Procion-Blue HERD-sepharose matrix equilibrated with KP20, pH 7.0, and washed with the same buffer. The hydrogenase was eluted with 1 M KCI in KP20, pH 7.0. Fractions with high hydrogenase activity were combined, concentrated by ultrafiltration in a diaflow chamber (Amicon Corp., USA) using a PM10 filter and layered on the top of a Sephacryl S300 column. KP20, pH 8.0, was used for pre-equilibration and for elution. The most active fractions were combined, concentrated by ultrafiltration as above and applied onto a column of DEAE-Sephacel. This latter column was washed with KP20, pH 8.0, and eluted with a linear gradient of 0 to 1 M KCl in the same buffer.
As common steps for the soluble enzymes, 0.03 mg protamine sulfate were dissolved in 2 mL buffer A per mg of total protein in the supernatant of the first ultracentrifugation step. This solution was added drop-wise with stirring to the supernatant, and stirring was continued for 20 min. The resulting precipitate was removed by centrifugation (20 min 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, Zeta 20). The supernatant was fractionated by addition of pulverized (NH4)2SO4 to give a 25% (w/v) saturated solution. After centrifugation (10 min 15
000 rpm, Zeta 20). The supernatant was fractionated by addition of pulverized (NH4)2SO4 to give a 25% (w/v) saturated solution. After centrifugation (10 min 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, Zeta 20) the precipitate was discarded. Then the supernatant solution was brought to 40% (w/v) saturation of ammonium sulfate and again centrifuged. The pellet contained the CO2-fixing enzymes, the supernatant the soluble hydrogenase.
000 rpm, Zeta 20) the precipitate was discarded. Then the supernatant solution was brought to 40% (w/v) saturation of ammonium sulfate and again centrifuged. The pellet contained the CO2-fixing enzymes, the supernatant the soluble hydrogenase.
For purification of the soluble hydrogenase, this supernatant was brought to 60% (w/v) saturation of (NH4)2SO4 and centrifuged. The supernatant was discarded, the sediment dissolved in KP20, pH 7.0, and the conductivity of the final solution adjusted to that of 100 mM KCl in the same buffer. This solution was applied onto a DEAE-Sephacel column. The column was washed with KP20, pH 7.0, and eluted with a linear gradient (0–300 mM KCl) in the same buffer. The most active fractions were combined, concentrated by ultrafiltration as described above and layered on the top of a Sephacryl S300 column. KP20, pH 6.2, was used for pre-equilibration and for elution. The active fractions were combined, concentrated by ultrafiltration and applied onto a Procion-Yellow HE6g column. This last column was washed with KP20, pH 6.2, and eluted with KP20, pH 7.0, containing 50 mM KCl.
The sediment of the second ammonium sulfate precipitation containing the ribulose 1,5-bisphosphate carboxylase and phosphoribulokinase was dissolved in buffer A. It was layered on top of a Sephacryl S300 column, buffer A was used for pre-equilibration and for elution. The fractions within a broad range of activity were collected, concentrated by ultrafiltration and applied onto a Procion-Red HE7b-sepharose column. The column was washed with buffer A and eluted with the same buffer containing 1 M KCl. The active fractions were collected, concentrated by ultrafiltration and again layered on top of a Sephacryl S300 column, which was also pre-equilibrated and eluted with buffer A to separate both enzymes. The most active fractions of both enzymes were collected and concentrated by ultrafiltration.
