Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
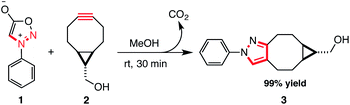
Strain-promoted sydnone bicyclo-[6.1.0]-nonyne cycloaddition†
Stephen
Wallace
and
Jason W.
Chin
*
Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge, CB2 0QH, UK. E-mail: chin@mrc-lmb.cam.ac.uk; Web: http://www.2.mrc-lmb.cam.ac.uk/ccsb/
First published on 5th February 2014
Abstract
The discovery and exploration of bioorthogonal chemical reactions and the biosynthetic incorporation of their components into biomolecules for specific labelling is an important challenge. Here we describe the reaction of a phenyl sydnone 1,3-dipole with a bicyclononyne dipolarophile. This strain-promoted reaction proceeds without transition metal catalysis in aqueous buffer, at physiological temperature, and pressure with a rate comparable to that of other bioorthogonal reactions. We demonstrate the quantitative and specific labelling of a genetically encoded bicyclononyne with a sydnone fluorophore conjugate, demonstrating the utility of this approach for bioorthogonal protein labelling.
Introduction
The discovery and exploration of bioorthogonal chemical reactions and the biosynthetic incorporation of their components into biomolecules for specific labelling is an important challenge. A variety of reactions have been described, including: the reactions of aldehydes and ketones with alpha-effected nucleophiles,1 traceless Staudinger ligations,2 copper-catalysed terminal azide–alkyne cycloadditions,3 strain-promoted azide–alkyne cycloadditions,4 strain-promoted alkyne–nitrone cycloadditions,5 variants of ruthenium-catalysed cross-metathesis6 and palladium-catalysed cross-couplings,7 photo-click chemistry,8 cyanobenzothiazole condensations with 1,2-aminothiols9 and inverse electron-demand Diels–Alder reactions between strained alkenes or alkynes and tetrazines.10 The rate constants for these reactions span approximately 10 orders of magnitude, and range from 10−5 M−1 s−1 to 105 M−1 s−1.11 While Hüisgen's seminal work on azide–alkyne 1,3-dipolar cycloadditions was crucial to the subsequent development of bioorthogonal chemistry and “click” chemistry,12 his concurrent work on the dipolar cycloaddition chemistry of aryl sydnones has received comparatively little attention.13 Sydnones typify a small class of mesoionic 5-membered heterocycles first discovered by Earl et al. in 1935,14 and can be considered as cyclic 1,5-dipolar aromatic azomethine imines.Sydnones undergo thermal [3 + 2] cycloaddition with a range of dipolarophiles to afford substituted pyrazole products. Hüisgen reported that N-phenylsydnone undergoes cycloaddition with acetylene after 25 h at 170 °C to afford N-phenyl pyrazole in 75% isolated yield. Very recently, a copper-catalyzed sydnone–alkyne cycloaddition reaction using a Cu–phenanthroline catalyst was reported.15 This complex catalyzes the cycloaddition of phenyl sydnone and a range of mono-substituted alkynes (20 mol% catalyst, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 tBuOH–H2O, triethanolamine, sodium ascorbate) in up to 99% yield after 16 h at 60 °C. Additionally, lysine residues in a bovine serum albumin (BSA) sample were reacted with a succinimidyl-activated sydnone derivative in vitro. The resulting protein adduct(s) reacted with a dansyl-conjugated propargylamine in the presence of CuSO4/ligand (pH 8.0) at 37 °C after 16 h to afford a mixture of protein species that are labelled (non-quantitatively) to various extents. Based on these precedents and the precedent for accelerating the azide–alkyne and alkyne–nitrone cycloaddition reaction by using pre-installed conformational strain4,5 we decided to investigate the potential reaction of a strained alkyne with phenyl sydnones for the site-specific labelling of proteins.
9 tBuOH–H2O, triethanolamine, sodium ascorbate) in up to 99% yield after 16 h at 60 °C. Additionally, lysine residues in a bovine serum albumin (BSA) sample were reacted with a succinimidyl-activated sydnone derivative in vitro. The resulting protein adduct(s) reacted with a dansyl-conjugated propargylamine in the presence of CuSO4/ligand (pH 8.0) at 37 °C after 16 h to afford a mixture of protein species that are labelled (non-quantitatively) to various extents. Based on these precedents and the precedent for accelerating the azide–alkyne and alkyne–nitrone cycloaddition reaction by using pre-installed conformational strain4,5 we decided to investigate the potential reaction of a strained alkyne with phenyl sydnones for the site-specific labelling of proteins.
