Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Engineering polypeptide folding through trans double bonds: transformation of miniature β-meanders to hybrid helices†
Mothukuri
Ganesh Kumar
a,
Sushil N.
Benke
a,
K. Muruga
Poopathi Raja
*b and
Hosahudya N.
Gopi
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research, Dr. Homi Bhabha Road, Pune-411 008, India. E-mail: hn.gopi@iiserpune.ac.in
bDepartment of Physical Chemistry, School of Chemistry, Madurai Kamaraj University, Madurai-625 021, India. E-mail: murugapoopathiraja@gmail.com
First published on 15th July 2015
Abstract
Utilization of conjugated double bonds to engineer the novel folded miniature β-meander type structures, single step transformation of miniature β-meanders into ααγ4-hybrid peptide 10/12-helices using catalytic hydrogenation, their solution and single crystal conformations are reported.
Engineering the folding of polypeptides into defined structures has a great significance from the perspective of medicinal chemistry, biology and materials science. Enormous efforts have been made in this regard to design novel folded architectures to mimic protein secondary structures using a variety of organic templates and synthetic backbone homologated β- and γ-amino acids.1,2 The homo and heterooligomers of β- and γ-amino acids have displayed a variety of helical patterns and based on the intramolecular H-bonding pseudocycles they are denoted as Cn-helices, where n is 14, 13, 12 etc.3 In addition to the helices, these unnatural oligomers have also been used to mimic β-sheets4 and turns.5 However, design of highly organized protein supersecondary structures such as helix-turn-helix, β-meanders, Greek key motifs etc. from unnatural oligomers has been scarcely investigated.
We have been interested in the utilization of naturally occurring functionalized γ-amino acids such as statines6 and α,β-unsaturated γ-amino acids7 in the design of foldamers. We recently showed stable β-hairpin8 and unusual planar structures in hybrid peptides containing α- and E-vinylogous amino acids.9 We foresee that the exceptional conformational properties of E-vinylogous amino acids can be explored to design highly organized protein supersecondary structure mimetics beyond the conventional secondary structures (helix, β-sheets and turns). We postulated that the β-sheet promoting8,10 and turn11 inducing properties of E-vinylogous amino acids can be readily exploited in the design of miniature β-meander mimetics. The β-meander is a common protein supersecondary structural motif consisting of two or more anti-parallel β-strands and each adjacent strand is connected by β-turns.12 The β-meander structural motif is frequently observed in many proteins including proteases and enzymes.13 Interestingly, β-barrel and β-propeller structural motifs are constituted from β-meander topology.14 We envisage that by introducing the conformational constraints through E-vinylogous amino acids after every two α-residues in a hybrid peptide sequence it is possible to construct sheet and reverse turns similar to that of β-meanders. Herein, we report the designed novel miniature β-meanders from ααE-vinylogous hybrid hexapeptides, single step transformation of miniature β-meanders into 10/12 ααγ-hybrid helices using catalytic hydrogenation, their solution and single crystal conformations.
To understand the conformational behavior of hybrid peptides composed of α- and α,β-unsaturated γ-amino acids, we designed two hexapeptides P1 and P2 composed of repeated ααE-vinylogous amino acid sequences (Scheme 1). Peptides P1 and P2 consist of α,β-unsaturated γ-phenylalanine and α,β-unsaturated γ-alanine, respectively. Peptides were synthesized using Boc-chemistry by a solution phase using DCC/HOBt coupling conditions. Pure peptides were subjected to conformational analysis in solution as well as to grow single crystals in the combination of various solvent mixtures.
