Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High resolution mass spectrometry imaging of plant tissues: towards a plant metabolite atlas†
Dhaka Ram
Bhandari
a,
Qing
Wang
b,
Wolfgang
Friedt
b,
Bernhard
Spengler
a,
Sven
Gottwald
*b and
Andreas
Römpp‡
*a
aJustus Liebig University Giessen, Institute of Inorganic and Analytical Chemistry, Schubertstrasse 60, Building 16, 35392 Giessen, Germany
bJustus Liebig University Giessen, Department of Plant Breeding, IFZ, Justus Liebig University Giessen, Heinrich-Buff-Ring 26–32, 35392 Giessen, Germany. E-mail: Sven.Gottwald@agrar.uni-giessen.de; Tel: +49641-9937426
First published on 16th September 2015
Abstract
Mass spectrometry (MS) imaging provides spatial and molecular information for a wide range of compounds. This tool can be used to investigate metabolic changes in plant physiology and environmental interactions. A major challenge in our study was to prepare tissue sections that were compatible with high spatial resolution analysis and therefore dedicated sample preparation protocols were established and optimized for the physicochemical properties of all major plant organs. We combined high spatial resolution (5 μm), in order to detect cellular features, and high mass accuracy (<2 ppm root mean square error), for molecular specificity. Mass spectrometry imaging experiments were performed in positive and negative ion mode. Changes in metabolite patterns during plant development were investigated for germination of oilseed rape. The detailed localization of more than 90 compounds allowed assignment to metabolic processes and indicated possible functions in plant tissues. The ‘untargeted’ nature of MS imaging allows the detection of marker compounds for the physiological status, as demonstrated for plant–pathogen interactions. Our images show excellent correlation with optical/histological examination. In contrast to previous MS imaging studies of plants, we present a complete workflow that covers multiple species, such as oilseed rape, wheat seed and rice. In addition, different major plant organs and a wide variety of compound classes were analyzed. Thus, our method could be used to develop a plant metabolite atlas as a reference to investigate systemic and local effects of pathogen infection or environmental stress.
1 Introduction
Plants are major suppliers of food and natural resources used in pharmaceuticals, cosmetics, and fine chemicals. As sessile organisms, plants cannot escape their environment, instead (progressive) acclimatization by specific metabolic adjustments is the only response to changing conditions.1 This resulted in a complex metabolism with over 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 known primary and secondary metabolites.2
000 known primary and secondary metabolites.2
As part of the ‘omics’ field, metabolic studies have been applied to a wide spectrum of topics in plant science. These include basic research in model plants such as Arabidopsis to discover fundamental biosynthetic processes and metabolic networks3 as well as applied research in crop plants such as wheat (Triticum aestivum) and oilseed rape (Brassica napus) to uncover biochemical mechanisms behind complex agronomical traits and phenotypes.4 Important current issues in crop plant research are developmental plasticity,5 and responses to abiotic6 and biotic stress.7 In contrast to genes, metabolites are the end products of cellular processes and therefore, can serve as direct signatures of biochemical activity.8 Metabolomics studies are typically based on mass spectrometry (MS), hyphenated with techniques such as gas and liquid chromatography, covering an extensive variety of compound classes and concentration ranges. In these studies, usually homogenized samples are used, resulting in a loss of spatial information. On the other hand, visualization has become an important topic in plant science, because detailed knowledge on metabolite distributions in plants is fundamental for the understanding of local regulatory networks, which underlay selected traits.9 Visualization is usually achieved by fluorescence microscopy, in situ hybridization or immunohistochemistry,10 techniques which provide high spatial resolution, but chemical information is typically limited to very few compounds per experiment.
Mass spectrometry (MS) imaging combines spatial information and molecular information for a wide range of compounds. This method can, therefore, complement the classical approaches of metabolomics studies and microscopic methods. MS imaging is the method of scanning a sample of interest and generating images of the intensity distribution of analyte ions. In contrast to classical histochemical methods, MS imaging is a label free technique and thus, can be used without prior knowledge of the analyte. Due to this untargeted nature, hundreds of compounds can be detected simultaneously. While numerous studies have been published in the field of clinical research, plant research is a relatively new application of MS imaging which, however, offers important advantages for the detailed investigation of metabolites from complex plant tissues.7,11
The first studies using MALDI imaging on plants analyzed the distribution of agrochemical compounds in soya bean (Glycine max)12 and carbohydrates in wheat (Triticum aestivum) stem.13 Subsequent studies included applications on rice (Oryza sativa) seeds,14,15 flowers and roots of the model plant Arabidopsis thaliana16–19 and barley (Hordeum vulgare) seeds.20 However, most MS imaging studies in plants were performed with MALDI-TOF instruments, which provide lower mass resolution data, possibly leading to ambiguous results. Only recently, high mass resolution orbital trapping mass analyzers were applied.16,21–24 A pixel size of 25 μm25,26 and 5 μm27 was obtained in selected experiments, but most studies were performed at 100 to 200 μm spatial resolution. A related method is laser desorption ionization (LDI) which was used for the analysis of plant tissue with a TOF mass spectrometer at a 10 μm pixel size.28 This approach does not require any matrix application, but is limited to compounds with UV-absorbing functional groups.
We have introduced a MS imaging method that combines for the first time high selectivity (1 ppm mass accuracy) and high spatial resolution (3 to 10 μm pixel size) in one experiment.29 This results in a significantly improved reliability of compound identification and provides molecular information on a cellular level. This technique was initially developed for mammalian tissues,30 but has now been further developed and optimized for non-mammalian tissues. Recently, we have demonstrated the applicability of MS imaging for metabolite characterization of wine (Vitis vinifera) grapes,31 the bird's nest fungus Cyathus striatus32 and the licorice (Glycyrrhiza glabra) rhizome33 at 20 μm, 15 μm and 10 μm pixel size, respectively.
Previous MS imaging studies of plants were limited to a selected organ in a particular species and typically focused on a certain class of compounds. In this publication we present new developments in MS imaging of plant tissues that enable a comprehensive investigation of the plant metabolome. These developments include the first MS imaging experiments at a 5 μm pixel size which reveal the detailed structure of metabolites in plant organs. We were able to image a wide range of compound classes including different phenolic compounds, phospholipids and other lipid species, carbohydrates, phenolic choline esters, glycosides, and glycerides in mature and germinating oilseed rape. These data probably represent the most comprehensive spatially resolved information about metabolites in plants, so far.
To establish a plant metabolome atlas, it is necessary to analyze all relevant plant organs and to cover different plant species. This primarily concerns the development of dedicated sample preparation techniques. In plants, different organs and tissues are specialized and modified for specific functions. Thus sample preparation techniques need to be designed according to the different plant organs and (metabolic) stages. Consequently, we also present developments in MS imaging, which concern sectioning procedures that were optimized for high spatial resolution analysis in a variety of tissues with specific physicochemical properties.
The methodological approach and obtained information of MS imaging is discussed in detail in a case study comparing the mature and germinating seeds of oilseed rape (section 2.1). Subsequently additional plant species and organs are discussed. This includes experiments in negative ion mode and with a 5 μm pixel size for wheat seed (section 2.2). Sample preparation is the focus for wheat rachis (section 2.3), wheat stem base (section 2.4) and rice root (section 2.5). Finally, we have provided an example in which a wheat seed infected by the fungal pathogen, Fusarium graminearum, is compared to an uninfected seed and cultured fungal mycelium (section 2.6). Details of the experimental procedure for each experiment, particularly sample preparation, are discussed in the Experimental section (section 3).