Here we describe the reaction of a phenyl sydnone 1,3-dipole with a bicyclononyne dipolarophile. This strain-promoted reaction proceeds without transition metal catalysis in aqueous buffer, at physiological temperature and pressure with a rate comparable to that of other useful bioorthogonal reactions. We demonstrate the quantitative and specific labelling of a genetically encoded bicyclononyne with a sydnone fluorophore conjugate, demonstrating the utility of this approach for bioorthogonal protein labelling.
Results and discussion
We first synthesized phenyl sydnone (1) and exo-((1R,8S)-bicyclo[6.1.0]non-4-yn-9-yl)methanol (BCN, 2). BCN was synthesized in 4 steps via cyclopropanation of 1,5-cyclooctadiene. Phenyl sydnone was synthesized in 2 steps from N-phenyl glycine via N-nitrosylation and intramolecular cyclization using acetic anhydride. The synthesis of 1 requires no column chromatography as the products of each synthetic step can be isolated by simple recrystallization, and 1 is a stable solid at room temperature.Next, we investigated the potential reaction of aryl sydnones with BCN. When equimolar amounts of phenyl sydnone and BCN were combined in methanol at room temperature we observed clean cycloaddition to the corresponding pyrazole in 30 min, with an isolated yield of 99%. This reaction appears to be much faster than the copper-catalysed cycloaddition of phenyl sydnones to terminal alkynes, which requires 20 mol% of a CuI catalyst and 16 h at 60 °C to achieve comparable yields.15 We suggest that the reaction proceeds via initial suprafacial [3 + 2] cycloaddition to afford a diaza-[2.2.1]-bicyclic lactone, which spontaneously undergoes a cycloreversion with the extrusion of carbon dioxide to afford a cyclooctane-fused N-phenyl pyrazole (see S1†) (Scheme 1).13
The rate constant for the cycloaddition was determined under pseudo-first order conditions by following the exponential decay in phenyl sydnone absorbance at 310 nm over time upon reaction with a 10–80 fold excess of BCN in 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 MeOH–H2O. The determined rate constant was 0.054 M−1 s−1 (±0.00067 M−1 s−1) at 21 °C, which is comparable to other strain-promoted [3 + 2] cycloadditions with demonstrated utility.4 The rate constant for the sydnone–BCN cycloaddition is greater than the rate constant reported for Staudinger ligations or uncatalysed ketone condensations with alpha-effected nucleophiles, and comparable to rate constants for some cross-metathesis reactions or strain-promoted azide–alkyne cycloadditions. However, the reaction is slower than the cyanobenzothiazole condensation with 1,2-aminothiols, strain-promoted alkyne–nitrone cycloadditions, photo-click chemistry, copper-catalysed azide–alkyne cycloadditions or inverse electron-demand Diels–Alder reactions between tetrazines and strained alkenes and alkynes.11a,11b
45 MeOH–H2O. The determined rate constant was 0.054 M−1 s−1 (±0.00067 M−1 s−1) at 21 °C, which is comparable to other strain-promoted [3 + 2] cycloadditions with demonstrated utility.4 The rate constant for the sydnone–BCN cycloaddition is greater than the rate constant reported for Staudinger ligations or uncatalysed ketone condensations with alpha-effected nucleophiles, and comparable to rate constants for some cross-metathesis reactions or strain-promoted azide–alkyne cycloadditions. However, the reaction is slower than the cyanobenzothiazole condensation with 1,2-aminothiols, strain-promoted alkyne–nitrone cycloadditions, photo-click chemistry, copper-catalysed azide–alkyne cycloadditions or inverse electron-demand Diels–Alder reactions between tetrazines and strained alkenes and alkynes.11a,11b
To demonstrate the biocompatibility of the reaction a BCN-containing unnatural amino acid (BCNK, 5) was site-specifically incorporated, via genetic code expansion, into a recombinant super-folder GFP protein.16 Labelling of the encoded unnatural amino acid with phenyl sydnone 1 and phenyl sydnone derived fluorophore 6 was characterised (Fig. 1).