Aut-negative mutants
Mutants were generated from strain AE104 that were no longer able to grow autotrophically in SGK medium. To isolate the mutants, strain AE104 was treated with the mutagenic agents nitrite, EMS (ethyl-methansulfonate), NMG (N-methyl-N′-nitro-N-nitroso-guanidine), or mitomycin C. For treatment with nitrite, a volume of 5 mL of an overnight NB (nutrient broth) culture were harvested by centrifugation, washed in 5 mL 0.1 M sodium acetate buffer, pH 4.6, and suspended in 1 mL of the same buffer. Solid NaNO2 was added to a final concentration of 50 mM. The cells were incubated 10 min at 30 °C, then 9 mL SGK medium were added. The cells were sedimented by low-spin centrifugation and re-suspended in 1 mL SGK. About 0.5 × 106 of the cells survived this treatment. For EMS mutagenesis, 2.5 mL of an overnight NB culture were added to 2.5 mL EMS-solution (2.4 mL SGK + 0.1 mL EMS). The solution was shaken for 2 h at 37 °C, then 45 mL SGK were added to stop EMD treatment. For NMG, 10 mL of an overnight NB culture were harvested by centrifugation and re-suspended in 1 mL TMA (1 L H2O + 6 g Tris + 5.8 g maleic acid + 1 g MgSO4·7H2O + 5 mg Ca(NO3)2). 100 μL NMG-solution (1 g L−1) were added, the cells were incubated for 30 min at 30 °C, sedimented and resuspended in 1 mL SGK. Finally, cells were grown in SGK containing 0.2% (w/v) sodium gluconate and different amounts of mitomycin C in the range of 0.2 to 50 mg L−1 were added.As a negative control, the cells not treated with any mutagenic agents were diluted and plated on SGK-agar containing 50 mg L−1 sodium pyruvate to allow heterotrophic growth of small pin-point colonies during incubation under air for 2 d. Subsequent cultivation for 3 d under an atmosphere of 80% (v/v) H2 + 10% O2 + 10% CO2 allowed autotrophic growth of parental cells while mutants ceased to grow and remained as pin-points. A total of 3200 of these small colonies were randomly selected and tested for their ability to grow autotrophically on SGK medium. The cells that had been treated with a mutagenic agent were also diluted and plated onto SGK-agar containing 50 mg L−1 sodium pyruvate, and the same procedure was followed. Again, 3200 pin-point colonies were tested. In a third independent experiment, the mutagenized cells were sedimented and resuspended in 1 mL SGK. This cell suspension was used as an inoculum for 19 mL SGK containing 9 g L−1 sodium gluconate and 3 g L−1 glycerol. The culture was shaken for 2 d at 30 °C, 2.5 mL of the culture were washed twice with SGK and resuspended in 20 mL SGK containing 3 g L−1L-histidine. The culture was incubated 2 d under air following 1.5 d under an atmosphere of 80% (v/v) H2 + 10% O2 + 10% CO2, D-cycloserine was added to a final concentration of 1 g L−1 to kill autotrophically growing cells, allowing enrichment of mutants. The incubation under a gas atmosphere of 80% (v/v) H2 + 10% O2 + 10% CO2 was continued for 5 h, the cells were washed twice with SGK and resuspended in 10 mL SGK. This suspension was used for pin-point colony selection as described above.
Characterization of the mutants
The mutant strains were grown in IND medium for 3 d, harvested by centrifugation, and disrupted by ultrasonication (3 min at 12.5 μ in a MSE ultrasonic des integrator MK2). The suspension was centrifuged twice (10 min 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, Zeta 20, and 35
000 rpm, Zeta 20, and 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 1 h, Omicron). In the supernatant the specific activity of the soluble hydrogenase, the ribulose 1,5-bisphosphate carboxylase, and the phosphoribulokinase was determined. The membranes were washed twice in KP50, pH 7.0, containing 10 mM MgCI2, dissolved in the same buffer and used for the determination of the activity of the membrane-bound hydrogenase.
000 rpm, 1 h, Omicron). In the supernatant the specific activity of the soluble hydrogenase, the ribulose 1,5-bisphosphate carboxylase, and the phosphoribulokinase was determined. The membranes were washed twice in KP50, pH 7.0, containing 10 mM MgCI2, dissolved in the same buffer and used for the determination of the activity of the membrane-bound hydrogenase.