In the Conclusions section (section 4) we have included measurements of additional plant organs (distal root, leaf and stem). We also discuss the idea of how our MS imaging approach could be used to build up an atlas of the plant metabolome as a reference to detect the systemic and local metabolic changes.
2 Results and discussion
2.1 Mapping of metabolic processes in seed development
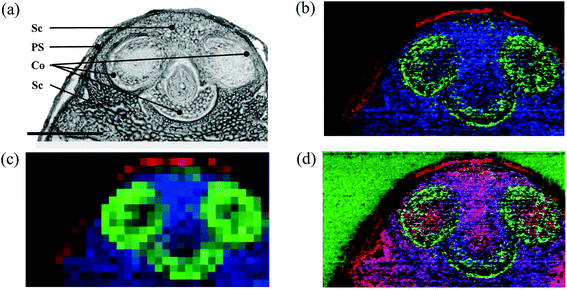
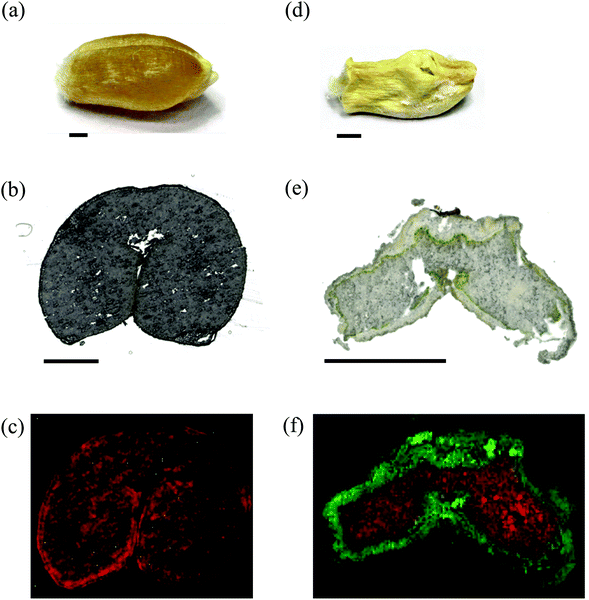
Maturity and germination are metabolically highly active stages during the development with significant implications for subsequent plant performance34,35 and nutritional quality of seed products.36 Therefore, these stages were chosen to establish high resolution MS imaging as a tool for mapping metabolites.Seeds of oilseed rape are 2–3 mm in diameter, which makes direct cryosectioning challenging. Therefore, cellulose (CMC) carboxymethyl cellulose solution was used as an embedding material as described in the Experimental section. The optical images of both the stages (Fig. 1a and b) can be directly correlated to the respective ion images (Fig. 1c, e, d and f). A mass spectrum acquired from a single 10 μm pixel in the shoot apical meristem region of a germinating seed is shown in Fig. 1g. The exact location of this pixel and the zoomed-in mass spectrum are provided in ESI, Fig. S1.† Additional single 10 μm spectra from cotyledon vasculature, nucellar tissue (putative), seed coat, and emerging radicle are shown in the ESI, Fig. S2–S5.† Several compounds such as sinapine, methyl sinapate, cyclic spermidine conjugate, triacylglycerols and phosphatidylcholines were detected. These compounds were identified based on accurate mass measurements. For example, sinapine (choline ester of sinapic acid) was detected with a mass error of −0.83 ppm (Fig. 1g) and the corresponding image is shown in Fig. 1c (blue). The root mean square error (RMSE), calculated from 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 523 spectra over the full image, was 1.0 ppm. The mass accuracy was better than 2 ppm for all compounds assigned in this article. This demonstrated a reliable detection of compounds also under imaging conditions. Using high resolution and high mass accuracy, it is possible to generate MS images with a narrow bin width of Δm/z = ±5 ppm, thus preventing interference from the neighboring peaks (an example is discussed for the MS imaging on a wheat seed). This bin width was used for all MS images presented in this study. A list of 93 compounds imaged in oilseed rape (including mass accuracy values) is given in ESI, Table S1,† and the respective MS images are compiled in ESI, Fig. S6 (germination) and Fig. S7† (maturation). The sequential numbering of compounds in Table S1† corresponds to the index numbers in Fig. S6 and S7.† For example, the MS images of sinapine (compound no. 1) are shown in Fig. S6.1 and S7.1.† These images are selected ion images of a single compound (allowing visualization of pixels with lower signal intensities), in contrast to the combined RGB images in Fig. 1.
523 spectra over the full image, was 1.0 ppm. The mass accuracy was better than 2 ppm for all compounds assigned in this article. This demonstrated a reliable detection of compounds also under imaging conditions. Using high resolution and high mass accuracy, it is possible to generate MS images with a narrow bin width of Δm/z = ±5 ppm, thus preventing interference from the neighboring peaks (an example is discussed for the MS imaging on a wheat seed). This bin width was used for all MS images presented in this study. A list of 93 compounds imaged in oilseed rape (including mass accuracy values) is given in ESI, Table S1,† and the respective MS images are compiled in ESI, Fig. S6 (germination) and Fig. S7† (maturation). The sequential numbering of compounds in Table S1† corresponds to the index numbers in Fig. S6 and S7.† For example, the MS images of sinapine (compound no. 1) are shown in Fig. S6.1 and S7.1.† These images are selected ion images of a single compound (allowing visualization of pixels with lower signal intensities), in contrast to the combined RGB images in Fig. 1.
The compound identification can be further assisted by on-tissue MS/MS experiments as demonstrated for sinapine in Fig. 1h, showing the single pixel on-tissue tandem mass spectrum. The fragment ion at m/z 251 corresponds to a loss of trimethylamine from the choline group. Additional fragment ions can be attributed to the sequential loss of C2H4O (m/z 207), CH3OH (m/z 175) and CO (m/z 147). All fragment ions were detected with a mass error of less than 1 ppm. This fragmentation pattern is also in accordance with electrospray-based MS/MS data,37 thus confirming the initial identification which was based on accurate mass data.
In the mature seed, sinapine was distributed throughout the entire embryo (Fig. 1d, blue) which consists of the cotyledons and the hypocotyl-radicle region (Fig. 1b). This broad distribution of sinapine was found to persist during the early germination (Fig. 1c, blue). In contrast, a cyclic spermidine conjugate showed a distinct distribution in the hypocotyl-radicle region in the mature seed (Fig. 1d, red, appears magenta due to overlay with sinapine in blue), and in the germinating seed their activities shifted to the emerging radicle (Fig. 1c, red). This demonstrates that MS imaging is able to detect shifts in metabolite distribution at different developmental stages. Since a second spermidine conjugate, the N1,N5,N10-tricaffeoyl spermidine, showed a similar spatial distribution (Fig. S6.28 and Fig. S7.28†), our results suggest that spermidine conjugates play a role in the development of the hypocotylradicle, which later differentiates into the plant root and stem (a more detailed discussion is given in the ESI, Notes S1†).