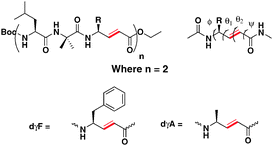 | ||
| Scheme 1 Sequences of P1 (Boc-Leu-Aib-dγF-Leu-Aib-dγF-OEt) and P2 (Boc-Leu-Aib-dγA-Leu-Aib-dγA-OEt). Local torsional variables of α,β-unsaturated γ-residues are also shown. | ||
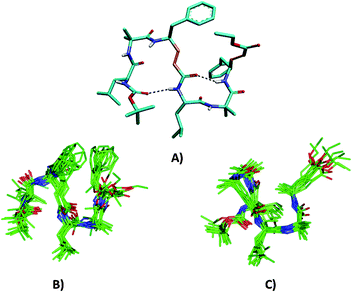
Single crystals of P1 obtained from the slow evaporation of a methanol/toluene solution yielded the structure shown in Fig. 1. Peptide P2 did not give X-ray quality single crystals. Instructively, P1 adopted a novel miniature β-meander type of structural motif with two reverse-turns. The analysis of the crystal structure of P1 reveals that two C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯NH intramolecular hydrogen bonds between the urethane carbonyl and the NH of Leu4 (1 ← 4) and between the carbonyl of dγF3 and NH of dγF6 (1 ← 4) stabilize the overall folding of the molecule. The torsional angles of α,β-unsaturated γ-amino acids were measured by introducing two new additional variables θ1 and θ2 along with ϕ and ψ (Scheme 1). The H-bond parameters and torsional angles of P1 are tabulated in the ESI.† Inspection of the crystal structure reveals that Aib2 and dγF3 adopted a reverse turn type of conformation stabilized by a 15-membered H-bond. Furthermore, analysis of the central segment, Leu4-Aib5, reveals that it adopted a type II β-turn conformation by displaying torsional values ϕ4 = −48°, ψ4 = 128°, ϕ5 = 68° and ψ5 = 11° [for a typical type II β-turn ϕ(i) = −60°, ψ(i) = 120, ϕ(i + 1) = 80° and ψ(i + 1) = 0°]. The type II β-turn is stabilized by the 1 ← 4 hydrogen bond between the carbonyl of dγF3 and NH of dγF6.
O⋯NH intramolecular hydrogen bonds between the urethane carbonyl and the NH of Leu4 (1 ← 4) and between the carbonyl of dγF3 and NH of dγF6 (1 ← 4) stabilize the overall folding of the molecule. The torsional angles of α,β-unsaturated γ-amino acids were measured by introducing two new additional variables θ1 and θ2 along with ϕ and ψ (Scheme 1). The H-bond parameters and torsional angles of P1 are tabulated in the ESI.† Inspection of the crystal structure reveals that Aib2 and dγF3 adopted a reverse turn type of conformation stabilized by a 15-membered H-bond. Furthermore, analysis of the central segment, Leu4-Aib5, reveals that it adopted a type II β-turn conformation by displaying torsional values ϕ4 = −48°, ψ4 = 128°, ϕ5 = 68° and ψ5 = 11° [for a typical type II β-turn ϕ(i) = −60°, ψ(i) = 120, ϕ(i + 1) = 80° and ψ(i + 1) = 0°]. The type II β-turn is stabilized by the 1 ← 4 hydrogen bond between the carbonyl of dγF3 and NH of dγF6.
In order to understand the conformations of P1 and P2 in solutions, we recorded 2D NMR (TOCSY, ROESY and COSY) in CDCl3. The 1H NMR reveals that all amide NHs are in the range of δ 6.5–7.3 ppm. The amino acid residues and their positions in the peptide sequences were identified using TOCSY and ROESY spectra, respectively. The strong sequential NH ↔ NH connectivity between Aib2 ↔ dγF3 and Aib5 ↔ dγF6 residues observed in P1 suggests the reverse-turn conformations of dipeptide segments in the hexapeptide. In addition, a strong NOE between the dγF3 CαH and Leu4 NH indicates the extended conformation of the conjugated amide. The solution conformation analysis of P2 also reveals a similar type of NOE pattern. Adding to the characteristic sequential NH ↔ NH NOEs between Leu1 ↔ Aib2, Aib2 ↔ dγA3 and Aib5 ↔ dγA6, the peptide also showed the NOEs between Aib2 side chains with both Leu1 NH as well as NH of dγA3. The long range strong NOE is observed between dγA3 βH and the dγA6 side-chain which further supports a well folded conformation of P2 in solution. Using unambiguous NOEs and variable temperature data, solution structure models of P1 and P2 are generated and shown in Fig. 1. Instructively, both peptides adopt distinctively different and a spectacular 3-dimentional folded miniature β-meander type of structures in solution as compared to the 2-dimentional flat crystal structure. We understand that the crystal packing forces possibly directed the 3-dimentional architecture of P1 in single crystals. Overall, the crystal structure and the NMR models suggested that both P1 and P2 adopted novel and distinct miniature β-meander type of structures.