Reversion-analysis
The mutant strains were grown overnight in NB, diluted and plated onto NB and SGK agar. The NB-plates were incubated under air, the SGK-plates under “Knallgas” conditions (80% (v/v) H2 + 10% O2 + 10% CO2). The spontaneous reversion rate was calculated as the ratio of the cells able to grow autotrophically divided by the total cell number. Treating the cells with the chemical procedure that had originally been used to produce the respective mutant strain also induced reversion, and subsequently the reversion rate was determined.RNA isolation
C. metallidurans CH34, AE104 and ΔzupT cells were cultivated heterotrophically in TMM as described above. For the main culture EDTA or zinc chloride were added. At a cell turbidity of 100 Klett, the cells were rapidly harvested at 4 °C and stored at −80 °C. Total RNA was isolated with RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. A second DNase treatment with a precipitation step was performed. To exclude experimental artifacts resulting from DNA contaminations, only RNA was used that did not generate products in a PCR reaction with chromosomal primers. RNA concentration was determined photometrically, and RNA quality was checked on formamide gels88 and measured as RNA integrity number (RIN) on an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany).Microarrays of C. metallidurans
C. metallidurans parental strain CH34 was cultivated without additions (three repeats). Strain AE104 (three per condition) and ΔzupT (two repeats per condition) were either grown for 10 min in the presence of 50 μM EDTA or 10 μM zinc chloride. Moreover, strain AE104 was cultivated with or without 100 μM zinc chloride (added for 10 min at a turbidity of 100 Klett). Isolated and quality-checked RNA were provided to IMGM Laboratories GmbH (Martinsried, Germany) for hybridization with a C. metallidurans Agilent Custom GE microarray (8 × 15 K) (Agilent Technologies, Waldbronn, Germany) with a single color (Cy3)-based protocol. Signals were detected using the Agilent DNA microarray Scanner. Software tool Feature Extraction 10.7.3.1. was used for raw data extraction.In the algorithm used, (i) the mean intensity of the pixels of the surrounding area was subtracted from the mean density of the pixels of the spots to give the signal strength. Its deviation was half of the sum of both intensity deviations. The distance (D) value was the distance between spot and background pixel intensities divided by the sum of the deviations. D was a more useful value than Student's t-test because non-touching deviation bars of two values (D > 1) at three repeats always indicates at least a significant (>95%) difference. Signals were further processed if D > 1. Subsequently, (ii) the mean values of the biological repeats were calculated plus their deviation and the smallest D value. Third, (iii) the values from different spots, positions or oligonucleotides assigned to the same gene were taken to calculate a gene-specific mean value and minimum D-value. Finally, (iv) the respective spot signals coming from various growth conditions were compared (Q- and D-values, respectively).
Microarray data accession number
The microarray data were deposited in the GEO database at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE64196.Conclusions
The aut region 1 on CMGI-3, which harbors genes for the soluble hydrogenase of C. metallidurans, and aut region 2 on CMGI-2, which harbors the genes encoding the membrane-bound hydrogenase, are silenced under conditions of a disturbed metal ion homeostasis or increased zinc availability. This happens when strain CH34 is confronted with a transition metal mixture or when the plasmid-free strain AE104, devoid of the sophisticated plasmid-encoded metal resistance determinants, is cultivated in a phosphate-poor Tris-buffered medium that does not complex transition metal cations very strongly and allows their rapid metal uptake. Histone-like proteins or sigma factors might be involved in this silencing process. Additionally, a global zinc-dependent regulatory process in C. metallidurans might be involved, which also controls metal uptake systems and acts in parallel with the Zur zinc uptake regulator. The presence of a dksA gene in the same operon region as zur and cobW2 indicates that an interplay between DksA proteins might be responsible for this global zinc-dependent regulatory process. In a second step, silencing of the genes in aut region 1 is reverted in the ΔzupT mutant of AE104, which has problems to allocate zinc efficiently to zinc-dependent proteins, or in a sigma factor triple mutant with a high cellular nickel content. Together, these data suggest that the zinc-dependent nickel discrimination proteins HypA and HypB are responsible for the “un-silencing” of aut region 1, as already discussed in detail elsewhere.8Acknowledgements
Funding for this work was provided by the Deutsche Forschungsgemeinschaft (Ni262/10). We thank Grit Schleuder for skillful technical assistance and Gary Sawers for helpful comments.Notes and references
- K. J. Waldron, J. C. Rutherford, D. Ford and N. J. Robinson, Nature, 2009, 460, 823–830 CrossRef CAS PubMed.