The distribution of the compounds mentioned above corresponds very well to the hitherto only tissue-specific metabolite mapping in oilseed rape, carried out by combining laser microdissection and HPLC-DAD/MS.38 This demonstrates both the ability of MS imaging to confirm the findings from alternative techniques and to significantly provide more details on spatial distributions. The approach applied by Fang et al.38 allowed differentiating between inner and outer cotyledon in the mature seed. MS imaging at high spatial resolution, however, enabled association of metabolites with more detailed morphological features. For instance, sinapoyl glucose, previously identified in a bulk analysis of oilseed rape,39 was mapped at both the seed developmental stages to a thin layer between the two cotyledons (Fig. 1c and d, green). This region corresponds to the adaxial (upper) cotyledon epidermis which also contained different kaempferol derivatives (Fig. S6.30–6.33 and S71.30–7.33†). Therefore, these compounds might contribute to the epidermal shield against various abiotic stress factors such as UV-B radiation.40,41 In contrast, the choline ester ‘UC#1’ was mapped at both seed stages to the outer layer of cotyledons, representing the abaxial (lower) cotyledon epidermis (Fig. 1e and f, blue). UC#1 belongs to a group of ‘unknown’ choline esters which were initially detected in mature oilseed rape.42,43 The possibility to map compounds even in the epidermis layers makes MS imaging an interesting tool to examine metabolomic processes.
Feruloylcholine guaiacyl (FC(4-O-8′)G) was distributed throughout the mature embryo (Fig. 1f, green), but demonstrated different and more specific accumulations in the germinating embryo, i.e. in the parenchyma layer of the emerging hypocotyl and shoot apical meristem (Fig. 1e, green). The general presence of feruloylcholine FC(4-O-8′)-guaiacyl in oilseed rape has previously been reported.42
MS imaging is a direct analysis, which does not require preliminary extraction to detect a wide range of compound classes from a single tissue section. In fact, the obtained results were in agreement with other untargeted hyphenated-MS techniques which used extraction of the whole seed42,43 or more specific analysis including laser microdissection and HPLC-DAD/MS.38 The high-resolution images generated by our MS imaging method provide much more detailed spatial distributions, which enable elucidation of biological metabolite functions.
This data can be used to investigate metabolomic networks. An example is the sinapate ester metabolism presented in Fig. 2, S8 and S9,† which is relevant for oil production.44 In this pathway not only abundant metabolites such as sinapine, coumaric acid and kaempferol compounds were mapped, but also less abundant compounds such as methyl sinapate with unknown functions. So far, methyl sinapate has rarely been reported.42,45 MS imaging has located this compound in the entire mature embryo as well as in the outer cotyledon, hypocotyl and radicle in the germination stage (Fig. 2). Since the biological function of methyl sinapate is still unknown, it was interesting to find them remarkably enriched in the central apical shoot meristem (Fig. 2), indicating a possible function in the developing hypocotyl. More details on sinapine esters and kaempferol glycosides are shown in the ESI, Notes S2.†
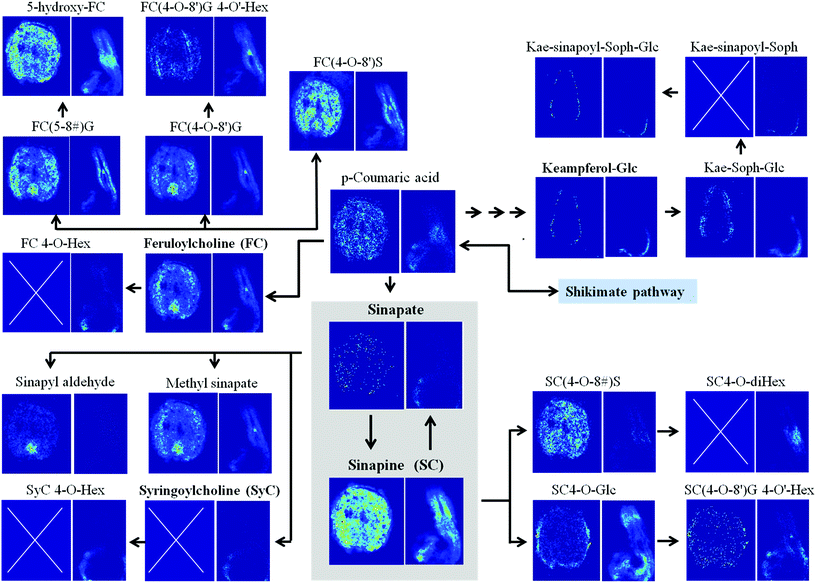 | ||
| Fig. 2 Scheme of metabolomic network associated with sinapate ester metabolism, localized by high-resolution MS imaging in mature and early germinating oilseed rape. Spatial distributions are shown in the ion images below the respective metabolites (left image: mature seed; right image early germinating seed). Blue boxes marked by a white cross indicate that the metabolite has not been detected at that developmental seed stage. Localized key metabolites are marked in bold letters. The sinapate ester metabolism is highlighted as a grey box. The shikimate pathway as a gateway to the displayed branch of phenylpropanoid metabolism is indicated as light blue box. The scheme was redrawn based on Clauss et al.42 Abbreviations: G, guaiacyl moiety; Glc, glucoside; Hex, unknown hexose moieties (commonly Glc); S, syringyl moiety; Soph, sophorose. | ||
2.2 Wheat seed
Wheat seeds at the soft dough stage of maturity are physiologically mature, but still characterized by rapid accumulation of starch and nutrients.46 In contrast to oilseed rape, wheat seeds (dough stage) have higher moisture content (about 40%), but are still elastic compared to the later stages, such as the postharvest state, which has been analyzed for oilseed rape. The size and the shape of the wheat seed allowed direct mounting in the cryomicrotome without embedding. This allows tissue section preparation by direct cryosectioning. The optical image (Fig. 3a) from a transverse section of the germ-region (embryo) can be directly correlated to MS images (Fig. 3b). In addition to sample preparation, spatial resolution and mass resolution are central parameters for generating high-quality images in untargeted metabolite screening. An important reason is the lack of chromatographic separation, which makes analyte assignments in MS imaging more complicated compared to metabolomic analysis based on GC/MS or LC/MS.16 We combined high mass resolution (which allows generating images with a narrow m/z bin width of ±5 ppm) with high spatial resolution (5 μm pixel size). This enabled us to represent the grain architecture with a reliable assignment of imaged metabolites to their respective in situ locations. For example, it was possible to demonstrate the specific location of a polyphenol glycoside in the protection sheath coleoptile/coleorhizae (Fig. 3b, green), suggesting a function in protecting the emerging shoot, plumules and/or radicle against UV-B radiation, pathogens or herbivores.47 In addition, a phosphatidylcholine, PC(36:4), was specifically located in the scutellum (Fig. 3b, blue), while a phosphatidylglycerol, PG(38:2), was restricted to the pericarp-seed coat region (Fig. 3b, red). PG membrane lipids are vital for normal embryo development as they are involved in formation of chloroplast thylakoid membranes, which are the primary site of photosynthesis.48 By contrast, a high mass resolution (±5 ppm) combined with a low spatial resolution results in a loss of spatial information (Fig. 3c), which shows the same measurement with a recalculated pixel size of 50 μm. A critical loss of information is also observed using a bin width of 0.1, typically used for MALDI-TOF imaging experiments. Here, the low mass resolution affects the image generation due to the interference of the neighboring peaks (Fig. 3d). Consequently, a combination of high mass resolution with high spatial resolution is needed to resolve the complexity of metabolites in plant tissue sections.Negative ion mode measurements provide a possibility to obtain complementary information on identification of the compounds. A section of the wheat germ similar to Fig. 3a was scanned in negative ion mode (Fig. 4a). For example, the polyphenol glycoside in Fig. 3b (green) was confirmed to have the same spatial distribution in negative ion mode (Fig. 4b, green). Similar distribution of additional polyphenol glycosides is demonstrated by comparing positive and negative ion measurements (Fig. S10†). Moreover, the negative mode experiment enables imaging of additional compound classes such as a phosphatidylinositol [PI(34:2)–H]− located in the scutellum and endosperm of the wheat seed (Fig. 4b, red). PI is a key membrane constituent and an important participant in signaling processes in seed and vegetative tissue,49,50 responsible for normal plant growth, stress response and seed germination.51,52