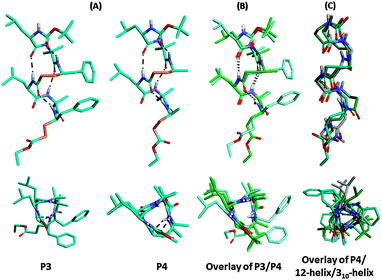
To appreciate the conformations of their saturated analogues, we subjected both P1 and P2 to catalytic hydrogenation using 20% Pd/C in ethanol. The complete conversion of unsaturated hybrid peptides P1 and P2 into their saturated analogues P3 [Boc-Leu-Aib-γ4Phe-Leu-Aib-γ4Phe-OEt] and P4 [Boc-Leu-Aib-γ4Ala-Leu-Aib-γ4Ala-OEt], respectively, was achieved within six hours and isolated with 92–95% yield (Scheme 2). This transformation also provides an alternative opportunity to synthesize hybrid γ-peptides without starting from saturated γ-amino acids. To understand their conformations, both peptides were subjected to crystallization in various solvent combinations. Both P3 and P4 gave X-ray quality single crystals in an aqueous methanol solution and their X-ray structures are shown in Fig. 2. Both peptides adopted helical conformations in single crystals. The release of the geometrical constraints of vinylogous double bonds in the miniature β-meanders leads to the hybrid helices. Analysis of the crystal structure of P3 reveals that the helical structure is stabilized by four intramolecular H-bonds between i and i + 3 residues similar to the 310-helix15 and the α,γ-hybrid peptide 12-helix,16 however with 10- and 12-membered H-bonds. The 10-membered H-bonds were observed at both N- and C-termini of the helix, while two consecutive 12-membered H-bonds were observed at the centre of the helix. Similar to P3, P4 also adopted a 10/12 helical structure. These helical structures of ααγ-hybrid peptides are consistent with the hybrid helices of –(αγα)n– sequences reported by Balaram and colleagues.17 The stereochemical analysis of γ4-residues in both P3 and P4 adopted gauche+, gauche+ (g+, g+, θ1 ≈ θ2 ≈ 60°) conformation about the Cβ–Cγ and Cα–Cβ bonds, while the C-terminal γ-residues displayed gauche+ (Cβ–Cγ), anti (Cα–Cβ) conformation due to the lack of a terminal H-bond donor (NH) to participate in the helix. Similar to the E-vinylogous amino acids the torsional angles of γ-amino acids were measured by introducing additional torsional variables θ1 and θ2 along with ϕ and ψ. The H-bond parameters and torsional angels of γ-residues are tabulated in the ESI.† Interestingly, the top view of the helical peptides in ααγ-hybrid peptides showed the projection of side-chains at three faces of the helical cylinder similar to the β-peptide 12-helices and 310-helix.18 In contrast, αγ-hybrid peptide 12-helices showed the projection of the side-chains at four corners of the helical cylinder.14c The backbone and side-chain correlation of 10/12 hybrid helices with 310-helix and β-peptide 12-helix is shown in Fig. 2C. Analysis suggests that the ααγ-hybrid peptides can mimic β-peptide 12-helices as well as 310-helices.