- S. Tottey, D. R. Harvie and N. J. Robinson, in Molecular microbiology of heavy metals, ed. D. H. Nies and S. Silver, Springer-Verlag, Berlin, 2007, vol. 6, pp. 3–36 Search PubMed.
- D. H. Nies, in Molecular microbiology of heavy metals, ed. D. H. Nies and S. Silver, Springer-Verlag, Berlin, 2007, vol. 6, pp. 118–142 Search PubMed.
- Z. Ma, P. Chandrangsu, T. C. Helmann, A. Romsang, A. Gaballa and J. D. Helmann, Mol. Microbiol., 2014, 94, 756–770 CrossRef CAS PubMed.
- K. Helbig, C. Bleuel, G. J. Krauss and D. H. Nies, J. Bacteriol., 2008, 190, 5431–5438 CrossRef CAS PubMed.
- C. Große, G. Schleuder, C. Schmole and D. H. Nies, Appl. Environ. Microbiol., 2014, 80, 7071–7078 CrossRef PubMed.
- J. A. Imlay, J. Biol. Chem., 2014, 289, 28121–28128 CrossRef CAS PubMed.
- M. Herzberg, D. Dobritzsch, S. Helm, S. Baginski and D. H. Nies, Metallomics, 2014, 6, 2157–2165 RSC.
- Y. Li and D. B. Zamble, Chem. Rev., 2009, 109, 4617–4643 CrossRef CAS PubMed.
- R. P. Hausinger and D. B. Zamble, in Molecular Microbiology of heavy metals, ed. D. H. Nies and S. Silver, Springer-Verlag, Berlin, 2007, pp. 287–320 Search PubMed.
- M. Mergeay, D. Nies, H. G. Schlegel, J. Gerits, P. Charles and F. van Gijsegem, J. Bacteriol., 1985, 162, 328–334 CAS.
- C. Schafer, B. Friedrich and O. Lenz, Appl. Environ. Microbiol., 2013, 79, 5137–5145 CrossRef PubMed.
- M. Bernhard, T. Buhrke, B. Bleijlevens, A. L. De Lacey, V. M. Fernandez, S. P. J. Albracht and B. Friedrich, J. Biol. Chem., 2001, 276, 15592–15597 CrossRef CAS PubMed.
- P. J. Janssen, R. Van Houdt, H. Moors, P. Monsieurs, N. Morin, A. Michaux, M. A. Benotmane, N. Leys, T. Vallaeys, A. Lapidus, S. Monchy, C. Medigue, S. Taghavi, S. McCorkle, J. Dunn, D. van der Lelie and M. Mergeay, PLoS One, 2010, 5, e10433 Search PubMed.
- A. Kirsten, M. Herzberg, A. Voigt, J. Seravalli, G. Grass, J. Scherer and D. H. Nies, J. Bacteriol., 2011, 193, 4652–4663 CrossRef CAS PubMed.
- D. H. Nies, FEMS Microbiol. Rev., 2003, 27, 313–339 CrossRef CAS PubMed.
- J. Scherer and D. H. Nies, Mol. Microbiol., 2009, 73, 601–621 CrossRef CAS PubMed.
- T. von Rozycki and D. H. Nies, Antonie van Leeuwenhoek, 2009, 96, 115–139 CrossRef CAS PubMed.
- T. von Rozycki, D. H. Nies and M. H. J. Saier, Comp. Funct. Genomics, 2005, 6, 17–56 CrossRef CAS PubMed.
- M. H. J. Saier, C. V. Tran and R. D. Barabote, Nucleic Acids Res., 2006, 34, D181–D186 CrossRef CAS PubMed.
- M. Herzberg, L. Bauer and D. H. Nies, Metallomics, 2014, 6, 421–436 RSC.