2.3 Wheat spike rachis
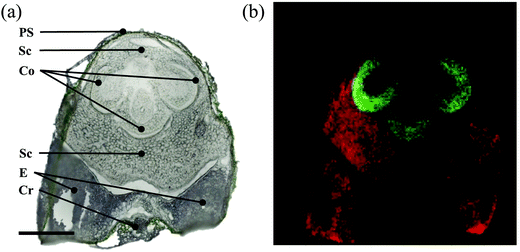
The rachis is the primary axis of a cereal spike, which bears the spikelets containing flowers. The spike rachis consists of low-density tissues, resulting in floating of the specimen in CMC solution (used for oilseed rape, Fig. 1). Therefore, an alternative embedding material, a paste of 15% (w/v) tragacanth gum in water was employed.53 The rachis is not only responsible for the translocation of assimilates into florets and developing seeds,54 but also the passage for systemic inflorescence colonization by fungal pathogens such as Fusarium graminearum in wheat55 or Aspergillus flavus in maize.56 Although recent studies have demonstrated the potential of the rachis as an effective defense structure, the molecular and cellular processes behind these defenses are still unknown.56,57 The optical image of a transverse rachis section (Fig. 5a) correlates well with the MS image. The MS image shows a polyphenol glycoside located in the epidermis (Fig. 5b, green) and two further metabolites, a lysophosphatidylcholine (Fig. 5b, blue) and a pheophytin (Fig. 5b, red), spatially distributed in the parenchyma cells, which include the photosynthetically active chlorenchyma. In fact, pheophytin is a chlorophyll derivative involved in the electron transfer pathway of photosystem II.58 In addition to the above mentioned nutritional aspects, compounds such as lysophosphatidylcholine are essentially involved in plant health by functioning in the synthesis of phyto-oxylipins which comprise, for example, antifungal peptides and defense-signaling molecules.592.4 Wheat stem base
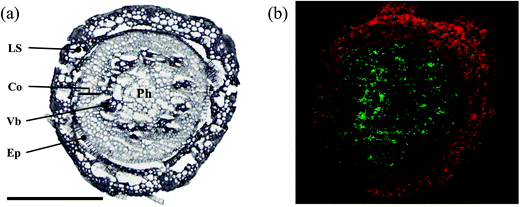
As the gateway from below- to above-ground plant parts, the stem base is a neuralgic point for the spread of soil-borne pathogens into the upper plant organs.60 The stem base of wheat seedlings has a relatively high water content compared to seed or rachis. Thus, a faster freezing method was obtained by using a coolant mixture of dry ice and hexane. The diameter of a stem base is relatively small for a low spatial resolution measurement. Therefore, a 5 μm step size was selected for MS imaging. As examples, the optical image (Fig. 6a) is well correlated with the ion image, where a polyphenol glycoside was mapped specifically to the leaf sheath (Fig. 6b, red), while a phospholipid was enriched in the stem tissue (Fig. 6b, green). Similarly to the lysophosphatidylcholine reported from the rachis, phosphatidylcholines are involved in the synthesis of phyto-oxylipins such as jasmonates59 which are currently discussed as essential signaling molecules in wheat defences against F. graminearum.61 Changes in these phospholipids occur at the site of fungal infection, since they are rapidly formed in response to pathogen attacks, which makes them useful markers for induced resistance.62 Therefore, spatial distributions in diseased tissues could facilitate the identification of resistance-related lipids.2.5 Plant roots
Roots remained largely unexplored for quite a long time.63 The soil micro-flora living directly at the surface or within roots facilitates pathogenic interactions and cooperative interactions. The latter improve nutrient uptake, stress resistance and yield performance.64 Root development and physiology are highly flexible in changing environments. However, the chemical signaling behind these changes, as well as behind the root–microbe interactions is largely unknown.65With this background, roots represent an important target for an MS imaging application. The first studies towards a procedure that meets the specific requirements of roots, have been conducted on in vitro-grown roots of wheat, barley and rice. Cereals have a fibrous system of thin seminal roots with a typically high water content and fragile structure, which are characteristics that challenge the preparation of tissue sections. The CMC embedding and sectioning in a cryomicrotome as described for the aerial plant parts was not successful for root tissue. The large vacuole in the root cell caused the cells to rupture due to the change in temperature (during the thawing process). Thin and uniform root cross sections were only obtainable with a common shaving blade.
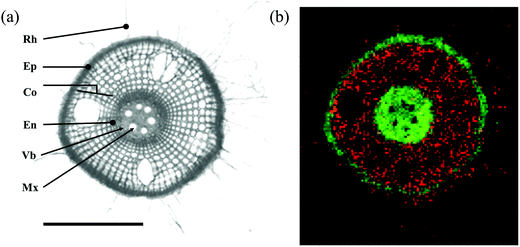
Seminal root tissues were found to be generally very fragile, and the structural integrity was easily lost. While rice root sections were on average 0.8 mm in diameter, wheat and barley root sections were not more than 0.5 mm in diameter. In the case of rice roots, it was possible to generate cross sections that were appropriate for MS imaging measurements (Fig. 7a), for wheat and barley the limiting factor was a high fragility together with a low diameter, which, therefore, hampers the preparation of cross sections with sufficient quality. Consequently, the MS imaging measurements resulted in low quality ion images, which were not suitable for fine-scale metabolite localization. However, we were able to image a cross section from the elongation zone of seminal rice roots, which had a diameter (ca. 500 μm) similar to wheat and barley roots (Fig. S11†). This is most likely related to the fact that root cells in this zone are in the state of elongation, and their vacuoles are less filled with water, leading to a more stable tissue structure.
The optical image of a rice root shows the main compartments: root hair, epidermis, cortex, endodermis, and stele containing the vascular bundles and metaxylem vessels (Fig. 7a). The example MS imaging measurement demonstrates the fine-scale localization of a penta-hexose carbohydrate in the root cortex (Fig. 7b, red), and of a phosphatidylcholine mapped to the epidermis and the central stele with a specific absence from metaxylem vessels (Fig. 7b, green). The detected hexose is likely part of the carbohydrate reserves in the root cortex. The carbon metabolism generally represents an essential sink–source interaction, in which root and shoot growth influence each other mutually. Consequently, the carbohydrate translocation from leaves and their storage in roots are very sensitive to stress, management practices and root–microbe interactions. Decreased root reserves are known to profoundly affect plant performance.66,67 Thus, carbon use and sugar signaling are of high scientific interest and represent a good example for the applicability of a plant metabolome atlas.