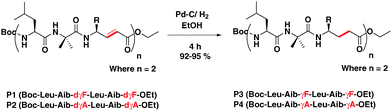 | ||
| Scheme 2 Direct transformation of ααE-vinylogous hybrid peptides (P1 and P2) into ααγ4-hybrid peptides (P3 and P4). | ||
In conclusion, we presented the utilization of geometrically constrained trans double bonds to engineer folding in peptides. The solution and crystal conformations suggested that the designed unsaturated hybrid peptides P1 and P2 adopted a novel miniature β-meander type of structure. The miniature β-meander type structure obtained from P1 and P2 offers a glimpse of the potential to generate higher order structures using α,β-unsaturated γ-amino acids as guests in host α-peptides. Furthermore, these peptides also provide a prospect to transform them into hybrid helices through catalytic hydrogenation. The structural analogy of ααγ-hybrid 10/12 helices suggests that they can be used to mimic β-peptide 12-helices as well as 310 helices. As peptide foldamers have been emerging as promising candidates in medicinal chemistry and chemical biology, novel miniature β-meanders and ααγ-hybrid peptide helices reported here can be further explored to design functional hybrid peptide foldamers.
K.M.P.R. acknowledges DST-SERB, Govt. of India, for the financial support. M.G.K. and S.N.B. are thankful to CSIR and UGC, respectively, for senior research fellowship.
Notes and references
- (a) G. Guichard and I. Huc, Chem. Commun., 2011, 47, 5933 RSC; (b) C. G. Cummings and A. D. Hamilton, Curr. Opin. Chem. Biol., 2010, 14, 341 CrossRef CAS PubMed.
- (a) D. Seebach, A. K. Beck and D. J. Bierbaum, Chem. Biodiversity, 2004, 1, 1111 CrossRef CAS PubMed; (b) R. P. Cheng, S. H. Gellman and W. F. DeGrado, Chem. Rev., 2001, 101, 3219 CrossRef CAS PubMed; (c) T. A. Martinek and F. Fulop, Chem. Soc. Rev., 2012, 41, 687 RSC; (d) P. G. Vasudev, S. Chatterjee, N. Shamala and P. Balaram, Chem. Rev., 2011, 111, 657 CrossRef CAS PubMed; (e) L. K. A. Pilsl and O. Reiser, Amino Acids, 2011, 41, 709 CrossRef CAS PubMed; (f) F. Bouillere, F. Thetiot-Laurent, C. Kouklovsky and V. Alezra, Amino Acids, 2011, 41, 687 CrossRef CAS PubMed.
- (a) W. S. Horne and S. H. Gellmann, Acc. Chem. Res., 2008, 41, 1399 CrossRef CAS PubMed; (b) G. V. M. Sharma, P. Nagendar, P. Jayaprakash, P. R. Krishna, K. V. S. Ramakrishna and A. C. Kunwar, Angew. Chem., Int. Ed., 2005, 44, 5878 CrossRef CAS PubMed; (c) N. Pendem, Y. R. Nelli, C. Douat, L. Fischer, M. Laguerre, E. Ennifar, B. Kauffmann and G. Guichard, Angew. Chem., Int. Ed., 2013, 52, 4147 CrossRef CAS PubMed; (d) S. Chatterjee, R. S. Roy and P. Balaram, J. R. Soc., Interface, 2007, 4, 587 CrossRef CAS PubMed.
- (a) I. L. Karle, H. N. Gopi and P. Balaram, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5160 CrossRef CAS PubMed; (b) G. A. Lengyel, R. C. Frank and W. S. Horne, J. Am. Chem. Soc., 2011, 133, 4246 CrossRef CAS PubMed; (c) L. Sebaoun, V. Maurizot, T. Granier, B. Kauffmann and I. Huc, J. Am. Chem. Soc., 2014, 136, 2168 CrossRef CAS PubMed.
- (a) D. Seebach, S. Abele, K. Gademann and B. Jaun, Angew. Chem., Int. Ed., 1999, 38, 1595 CrossRef CAS; (b) S. Chatterjee, P. G. Vasudev, S. Raghothama, C. Ramakrishnan, N. Shamala and P. Balaram, J. Am. Chem. Soc., 2009, 131, 5956 CrossRef CAS PubMed.
- (a) B. M. Dunn, Chem. Rev., 2002, 102, 4431 CrossRef CAS PubMed; (b) D. Leung, G. Abbenante and D. P. Fairlie, J. Med. Chem., 2000, 43, 305 CrossRef CAS; (c) A. Bandyopadhyay, A. Malik, M. Ganesh Kumar and H. N. Gopi, Org. Lett., 2014, 16, 294 CrossRef CAS PubMed.