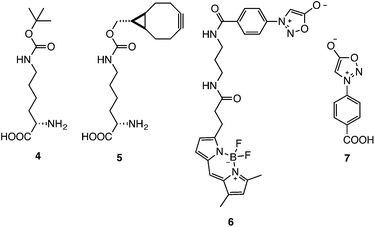 | ||
| Fig. 1 The structures of the unnatural amino acids 4/5 and the sydnone derivatives 6/7 used in this study. | ||
The BCNRS/tRNACUA pair (a mutant of the orthogonal Methanosarcina barkeri pyrrolysyl-tRNA synthetase (MbPylRS)/tRNACUA pair that directs the incorporation of 5 containing three mutations in the enzyme's active site (Y271M, L274G and C313A))10e and a gene encoding a C-terminally hexahistidine-tagged sfGFP containing an amber codon (TAG) at position 150 (sfGFP150TAGHis6) were introduced into E. coli. Addition of 5 (2 mM) led to the amino acid dependent synthesis of full-length sfGFP in good yield (5 mg L−1 of culture). Recombinant sfGFP bearing 5 at position 150 (sfGFP-5150) was purified using Ni–NTA chromatography and ESI-MS confirmed the genetically-directed incorporation of 5. To demonstrate the utility of the reaction for site-specific protein labelling we incubated sfGFP-5150 (4 pmol) with 50 eq. of phenyl sydnone 1 in aqueous buffer (20 mM Tris–HCl, 220 mM imidazole, 300 mM NaCl, pH 8.0) at 37 °C. After 6 h we observed a single product, corresponding to quantitative cycloaddition of the sydnone to the encoded BCN by mass spectrometry (Fig. 2).
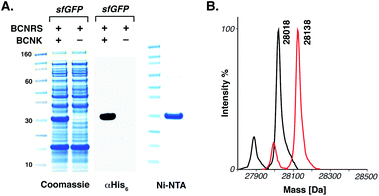 | ||
Fig. 2 The genetic incorporation of 5 in E. coli. (A) Amino acid dependent overexpression of sfGFP-5150. Protein was detected in lysate using an anti-His6 antibody and by Coomassie staining. sfGFP-5150 was purified using Ni–NTA beads. (B) ESI-MS data for amino acid incorporation and quantitative site-specific labelling of sfGFP-5150 with N-phenyl sydnone 2. For sfGFP-5150: calculated 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018, found 28 018, found 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018. For sfGFP-5150 labelled with 1: calculated 28 018. For sfGFP-5150 labelled with 1: calculated 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 136, found 28 136, found 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 138. Minor mass peaks represent proteolysis of the N-terminal methionine (see S4 and S5†). 138. Minor mass peaks represent proteolysis of the N-terminal methionine (see S4 and S5†). | ||
To further demonstrate the specificity of the reaction for protein labelling and to allow visualization of the cycloaddition product on proteins a fluorescent sydnone–BODIPY conjugate (PheSyd–BODIPY-FL, 6) was synthesized in three steps from p-carboxylphenyl sydnone 7 in 52% overall yield. When 4 pmol of purified sfGFP-5150 was incubated with 0.2 nmol 6 (20 mM Tris–HCl, 220 mM imidazole, 300 mM NaCl, pH 8.0, 37 °C, 6 h) the reaction leads to a mobility shift of the protein in Coomassie-stained SDS-PAGE. Fluorescent imaging reveals that the protein is fluorescently labelled. Control experiments in which BocK (4) is incorporated into the protein in place of 5 (sfGFP-4150) demonstrate that the labelling reaction is dependent on the presence of BCNK in the protein. ESI-MS demonstrates quantitative labelling of sfGFP-5150 with 6. To further demonstrate the specificity of the reaction we carried out labelling in an E. coli lysate in which sfGFP-5150 was present at a comparable level to many E. coli proteins. Despite the presence of a number of proteins at comparable levels to sfGFP-5150 we observe clear and selective fluorescent labelling of sfGFP-5150 with 6 (PBS, pH 7.4, 37 °C, 6 h) (Fig. 3).