2.6 Markers for Fusarium infection
In cereal crops, F. graminearum (Fg) is a major agent of fungal diseases which causes severe yield reductions and quality deterioration, especially due to mycotoxin accumulation in seeds which are toxic to farm animals and humans.68 Both the healthy and infected wheat seeds have a low moisture content (<20%). An infected wheat seed demonstrates the characteristic weight reduction, shriveled and light-brown appearance due to colonization by Fg69 as shown in the photographic (Fig. 8a and d) and microscopic images (Fig. 8b and e). The characteristic low moisture content in healthy wheat seeds and additionally the substantial disease-related physical changes represent a specific challenge for the preparation of thin uniform sections from wheat seeds.To overcome this intrinsic problem, a tape-based method70 was adapted, which assists the generation of thin complete sections from hard specimens. A specially prepared adhesive tape, functioning at low temperatures, was attached to the cut surface of the sample to support the sectioning. The seed, surrounded by CMC as a block, was formed by freezing CMC in a mold using a cooling mixture. For a comparative MS imaging, healthy and diseased seeds of the highly Fusarium head blight (FHB) susceptible wheat cultivar Florence-Aurore were used. The overlay ion images of a healthy (Fig. 8c) and an infected wheat bran (Fig. 8f) show uniform distributions of a tetra-hexose metabolite in the endosperm (Fig. 8f, red). However, several compounds were specifically detected in the bran of the infected seed, e.g. 4-(trimethylammonio)but-2-enoate (Fig. 8f, green). This distribution indicates a relationship with the pathogen infection since Fg preferably colonizes this peripheral tissue in mature seeds.71 The localization of Fg in this particular sample was confirmed by fluorescence microscopy (Fig. S12†). The chemical identity of the fungus-related metabolite (4-(trimethylammonio)but-2-enoate) was confirmed by MS/MS analysis (Fig. S13f†). MALDI MS spectra of Fg hyphae also resulted in the same metabolite signals (Fig. S13e†). In addition, a carnitine metabolite was detected in the infected wheat bran (Fig. S13a and b†) and in fungal hyphae (Fig. S13e†). Both metabolites are a result of the carnitine metabolism, which is quite similar in plants and fungi. However, they were identified from hyphae in planta (diseased wheat seeds) and from hyphae in vitro (culture medium). This indicates a fungal origin and thus, both were denoted as pathogenesis-related metabolites.
As demonstrated here, MS imaging can be applied to detect candidates for pathogenesis-related metabolites solely by their spatial distribution within the infected parts of a tissue. Such “biomarkers” could be applied to visualize unknown sites of host–pathogen interactions. The few available metabolomic studies on interactions between biotic stressors and plants have already demonstrated that those metabolic processes are highly specific for a given tissue, species and pathosystem. Therefore, it is conceivable that untargeted MS visualization of metabolomic plant–pathogen interactions will lead to new insights into chemical plant defense mechanisms.
3 Experimental
Detailed information on chemicals and origin of plant material can be found in the ESI, Methods S1, and Methods S2,† respectively.3.1 Sample preparation for MALDI imaging
We optimized sectioning procedures like embedding in different media, snapshot freezing, use of adhesive tape and manual sectioning to obtain thin uniform sections. For cryosectioning, water (ice) was used as an adhesive to hold the specimen on a sample holder of a cryomicrotome (HM 525 cryostat, Thermo Scientific, Dreieich, Germany). Sections of 10–20 μm thickness were cut at −15 to −25 °C. The sections were thaw-mounted on microscope glass slides (75 × 25 × 1 mm), which were stored at −80 °C until analysis. Prior to matrix application, sections were brought to room temperature in a desiccator to avoid condensation of humidity on the sample surface. An Olympus BX-41 (Olympus Europa GmbH, Hamburg, Germany) microscope was used to capture optical images of the sections before and after matrix application. In the case of positive ion mode, a solution of 200 μL 2,5-dihydroxybenzoic acid (DHB) (30 mg mL−1 in acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (50
water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v), 0.1% trifluoroacetic acid), was applied. For negative ion mode, a solution of 200 μL 4-nitroaniline (10 mg mL−1 in acetone
50, v/v), 0.1% trifluoroacetic acid), was applied. For negative ion mode, a solution of 200 μL 4-nitroaniline (10 mg mL−1 in acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (50
water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v)) was sprayed with a pneumatic sprayer.72 Specific details on sample preparation for individual specimen are given in section 3.3.
50, v/v)) was sprayed with a pneumatic sprayer.72 Specific details on sample preparation for individual specimen are given in section 3.3.
3.2 Instrumentation for MALDI imaging
Highly resolved mass spectra were generated with a Fourier transform orbital trapping mass spectrometer (Exactive or Q Exactive, Thermo Fisher Scientific GmbH, Bremen, Germany) coupled to an atmospheric-pressure scanning-microprobe matrix assisted laser desorption/ionization imaging source (AP-SMALDI10, TransMIT GmbH, Giessen, Germany).29,73 For desorption/ionization of the analyte a nitrogen laser (LTB MNL-106, LTB, Berlin, Germany) with a repetition rate of 60 Hz and wavelength of 337 nm was used. The laser beam was focused perpendicular to the sample to a laser ablation spot size of 5 μm. The samples were scanned with 5 to 25 μm step size and the target voltage was set to 4.3 kV. The mass spectrometer was operated in positive ion/negative ion mode and spectra were scanned in different mass-to-charge (m/z) ranges and mass resolutions (R), with m/z = 100–1500 and R = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 140
000 to 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 respectively. Automatic gain control was disabled and the ion injection time was set to 500 ms. Internal calibration was achieved by using lock masses from matrix clusters. The cycle time for one pixel at 140
000 respectively. Automatic gain control was disabled and the ion injection time was set to 500 ms. Internal calibration was achieved by using lock masses from matrix clusters. The cycle time for one pixel at 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolving power was 1.3 s. A detailed description of the measurement parameters for each sample is given below.
000 resolving power was 1.3 s. A detailed description of the measurement parameters for each sample is given below.
3.3 Experimental details for individual specimen
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ m/z 200 for a mass range of m/z 150–1000 in positive ion mode.
000 @ m/z 200 for a mass range of m/z 150–1000 in positive ion mode.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ m/z 200 for a mass range of m/z 400–1000 in positive ion mode. For the negative ion mode measurement, an area of 2250 × 2250 μm2 (150 × 150 pixels) was scanned with a pixel size of 15 μm (Fig. 4b). The mass spectrometer was set to a resolution of 100
000 @ m/z 200 for a mass range of m/z 400–1000 in positive ion mode. For the negative ion mode measurement, an area of 2250 × 2250 μm2 (150 × 150 pixels) was scanned with a pixel size of 15 μm (Fig. 4b). The mass spectrometer was set to a resolution of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ m/z 200 for a mass range of m/z 200–1000.
000 @ m/z 200 for a mass range of m/z 200–1000.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ m/z 200 for a mass range of m/z 100–1000 in positive ion mode.
000 @ m/z 200 for a mass range of m/z 100–1000 in positive ion mode.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ m/z 200 for a mass range of m/z 250–1000 in positive ion mode.
000 @ m/z 200 for a mass range of m/z 250–1000 in positive ion mode.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ m/z 200 for a mass range of m/z 100–1200 in positive ion mode.
000 @ m/z 200 for a mass range of m/z 100–1200 in positive ion mode.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ m/z 200 for a mass range of m/z 100–1000 in positive ion mode.
000 @ m/z 200 for a mass range of m/z 100–1000 in positive ion mode.