- S. P. Ballantine and D. H. Boxer, J. Bacteriol., 1985, 163, 454–459 CAS.
- I. H. Segel, Enzyme kinetics, John Wiley and Sons, New York, 1975 Search PubMed.
- S. Frielingsdorf, J. Fritsch, A. Schmidt, M. Hammer, J. Lowenstein, E. Siebert, V. Pelmenschikov, T. Jaenicke, J. Kalms, Y. Rippers, F. Lendzian, I. Zebger, C. Teutloff, M. Kaupp, R. Bittl, P. Hildebrandt, B. Friedrich, O. Lenz and P. Scheerer, Nat. Chem. Biol., 2014, 10, U378–U392 CrossRef PubMed.
- J. Fritsch, P. Scheerer, S. Frielingsdorf, S. Kroschinsky, B. Friedrich, O. Lenz and C. M. T. Spahn, Nature, 2011, 479, 249–252 CrossRef CAS PubMed.
- A. Toussaint, C. Merlin, S. Monchy, M. A. Benotmane, R. Leplae, M. Mergeay and D. Springael, Appl. Environ. Microbiol., 2003, 69, 4837–4845 CrossRef CAS PubMed.
- R. Van Houdt, S. Monchy, N. Leys and M. Mergeay, Antonie van Leeuwenhoek, 2009, 96, 205–226 CrossRef CAS PubMed.
- R. Antoine, F. Jacob-Dubuisson, H. Drobecq, E. Willery, S. Lesjean and C. Locht, J. Bacteriol., 2003, 185, 1470–1474 CrossRef CAS PubMed.
- Y. Taniguchi, P. J. Choi, G.-W. Li, H. Chen, M. Babu, J. Hearn, A. Emili and X. S. Xie, Science, 2010, 329, 533–538 CrossRef CAS PubMed.
- M. L. Coleman, M. B. Sullivan, A. C. Martiny, C. Steglich, K. Barry, E. F. DeLong and S. W. Chisholm, Science, 2006, 311, 1768–1770 CrossRef CAS PubMed.
- M. Gaillard, T. Vallaeys, F. J. Vorholter, M. Minoia, C. Werlen, V. Sentchilo, A. Puhler and J. R. van der Meer, J. Bacteriol., 2006, 188, 1999–2013 CrossRef CAS PubMed.
- J. E. Peters, A. D. Fricker, B. J. Kapili and M. T. Petassi, Mol. Microbiol., 2014, 93, 1084–1092 CAS.
- C. Dressler, U. Kües, D. H. Nies and B. Friedrich, Appl. Environ. Microbiol., 1991, 57, 3079–3085 CAS.
- S. Kar, R. Edgar and S. Adhya, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 16397–16402 CrossRef CAS PubMed.
- M. Macvanin and S. Adhya, Biochim. Biophys. Acta, Gene Regul. Mech., 2012, 1819, 830–835 CrossRef CAS PubMed.
- M. Macvanin, R. Edgar, F. Cui, A. Trostel, V. Zhurkin and S. Adhya, J. Bacteriol., 2012, 194, 6046–6055 CrossRef CAS PubMed.
- J. Oberto, S. Nabti, V. Jooste, H. Mignot and J. Rouviere-Yaniv, PLoS One, 2009, 4, e4367 Search PubMed.
- A. Balandina, L. Claret, R. Hengge-Aronis and J. Rouviere-Yaniv, Mol. Microbiol., 2001, 39, 1069–1079 CrossRef CAS PubMed.
- A. Kuroda, K. Nomura, N. Takiguchi, J. Kato and H. Ohtake, Cell. Mol. Biol., 2006, 52, 23–29 Search PubMed.
- A. Ishihama, A. Kori, E. Koshio, K. Yamada, H. Meada, T. Shimada, H. Makinoshima, A. Iwata and N. Fujita, J. Bacteriol., 2014, 196, 2718–2727 CrossRef PubMed.