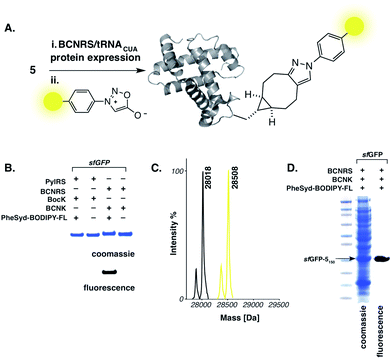 | ||
Fig. 3 (A) Genetic encoding and fluorogenic labelling of 5via the strain-promoted sydnone–bicyclononyne cycloaddition. (B) Specific labelling of sfGFP-5150 with fluorescent sydnone conjugate 6 using Ni–NTA-purified sfGFP-5150. (C) ESI-MS data for quantitative labelling of sfGFP-5150 with PheSyd–BODIPY-FL 6. For sfGFP-5150: calculated 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018, found 28 018, found 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018. For sfGFP-5150 labelled with 6: calculated 28 018. For sfGFP-5150 labelled with 6: calculated 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510, found 28 510, found 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 508. The minor mass peaks represent proteolysis of the N-terminal methionine (see S4 and S6†). (D) Specific fluorescent labelling of sfGFP-5150 with 6 in E. coli cell lysate. 508. The minor mass peaks represent proteolysis of the N-terminal methionine (see S4 and S6†). (D) Specific fluorescent labelling of sfGFP-5150 with 6 in E. coli cell lysate. | ||
Conclusions
In conclusion, we report a new strain-promoted bioorthogonal reaction between N-arylated sydnones and bicyclo-[6.1.0]-nonyne (BCN, 2). The uncatalysed cycloaddition of N-phenyl sydnone and BCN proceeds to completion at ambient temperature in organic solvent and in aqueous buffer. We have demonstrated the quantitative site-specific labelling of proteins bearing a genetically incorporated BCN with phenyl sydnone 1 and PheSyd–BODIPY-FL 6 in aqueous buffer at 37 °C and demonstrated that the reaction is chemoselective with respect to the E. coli proteome. Future work will explore the scope of this reaction for biomolecule labelling and the effect of substituents on the components of the reaction for tuning its properties in vivo.Acknowledgements
This work was supported by Medical Research Council, UK (grants U105181009 and UD99999908). S.W is the recipient of a Career Development Fellowship from the Medical Research Council. We are grateful to Kathrin Lang for experimental assistance.Notes and references
- (a) D. Rideout, Science, 1986, 233, 561 CAS; (b) L. K. Mahal, et al. , Science, 1997, 278, 112 Search PubMed; (c) E. M. Brustad, et al. , J. Am. Chem. Soc., 2008, 130, 17664 CrossRef CAS PubMed; (d) I. Chem, et al. , Nat. Methods, 2005, 2, 99 CrossRef PubMed; (e) P. Agarwal, et al. , Proc. Nat. Acad. Sci. U. S. A., 2013, 110(1), 46 CrossRef PubMed.
- (a) E. Saxon, et al. , Science, 2000, 287, 2007 CrossRef CAS; (b) E. Saxon, et al. , Org. Lett., 2000, 2, 2141 CrossRef CAS PubMed; (c) J. A. Prescher, et al. , Nature, 2004, 430, 873 CrossRef CAS PubMed; (d) M. L. Tsao, et al. , ChemBioChem, 2005, 6, 2147 CrossRef CAS PubMed; (e) K. L. Kiick, et al. , Proc. Nat. Acad. Sci. U. S. A., 2002, 99, 19 CrossRef CAS PubMed.
- (a) Q. Wang, et al. , J. Am. Chem. Soc., 2003, 125, 3192 CrossRef CAS PubMed; (b) D. P. Nguyen, et al. , J. Am. Chem. Soc., 2009, 131, 8720 CrossRef CAS PubMed; (c) S. I. Presolski, et al. , J. Am. Chem. Soc., 2010, 132, 14570 CrossRef CAS PubMed; (d) C. Uttamapinant, et al. , Angew. Chem., Int. Ed., 2012, 51, 5852 CrossRef CAS PubMed.