3.4 Data processing and image generation
Ion images of selected m/z values were generated using the in-house developed MIRION software package74 with a m/z bin width of Δm/z = ± 5 ppm. The ion images were normalized to the highest intensity for each ion species separately. RGB images of three different m/z values were overlaid and demonstrated simultaneously. Other data processing steps like interpolation, smoothing or normalization to the matrix signals were not needed and were not applied during the image generation process. The MS images in ESI, Fig. S6 and Fig S7,† were generated by converting the raw files from the Orbitrap instrument to the imzML format.75 Images were then generated with an MSiReader version 0.0476 in the batch processing option, based on a predefined theoretical m/z value list. Mass accuracies for Table S1† were calculated by77 using intensity-weighted average values from all spectra of one image.Literature and METLIN search was used to identify compounds. The compounds were identified using high mass accuracy (<2 ppm). For subsequent biological interpretation, annotated metabolites were assigned to appropriate metabolome categories and pathways by consulting the web applications, KEGG Pathway Maps,78 MetaCyc Pathway,79 and LIPID MAPS.80
4 Conclusions
In this study, we present an approach for MS imaging of plant metabolites that cover multiple mono- and dicot species, major plant organs and a wide variety of compound classes. The results were in excellent agreement with previous reports including bulk sample MS studies and in addition provided new information on the detailed spatial distribution of important plant metabolites. In contrast to classical imaging methods, the analytes in MS imaging do not have to be known upfront. All presented MS imaging measurements are based on accurate mass determination (mass accuracy better than 2 ppm) for reliable identification, and were performed at 5 to 25 μm pixel size in order to visualize small spatial features.Previous MS imaging studies of plants were limited to a selected organ in a particular species and focused on a certain compound class. In order to cover a wider range of plant organs, we optimized different preparation methods, which were designed according to the physicochemical properties of the varying plant organs and tissues. Fig. 9 summarizes the results presented in this manuscript and also includes additional plant tissues (distal root, leaf and stem) which are discussed in the ESI (Fig. S11, S14 and S15†). This demonstrates that MS imaging is now applicable to the entire plant from head to roots. It covers all the plant organs with relevance for current and future studies towards a comprehensive understanding of physiological and molecular mechanisms behind complex traits and environmental adaptations. One application is to investigate the changes in metabolites during plant development as shown for mature and germinating seeds of oilseed rape. The detailed localization of more than 90 compounds allowed assignments to certain metabolic processes and the first clues to functions in plant tissues. The wide range of analyzed compounds also enabled us to describe metabolic pathways, as demonstrated for example by the hitherto unseen spatio-temporal distributions of 22 phenylpropanoids representing a metabolomic network associated with sinapate esters which is considered a major component of oilseed rape meal quality.
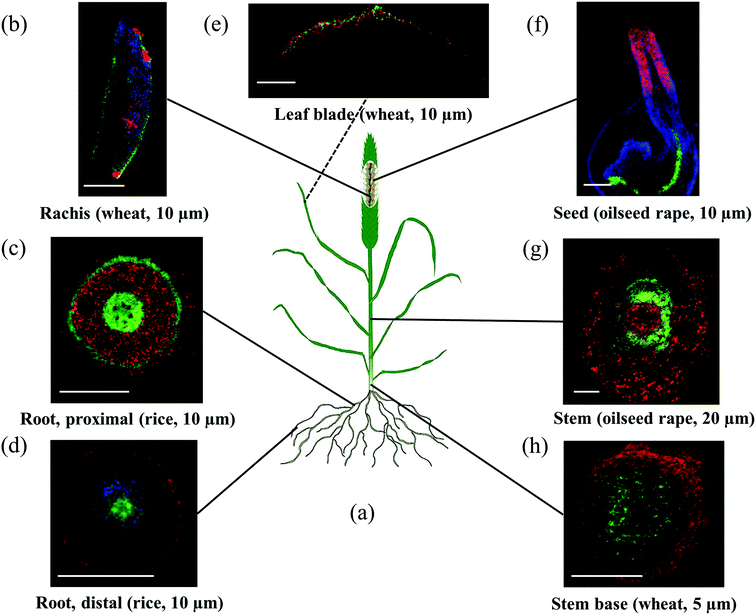 | ||
| Fig. 9 Overview on the different plant structures which have been made available for high resolution MS imaging. Scale bars in all images 500 μm. (a) Sketch of a plant. (b) MS image of wheat inflorescence rachis measured at 10 μm pixel size, details are given in Fig. 5. (c) MS image of a seminal proximal rice root (maturation zone) measured at 10 μm pixel size; details are given in Fig. 7. (d) MS image of a seminal distal rice root (elongation zone) measured at 10 μm pixel size, details are given in ESI, Fig. S11.† (e) MS image of a wheat leaf blade measured at 10 μm pixel size, details are given in ESI, Fig. S14.† (f) MS image of germinating oilseed rape at 10 μm pixel size, details are given in Fig. 1. (g) MS image of oilseed rape stem at 20 μm pixel size, details are given in ESI, Fig. S15.† (h) MS image of wheat stem base at 5 μm pixel size, details are given in Fig. 6. | ||
The ‘untargeted’ nature of MS imaging allows the detection of marker compounds for a specific physiological status, for example to investigate or predict the quality of plant–pathogen interactions. For Fusarium head blight, one of the most devastating diseases affecting grain crops, fungus-specific compounds were detected in the infected wheat seeds and their distribution matched with florescence labeling.
These examples demonstrate the wealth of information that can be obtained from high resolution MS imaging of plant metabolites. If this approach is combined with the classical LC-MS/MS approaches and applied on a larger scale it could be used to build up an inventory of the plant metabolome. The long-term goal would be to establish a plant metabolome atlas for different species similar to the human proteome atlas81 or the Drosophila melanogaster metabolome atlas.82 Similar to the insect field, the first step would be to establish a metabolome atlas for selected species, e.g. wheat as an important crop plant and Arabidopsis thaliana as a frequently used plant model. The initial generation of a baseline tissue map as initiated with the presented study would be an important step in this direction. Metabolites are the end products of cellular processes and therefore, can provide comprehensive information on a plant's physiological status. Therefore an atlas of biochemical networks can lead to novel insights into metabolic capacities of cells,83 or can be used as reference for profiling studies. For example, in pathogen-interaction studies MS imaging data from the infected plants (as shown in section 2.6) could be compared with the atlas data to identify compounds which are specific for this plant physiological status. In general this kind of information can be used as a reference to investigate systemic and local effects of stress (biotic or abiotic), developmental stages as well as wildtype/genotype studies. In addition, the distribution of these markers could give a first indication about their functional role.
Therefore the authors are convinced that high resolution MS imaging will provide new valuable insights into many areas of plant research in the future.
Acknowledgements
The authors would like to acknowledge Fateme Mirzajani (Department of Phytochemistry, Shahid Beheshti University, Iran) for the rice root samples and Christian Obermeier (Department of Plant Breeding, IFZ, Justus Liebig University Giessen, Germany) for providing seeds and stem of oilseed rape. This work was funded by the Hessian Ministry of Science and Arts (HMWK) through LOEWE focus “Ambiprobe” and by the Deutsche Forschungsgemeinschaft DFG Sp314/13-1.References
- R. M. Pérez-Clemente, V. Vives, S. I. Zandalinas, M. F. López-Climent, V. Muñoz and A. Gómez-Cadenas, BioMed Res. Int., 2013, 2013, 654120 CrossRef PubMed.
- R. A. Dixon and D. Strack, Phytochemistry, 2003, 62, 815–816 CrossRef CAS PubMed.
- R. Nakabayashi and K. Saito, Anal. Bioanal. Chem., 2013, 405, 5005–5011 CrossRef CAS PubMed.
- N. Carreno-Quintero, H. J. Bouwmeester and J. J. Keurentjes, Trends Genet., 2013, 29, 41–50 CrossRef CAS PubMed.
- C. Brunetti, R. M. George, M. Tattini, K. Field and M. P. Davey, J. Exp. Bot., 2013, 64, 4011–4020 CrossRef CAS PubMed.
- T. Obata and A. R. Fernie, Cell. Mol. Life Sci., 2012, 69, 3225–3243 CrossRef CAS PubMed.