- W. W. Navarre, S. Porwollik, Y. P. Wang, M. McClelland, H. Rosen, S. J. Libby and F. C. Fang, Science, 2006, 313, 236–238 CrossRef CAS PubMed.
- S. T. Arold, P. G. Leonard, G. N. Parkinson and J. E. Ladbury, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15728–15732 CrossRef CAS PubMed.
- J. D. Helmann, Adv. Microb. Phys., 2002, 46, 47–110 CAS.
- M. A. Lonetto, K. L. Brown, K. E. Rudd and M. J. Buttner, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 7573–7577 CrossRef CAS.
- C. Große, S. Friedrich and D. H. Nies, J. Mol. Microbiol. Biotechnol., 2007, 12, 227–240 CrossRef PubMed.
- D. H. Nies, Arch. Microbiol., 2004, 181, 255–268 CrossRef CAS PubMed.
- D. H. Nies, G. Rehbein, T. Hoffmann, C. Baumann and C. Grosse, J. Mol. Microbiol. Biotechnol., 2006, 11, 82–93 CrossRef CAS PubMed.
- C. Tibazarwa, S. Wuertz, M. Mergeay, L. Wyns and D. van der Lelie, J. Bacteriol., 2000, 182, 1399–1409 CrossRef CAS PubMed.
- G. Grass, B. Fricke and D. H. Nies, BioMetals, 2005, 18, 437–448 CrossRef CAS PubMed.
- G. Grass, C. Große and D. H. Nies, J. Bacteriol., 2000, 182, 1390–1398 CrossRef CAS PubMed.
- S. Leonhartsberger, A. Huber, F. Lottspeich and A. Böck, J. Mol. Biol., 2001, 307, 93–105 CrossRef CAS PubMed.
- R. C. Weast, CRC handbook of chemistry and physics, CRC Press, Inc., Boca Raton, Florida, USA, 64th edn, 1984 Search PubMed.
- H. G. Schlegel, G. Gottschalk and H. Kaltwasser, Arch. Microbiol., 1961, 38, 209–222 CAS.
- R. M. C. Dawson, D. C. Elliott, W. H. Elliott and K. M. Jones, Data for biochemical research, Clarendon Press, Oxford, 2nd edn, 1969 Search PubMed.
- R. J. Jackson, M. R. B. Binet, L. J. Lee, R. Ma, A. I. Graham, C. W. McLeod and R. K. Poole, FEMS Microbiol. Lett., 2008, 289, 219–224 CrossRef CAS PubMed.
- S. McCarthy, C. Ai, G. Wheaton, R. Tevatia, V. Eckrich, R. Kelly and P. Blum, J. Bacteriol., 2014, 196, 3562–3570 CrossRef PubMed.
- C. Schmidt, C. Schwarzenberger, C. Grosse and D. H. Nies, J. Bacteriol., 2014, 196, 3461–3471 CrossRef PubMed.
- M. Napolitano, M. A. Rubio, J. Santamaria-Gomez, E. Olmedo-Verd, N. J. Robinson and I. Luque, J. Bacteriol., 2012, 194, 2426–2436 CrossRef CAS PubMed.
- B. Thony, G. Auerbach and N. Blau, Biochem. J., 2000, 347, 1–16 CrossRef CAS PubMed.
- D. C. Brown and K. D. Collins, J. Biol. Chem., 1991, 266, 1597–1604 CAS.
- C. M. Zhang, T. Christian, K. J. Newberry, J. J. Perona and Y. M. Hou, J. Mol. Biol., 2003, 327, 911–917 CrossRef CAS PubMed.
- G. Auerbach, A. Herrmann, A. Bracher, G. Bader, M. Gutlich, M. Fischer, M. Neukamm, M. Garrido-Franco, J. Richardson, H. Nar, R. Huber and A. Bacher, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 13567–13572 CrossRef CAS PubMed.
- C. M. Zhang, J. J. Perona and Y. M. Hou, Biochemistry, 2003, 42, 10931–10937 CrossRef CAS PubMed.