- (a) N. J. Agard, et al. , J. Am. Chem. Soc., 2004, 126, 15046 CrossRef CAS PubMed; (b) J. M. Baskin, et al. , Proc. Nat. Acad. Sci. U. S. A., 2007, 104, 16793 CrossRef CAS PubMed; (c) X. Ning, et al. , Angew. Chem., Int. Ed., 2008, 47, 2253 CrossRef CAS PubMed; (d) T. Plass, et al. , Angew. Chem., Int. Ed., 2011, 50, 3878 CrossRef CAS PubMed; (e) J. Dommerholt, et al. , Angew. Chem., Int. Ed., 2010, 49, 9422 CrossRef CAS PubMed.
- (a) X. Ning, et al. , Angew. Chem., Int. Ed., 2010, 49, 3065 CrossRef CAS PubMed; (b) C. S. McKay, et al. , Chem. Commun., 2011, 47, 10040 RSC.
- (a) Y. A. Lin, et al. , J. Am. Chem. Soc., 2008, 130, 9642 CrossRef CAS PubMed; (b) Y. A. Lin, et al. , J. Am. Chem. Soc., 2010, 132, 16805 CrossRef CAS PubMed.
- (a) J. M. Chalker, et al. , J. Am. Chem. Soc., 2009, 131, 16346 CrossRef CAS PubMed; (b) C. D. Spicer, et al. , J. Am. Chem. Soc., 2012, 134, 800 CrossRef CAS PubMed.
- (a) W. Song, et al. , J. Am. Chem. Soc., 2008, 130, 9654 CrossRef CAS PubMed; (b) W. Song, et al. , Angew. Chem., Int. Ed., 2008, 47, 2832 CrossRef CAS PubMed; (c) Y. Zhipeng, et al. , Angew. Chem., Int. Ed., 2012, 51, 10600 CrossRef PubMed.
- (a) G. Liang, et al. , Nat. Chem., 2010, 2, 54 CrossRef CAS PubMed; (b) D. P. Nguyen, et al. , J. Am. Chem. Soc., 2011, 133, 11418 CrossRef CAS PubMed.
- (a) M. L. Blackman, et al. , J. Am. Chem. Soc., 2008, 130, 13518 CrossRef CAS PubMed; (b) N. K. Devaraj, et al. , Bioconjugate Chem., 2008, 19, 2297 CrossRef CAS PubMed; (c) K. Lang, et al. , Nat. Chem., 2012, 4, 297 CrossRef PubMed; (d) T. Plass, et al. , Angew. Chem., Int. Ed., 2012, 51, 4166 CrossRef CAS PubMed; (e) K. Lang, et al. , J. Am. Chem. Soc., 2012, 134, 10317 CrossRef CAS PubMed; (f) J. Yang, et al. , Angew. Chem., Int. Ed., 2012, 51, 7476 CrossRef CAS PubMed; (g) J. L. Seitchik, et al. , J. Am. Chem. Soc., 2012, 134, 2898 CrossRef CAS PubMed; (h) E. Kaya, et al. , Angew. Chem., Int. Ed., 2012, 51, 4462 Search PubMed.
- (a) K. Lang, et al. , ACS Chem. Biol., 2014, 9, 16–20 CrossRef CAS PubMed; (b) K. Lang, et al. , Chem. Rev., 2014 DOI:10.1021/cr400355w.
- (a) R. Hüisgen, Angew. Chem., Int. Ed., 1963, 2, 565 CrossRef; (b) E. Lallana, et al. , Angew. Chem., Int. Ed., 2011, 50, 8794 CrossRef CAS PubMed.
- R. Hüisgen, et al. , Angew. Chem., Int. Ed., 1962, 1, 48 CrossRef.
- J. C. Earl, et al. , J. Chem. Soc., 1935, 899 RSC.
- S. Kolodych, et al. , Angew. Chem., Int. Ed., 2013, 52, 12056 CrossRef CAS PubMed.
- J. D. Pedelacq, et al. , Nat. Biotechnol., 2006, 24, 79 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental details, 1H/13C NMR spectral data, protein synthesis and purification. See DOI: 10.1039/c3sc53332h |
| This journal is © The Royal Society of Chemistry 2014 |