- D. Balmer, V. Flors, G. Glauser and B. Mauch-Mani, Front. Plant Sci., 2013, 4, 82 Search PubMed.
- G. J. Patti, O. Yanes and G. Siuzdak, Nat. Rev. Mol. Cell Biol., 2012, 13, 263–269 CrossRef CAS PubMed.
- T. Neuberger, H. Rolletschek, A. Webb and L. Borisjuk, in Lipidomics, ed. D. Armstrong, Humana Press, 2009, vol. 579, ch. 24, pp. 485–496 Search PubMed.
- S. L. Shaw and D. W. Ehrhardt, Annu. Rev. Plant Biol., 2013, 64, 351–375 CrossRef CAS PubMed.
- Y. J. Lee, D. C. Perdian, Z. Song, E. S. Yeung and B. J. Nikolau, Plant J., 2012, 70, 81–95 CrossRef CAS PubMed.
- A. K. Mullen, M. R. Clench, S. Crosland and K. R. Sharples, Rapid Commun. Mass Spectrom., 2005, 19, 2507–2516 CrossRef CAS PubMed.
- S. Robinson, K. Warburton, M. Seymour, M. Clench and J. Thomas-Oates, New Phytol., 2007, 173, 438–444 CrossRef CAS PubMed.
- Y. Yoshimura, N. Zaima, T. Moriyama and Y. Kawamura, PLoS One, 2012, 7, e31285 CrossRef CAS PubMed.
- N. Zaima, N. Goto-inoue, T. Hayasaka and M. Setou, Rapid Commun. Mass Spectrom., 2010, 2723–2729 CrossRef CAS PubMed.
- J. H. Jun, Z. Song, Z. Liu, B. J. Nikolau, E. S. Yeung and Y. J. Lee, Anal. Chem., 2010, 82, 3255–3265 CrossRef CAS PubMed.
- R. Shroff, F. Vergara, A. Muck, A. Svatos and J. Gershenzon, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 6196–6201 CrossRef CAS PubMed.
- V. Vrkoslav, A. Muck, J. Cvačka and A. Svatoš, J. Am. Soc. Mass Spectrom., 2010, 21, 220–231 CrossRef CAS PubMed.
- J. Sarsby, M. W. Towers, C. Stain, R. Cramer and O. A. Koroleva, Phytochemistry, 2012, 77, 110–118 CrossRef CAS PubMed.
- M. Peukert, A. Matros, G. Lattanzio, S. Kaspar, J. Abadía and H.-P. Mock, New Phytol., 2012, 193, 806–815 CrossRef CAS PubMed.
- P. Franceschi, Y. Dong, K. Strupat, U. Vrhovsek and F. Mattivi, J. Exp. Bot., 2012, 63, 1123–1133 CrossRef CAS PubMed.
- P. J. Horn, A. R. Korte, P. B. Neogi, E. Love, J. Fuchs, K. Strupat, L. Borisjuk, V. Shulaev, Y. J. Lee and K. D. Chapman, Plant Cell Online, 2012, 24, 622–636 CrossRef CAS PubMed.
- A. R. Korte, Z. Song, B. J. Nikolau and Y. J. Lee, Anal. Methods, 2012, 4, 474 RSC.
- R. J. R. Jaeger, M. Lamshöft, S. Gottfried, M. Spiteller and P. Spiteller, J. Nat. Prod., 2013, 76, 127–134 CrossRef CAS PubMed.
- N. Goto-Inoue, M. Setou and N. Zaima, Anal. Sci., 2010, 26, 821–825 CrossRef CAS PubMed.
- P. J. Horn, J. E. Silva, D. Anderson, J. Fuchs, L. Borisjuk, T. J. Nazarenus, V. Shulaev, E. B. Cahoon and K. D. Chapman, Plant J., 2013, 76, 138–150 CAS.
- A. Korte, M. Yandeau-Nelson, B. Nikolau and Y. Lee, Anal. Bioanal. Chem., 2015, 407, 2301–2309 CrossRef CAS PubMed.
- D. Holscher, R. Shroff, K. Knop, M. Gottschaldt, A. Crecelius, B. Schneider, D. G. Heckel, U. S. Schubert and A. Svatos, Plant J., 2009, 60, 907–918 CrossRef PubMed.
- A. Römpp, S. Guenther, Y. Schober, O. Schulz, Z. Takats, W. Kummer and B. Spengler, Angew. Chem., Int. Ed., 2010, 49, 3834–3838 CrossRef PubMed.
- A. Römpp and B. Spengler, Histochem. Cell Biol., 2013, 139, 759–783 CrossRef PubMed.
- A. Berisha, S. Dold, S. Guenther, N. Desbenoit, Z. Takats, B. Spengler and A. Römpp, Rapid Commun. Mass Spectrom., 2014, 28, 1779–1791 CrossRef CAS PubMed.
- D. Bhandari, T. Shen, A. Römpp, H. Zorn and B. Spengler, Anal. Bioanal. Chem., 2014, 406, 695–704 CrossRef CAS PubMed.
- B. Li, D. R. Bhandari, C. Janfelt, A. Römpp and B. Spengler, Plant J., 2014, 80, 161–171 CrossRef CAS PubMed.
- M. J. Holdsworth, W. E. Finch-Savage, P. Grappin and D. Job, Trends Plant Sci., 2008, 13, 7–13 CrossRef CAS PubMed.
- R. Angelovici, G. Galili, A. R. Fernie and A. Fait, Trends Plant Sci., 2010, 15, 211–218 CrossRef CAS PubMed.
- P. Koehler, G. Hartmann, H. Wieser and M. Rychlik, J. Agric. Food Chem., 2007, 55, 4678–4683 CrossRef CAS PubMed.
- U. Thiyam, P. Claudia, U. Jan and B. Alfred, Eur. Food Res. Technol., 2009, 229, 825–831 CrossRef CAS.
- J. Fang, M. Reichelt, W. Hidalgo, S. Agnolet and B. Schneider, PloS One, 2012, 7, e48006 CrossRef CAS PubMed.
- A. Baumert, C. Milkowski, J. Schmidt, M. Nimtz, V. Wray and D. Strack, Phytochemistry, 2005, 66, 1334–1345 CrossRef CAS PubMed.
- D. Meissner, A. Albert, C. Böttcher, D. Strack and C. Milkowski, Planta, 2008, 228, 663–674 CrossRef CAS PubMed.
- P. Burchard, W. Bilger and G. Weissenböck, Plant Cell Environ., 2000, 23, 1373–1380 CrossRef CAS.
- K. Clauss, E. von Roepenack-Lahaye, C. Bottcher, M. R. Roth, R. Welti, A. Erban, J. Kopka, D. Scheel, C. Milkowski and D. Strack, Plant Physiol., 2011, 155, 1127–1145 CrossRef CAS PubMed.
- J. Mittasch, C. Böttcher, A. Frolov, D. Strack and C. Milkowski, Plant Physiol., 2013, 161, 1656–1669 CrossRef CAS PubMed.
- A. Hüsken, A. Baumert, D. Strack, H. C. Becker, C. Möllers and C. Milkowski, Mol. Breed., 2005, 16, 127–138 CrossRef.
- M. Noda and M. Matsumoto, Biochim. Biophys. Acta, 1971, 231, 131–133 CrossRef CAS.
- B. Manning, K. Schulze and T. McNee, in Wheat growth & development, ed. J. White and J. Edwards, NSW Department of Primary Industries, 2007, pp. 71–86 Search PubMed.