- C. E. Blaby-Haas, J. A. Flood, V. de Crecy-Lagard and D. B. Zamble, Metallomics, 2012, 4, 488–497 RSC.
- C. E. Haas, D. A. Rodionov, J. Kropat, D. Malasarn, S. S. Merchant and V. de Crecy-Lagard, BMC Genomics, 2009, 10, 1–21 CrossRef PubMed.
- P. Chandrangsu, J. J. Lemke and R. L. Gourse, Mol. Microbiol., 2011, 80, 1337–1348 CrossRef CAS PubMed.
- A. Perederina, V. Svetlov, M. N. Vassylyeva, T. H. Tahirov, S. Yokoyama, I. Artsimovitch and D. G. Vassylyev, Cell, 2004, 118, 297–309 CrossRef CAS PubMed.
- M. N. Vassylyeva, A. A. Perederina, V. Svetlov, S. Yokoyama, I. Artsimovitch and D. G. Vassylyev, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 1611–1613 CrossRef PubMed.
- N. Pfennig, Arch. Microbiol., 1974, 100, 197–206 CrossRef CAS.
- C. N. Pace and M. J. Scholtz, in Protein structure: a practical approach, ed. T. Creighton, IRL press, Oxford, UK, 1997, pp. 299–322 Search PubMed.
- A. Anton, A. Weltrowski, J. H. Haney, S. Franke, G. Grass, C. Rensing and D. H. Nies, J. Bacteriol., 2004, 186, 7499–7507 CrossRef CAS PubMed.
- D. H. Nies, J. Bacteriol., 1992, 174, 8102–8110 CAS.
- M. Kaltwasser, T. Wiegert and W. Schumann, Appl. Environ. Microbiol., 2002, 68, 2624–2628 CrossRef CAS PubMed.
- M. E. Kovach, P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, 2nd and K. M. Peterson, Gene, 1995, 166, 175–176 CrossRef CAS PubMed.
- C. Pinske, S. Kruger, B. Soboh, C. Ihling, M. Kuhns, M. Braussemann, M. Jaroschinsky, C. Sauer, F. Sargent, A. Sinz and R. G. Sawers, Arch. Microbiol., 2011, 193, 893–903 CrossRef CAS PubMed.
- K. Schneider and H. G. Schlegel, Biochim. Biophys. Acta, 1976, 452, 66–80 CrossRef CAS.
- B. Schink and H. G. Schlegel, Biochim. Biophys. Acta, 1979, 567, 315–324 CrossRef CAS.
- B. Bowien, F. Mayer, G. A. Codd and H. G. Schlegel, Arch. Microbiol., 1976, 110, 157–166 CrossRef CAS PubMed.
- K. Siebert, P. Schobert and B. Bowien, Biochim. Biophys. Acta, 1981, 658, 35–44 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- H. Stegemann, H. Francksen and V. Macko, Z. Naturforsch., 1973, 28C, 722–732 Search PubMed.
- M. Pinkwart, K. Schneider and H. G. Schlegel, Biochim. Biophys. Acta, 1983, 745, 262–278 Search PubMed.
- K. Weber and M. Osborn, J. Biol. Chem., 1969, 244, 4406–4412 CAS.
- K. Weber, J. R. Pringle and M. Osborn, Methods Enzymol., 1972, 26, 3–27 CAS.
- P. Andrews, Biochem. J., 1964, 91, 222–233 CrossRef CAS PubMed.
- R. G. Martin and B. N. Ames, J. Biol. Chem., 1961, 236, 1372–1379 CAS.
- T. Atkinson, P. M. Hammond, R. D. Hartwell, P. Hughes, M. D. Scawen, R. F. Sherwood, D. A. P. Small, C. J. Bruton, M. I. Harvey and C. R. Lowe, Biochem. Soc. Trans., 1981, 9, 290–293 CrossRef CAS PubMed.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular cloning, a laboratory manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y., 2nd edn, 1989 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mt00297k |
| ‡ (1924–2013). |
| This journal is © The Royal Society of Chemistry 2015 |