- (a) R. G. Linington, B. R. Clark, E. E. Trimble, A. Almanza, L.-D. Urena, D. E. Kyle and W. H. Gerwick, J. Nat. Prod., 2009, 72, 14 CrossRef CAS PubMed; (b) M. Hagihara and S. L. Schreiber, J. Am. Chem. Soc., 1992, 114, 6570 CrossRef CAS.
- A. Bandyopadhyay, S. M. Mali, P. Lunawat, K. M. P. Raja and H. N. Gopi, Org. Lett., 2011, 13, 4482 CrossRef CAS PubMed.
- A. Bandyopadhyay and H. N. Gopi, Org. Lett., 2012, 14, 2770 CrossRef CAS PubMed.
- M. Hagihara, N. J. Anthony, T. J. Stout, J. Clardy and S. L. Schreiber, J. Am. Chem. Soc., 1992, 114, 6568 CrossRef CAS.
- (a) T. K. Chakraborty, A. Ghosh, S. K. Kumar and A. C. Kunwar, J. Org. Chem., 2003, 68, 6459 CrossRef CAS PubMed; (b) C. Grison, P. Coutrot, S. Geneve, C. Didierjean and M. Marraud, J. Org. Chem., 2005, 70, 10753 CrossRef CAS PubMed.
- T. E. Creighton, Proteins: structure and molecular properties, 2nd edn, W. H. Freeman and Co., New York, 1993, p. 227 Search PubMed.
- D. N. Woolfson, G. J. Bartlett, A. J. Burton, J. W. Heal, A. Niitsu, A. R. Thomson and C. W. Wood, Curr. Opin. Struct. Biol., 2015, 33, 16 CrossRef CAS PubMed.
- (a) S. Balaji, Curr. Opin. Struct. Biol., 2015, 32, 156 CrossRef CAS PubMed; (b) W. C. Wimley, Curr. Opin. Struct. Biol., 2003, 13, 404 CrossRef CAS; (c) G. E. Schulz, Curr. Opin. Struct. Biol., 2000, 10, 443 CrossRef CAS; (d) C. Zhang and S.-H. Kim, J. Mol. Biol., 2000, 299, 1075 CrossRef CAS PubMed.
- (a) I. L. Karle and P. Balaram, Biochemistry, 1990, 29, 6747 CrossRef CAS; (b) C. Toniolo, M. Crisma, F. Formaggio and C. Peggion, Biopolymers, 2001, 60, 396 CrossRef CAS.
- (a) S. Chatterjee, P. G. Vasudev, S. Raghothama, C. Ramakrishnan, N. Shamala and P. Balaram, J. Am. Chem. Soc., 2009, 131, 5956 CrossRef CAS PubMed; (b) L. Guo, Y. Chi, A. M. Almeida, I. A. Guzei, B. K. Parker and S. H. Gellman, J. Am. Chem. Soc., 2009, 131, 16018 CrossRef CAS PubMed; (c) A. Bandyopadhyay, S. V. Jadhav and H. N. Gopi, Chem. Commun., 2012, 48, 7170 RSC.
- K. Basuroy, B. Dinesh, N. Shamala and P. Balaram, Angew. Chem., Int. Ed. Engl., 2013, 52, 3136 CrossRef CAS PubMed.
- (a) C. Crisma, M. Saviano, A. Moretto, Q. B. Broxterman, B. Kaptein and C. Toniolo, J. Am. Chem. Soc., 2007, 129, 15471 CrossRef PubMed; (b) S. H. Choi, I. A. Guzei, L. C. Spencer and S. H. Gellman, J. Am. Chem. Soc., 2010, 132, 13879 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, 1H NMR; 2D-NMR, and crystallographic information. CCDC 1063358 (P1), 1060827 (P3) and 1060826 (P4). See DOI: 10.1039/c5cc04523a |
| This journal is © The Royal Society of Chemistry 2015 |