- J. B. Harborne and C. A. Williams, Phytochemistry, 2000, 55, 481–504 CrossRef CAS PubMed.
- R. Tanoue, M. Kobayashi, K. Katayama, N. Nagata and H. Wada, FEBS Lett., 2014, 588, 1680–1685 CrossRef CAS PubMed.
- X. Wang, Curr. Opin. Plant Biol., 2004, 7, 329–336 CrossRef CAS PubMed.
- H. Xue, X. Chen and G. Li, Current Opin. Plant Biol., 2007, 10, 483–489 CrossRef CAS PubMed.
- J. A. Engelman, J. Luo and L. C. Cantley, Nat. Rev. Genet., 2006, 7, 606–619 CrossRef CAS PubMed.
- J. Liu, J. Zhou and D. Xing, PLoS One, 2012, 7, e33817 CrossRef CAS PubMed.
- F. Brignole-Baudouin, N. Desbenoit, G. Hamm, H. Liang, J.-P. Both, A. Brunelle, I. Fournier, V. Guerineau, R. Legouffe, J. Stauber, D. Touboul, M. Wisztorski, M. Salzet, O. Laprevote and C. Baudouin, PLoS One, 2012, 7, e50180 CrossRef CAS PubMed.
- R. Metzner, M. R. Thorpe, U. Breuer, P. Blumler, U. Schurr, H. U. Schneider and W. H. Schroeder, Plant Cell Environ., 2010, 33, 1393–1407 CAS.
- N. A. Brown, C. Bass, T. K. Baldwin, H. Chen, F. Massot, P. W. Carion, M. Urban, A. M. van de Meene and K. E. Hammond-Kosack, J. Pathog., 2011, 2011, 626345 Search PubMed.
- Z. Magbanua, W. Williams and D. Luthe, Maydica, 2013, 58, 182–188 Search PubMed.
- N. A. Brown, M. Urban, A. M. van de Meene and K. E. Hammond-Kosack, Fungal Biol., 2010, 114, 555–571 CrossRef PubMed.
- Y. Saga and H. Tamiaki, Chem. Biodiversity, 2012, 9, 1659–1683 Search PubMed.
- L. C. van Loon, M. Rep and C. M. J. Pieterse, Annu. Rev. Phytopathol., 2006, 44, 135–162 CrossRef CAS PubMed.
- A. M. Mudge, R. Dill-Macky, Y. Dong, D. M. Gardiner, R. G. White and J. M. Manners, Physiol. Mol. Plant Pathol., 2006, 69, 73–85 CrossRef CAS.
- S. Gottwald, B. Samans, S. Lück and W. Friedt, BMC Genomics, 2012, 13, 1–22 CrossRef PubMed.
- C. Tayeh, B. Randoux, F. Laruelle, N. Bourdon, D. Renard-Merlier and P. Reignault, Lipids as Markers of Induced Resistance in Wheat: A Biochemical and Molecular Approach, 2013 Search PubMed.
- J. Raaijmakers, T. Paulitz, C. Steinberg, C. Alabouvette and Y. Moënne-Loccoz, Plant Soil, 2009, 321, 341–361 CrossRef CAS.
- J. S. Singh, V. C. Pandey and D. P. Singh, Agric. Ecosyst. Environ., 2011, 140, 339–353 CrossRef.
- J. Shen, C. Li, G. Mi, L. Li, L. Yuan, R. Jiang and F. Zhang, J Exp. Bot., 2013, 64, 1181–1192 CrossRef CAS PubMed.
- Y. L. Ruan, Annu. Rev. Plant Biol., 2014, 65, 33–67 CrossRef CAS PubMed.
- L. V. Kravchenko, A. I. Shapozhnikov, N. M. Makarova, T. S. Azarova, K. A. L'vova, I. I. Kostyuk, O. A. Lyapunova and I. A. Tikhonovich, Russ. J. Plant Physiol., 2011, 58, 936–940 CrossRef CAS.
- K. Kazan, D. M. Gardiner and J. M. Manners, Mol. Plant Pathol., 2012, 13, 399–413 CrossRef CAS PubMed.
- S. Walter, P. Nicholson and F. M. Doohan, New Phytol., 2010, 185, 54–66 CrossRef CAS PubMed.
- T. Kawamoto, Arch. Histol. Cytol., 2003, 66, 123–143 CrossRef PubMed.
- E. M. Del Ponte, J. M. C. Fernandes and G. C. Bergstrom, J. Phytopathol., 2007, 155, 577–581 CrossRef CAS.
- W. Bouschen, O. Schulz, D. Eikel and B. Spengler, Rapid Commun. Mass Spectrom., 2010, 24, 355–364 CrossRef CAS PubMed.
- M. Koestler, D. Kirsch, A. Hester, A. Leisner, S. Guenther and B. Spengler, Rapid Commun. Mass Spectrom., 2008, 22, 3275–3285 CrossRef CAS PubMed.
- C. Paschke, A. Leisner, A. Hester, K. Maass, S. Guenther, W. Bouschen and B. Spengler, J. Am. Soc. Mass Spectrom., 2013, 24, 1296–1306 CrossRef CAS PubMed.
- T. Schramm, A. Hester, I. Klinkert, J.-P. Both, R. M. A. Heeren, A. Brunelle, O. Laprévote, N. Desbenoit, M.-F. Robbe, M. Stoeckli, B. Spengler and A. Römpp, J. Proteomics, 2012, 75, 5106–5110 CrossRef CAS PubMed.
- G. Robichaud, K. P. Garrard, J. A. Barry and D. C. Muddiman, J. Am. Soc. Mass Spectrom., 2013, 24, 718–721 CrossRef CAS PubMed.
- A. G. Brenton and A. R. Godfrey, J. Am. Soc. Mass Spectrom., 2010, 21, 1821–1835 CAS.
- M. Kanehisa, S. Goto, S. Kawashima, Y. Okuno and M. Hattori, Nucleic Acids Res., 2004, 32, D277–D280 CrossRef CAS PubMed.
- R. Caspi, T. Altman, K. Dreher, C. A. Fulcher, P. Subhraveti, I. M. Keseler, A. Kothari, M. Krummenacker, M. Latendresse, L. A. Mueller, Q. Ong, S. Paley, A. Pujar, A. G. Shearer, M. Travers, D. Weerasinghe, P. Zhang and P. D. Karp, Nucleic Acids Res., 2012, 40, D742–D753 CrossRef CAS PubMed.
- E. Fahy, M. Sud, D. Cotter and S. Subramaniam, Nucleic Acids Res., 2007, 35, W606–W612 CrossRef PubMed.
- M. Uhlen, P. Oksvold, L. Fagerberg, E. Lundberg, K. Jonasson, M. Forsberg, M. Zwahlen, C. Kampf, K. Wester, S. Hober, H. Wernerus, L. Bjorling and F. Ponten, Nat. Biotechnol., 2010, 28, 1248–1250 CrossRef CAS PubMed.
- V. R. Chintapalli, M. Al Bratty, D. Korzekwa, D. G. Watson and J. A. Dow, PLoS One, 2013, 8, e78066 CrossRef CAS PubMed.
- M. Arita, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1543–1547 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5an01065a |
| ‡ Current address: University of Bayreuth, Department of Food Analysis E.-C.-Baumann-Str. 20, 95326 Kulmbach, Germany. E-mail: E-mail: andreas.roempp@uni-bayreuth.de, Tel: 09221/87803111. |
| This journal is © The Royal Society of Chemistry 2015 |