 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Functional implications of unusual NOS and SONOS covalent linkages found in proteins
Matthew D.
Lloyd
 *,
Kyle S.
Gregory
*,
Kyle S.
Gregory
 and
K. Ravi
Acharya
and
K. Ravi
Acharya

Department of Life Sciences, University of Bath, Claverton Down, Bath BA2 7AY, UK. E-mail: M.D.Lloyd@bath.ac.uk; Tel: +44-1225-386786
First published on 2nd August 2024
Abstract
The tertiary and quaternary structures of many proteins are stabilized by strong covalent forces, of which disulfide bonds are the most well known. A new type of intramolecular and intermolecular covalent bond has been recently reported, consisting of the Lys and Cys side-chains linked by an oxygen atom (NOS). These post-translational modifications are widely distributed amongst proteins, and are formed under oxidative conditions. Similar linkages are observed during antibiotic biosynthesis, where hydroxylamine intermediates are tethered to the sulfur of enzyme active site Cys residues. These linkages open the way to understanding protein structure and function, give new insights into enzyme catalysis and natural product biosynthesis, and offer new strategies for drug design.
Introduction
Protein tertiary and quaternary structures are often stabilized by covalent bonds which cross-link amino acid side-chains which are non-adjacent in the primary sequence but close together in the folded structure. In peptides, these cross-links can be used to stabilize a desired conformation. Such peptides are typically referred to as “stapled”.1By far the best known of these cross-links are disulfide bonds between two cysteine residues, which form under oxidizing conditions.2 Disulfide bonds are prevalent in extracellular proteins, as these encounter more oxidizing conditions than in the cell. Disulfide bonds have also been implicated in protein folding.3 Disulfide bonds can be readily reduced to cysteine residues in the sulfhydryl form. Isopeptide bonds have also been described, where the Lys and Asn amino acid residue side-chains4,5 are covalently joined together by a peptide (amide) bond or where Lys side-chains are covalently linked with the side-chains of Glu residues5 or the C-terminus.6,7 Important examples of this process are ubiquitination6,7 and cross-linking of proteins upon exposure to organophosphorus nerve agents and pesticides.5
In 2016 a new type of protein covalent bond was described by Ruszkowski and Dauter,8 in which a cysteine sidechain was covalently linked to that of a lysine residue via a short spacer. This was identified using X-ray crystallography in a histidinol phosphate phosphatase structure at 1.32 Å resolution. This new covalent bond stabilized dimer assembly, and the short spacer was proposed to be a methylene group based on the high resolution crystal structure and mass spectrometric analysis.8 At the time it was speculated that this linker arose by reaction of carbon dioxide with the Lys side-chain followed by cross-linking and reduction,8 but this is extremely unlikely. Furthermore, an examination of the Protein Data Bank (PDB) in 20159 showed that there were 14 examples where a methylene bridge could be modelled into positive electron density between a Cys and Lys residue. Many subsequent reports have also observed clear electron density between Lys and Cys sidechains in different high resolution crystal structures, and they propose an oxygen atom as the linker (Table 1).2,10–14 These covalent linkers have also been reported in Clostridium botulinum neurotoxin structures (Fig. 1) (specifically in the cell binding domain) determined by the Acharya laboratory.12–15 Differentiating between a methylene and an oxygen bridge in proteins is non-trivial since these cannot be easily distinguished by mass spectrometry, given the unknown protonation states, or by electron density maps obtained by X-ray crystallography.10,16,17 The identity of the bridge can only be inferred indirectly18 unless very high-resolution crystallographic electron density maps (ideally beyond 1.0 Å resolution) are available17,19 or by sulfur K-edge X-ray absorption spectroscopy data.20
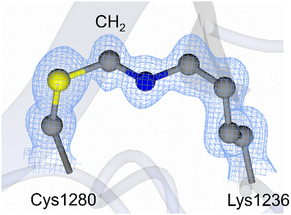 | ||
| Fig. 1 Methylene bridge reported in the 1.15 Å crystal structure of Clostridium botulinum neurotoxin (BoNT) subtype A5 cell binding domain (PDB code 6TWP).15 The sulfur atom is shown in yellow, carbon atoms in grey, and nitrogen in blue. The 2Fo-Fc electron density map (blue mesh) is contoured to 1σ. In the NOS linkage, the CH2 group is substituted with an oxygen atom. | ||
Methylene bridged species
Methanal (Scheme 1, 1) is a ubiquitous environmental pollutant and is produced as a metabolic intermediate in normal cells and cells undergoing oxidative stress.21 The earliest observation of methylene bridge 5 formation by X-ray crystallography was in histidinol phosphate phosphatase crystals treated with methanal 1.8 They proposed formation by the well-known Mannich reaction9 (Scheme 1) in which methanal 1 reacts with a deprotonated Lys 2 side-chain to generate an iminium ion 4, which readily undergoes nucleophilic attack by the deprotonated Cys 3 sulfur. The formation of methylene bridges 5 in other enzymes has also been proposed.15,16 However, it has been argued that histidinol phosphate phosphatase is a special case because the crystals are treated with methanal 1;8,16,18 this is not the case for the majority of proteins where these bridges are observed.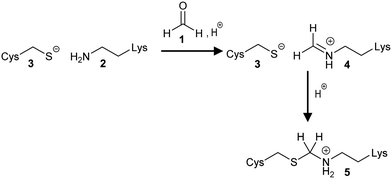 | ||
| Scheme 1 Formation of the methylene bridge 5 using methanal 1. The Mannich reaction proceeds via formation of an imine intermediate 4. | ||
Several studies using model peptides have been carried out.1,22 Metz et al., demonstrated that methanal 1 reacts with the Lys 2 amino side-chain to give the corresponding hydroxymethyl derivative, although some Lys residues were remarkably difficult to derivatize for reasons that are unclear. Methanal 1 also derivatized the N-terminal amino group of peptides as well as the sidechains of Cys 3, Arg, His or Trp residues to give hydroxymethyl derivatives (Cys 3 residues) or in some cases the corresponding imine (Lys 2 and Trp residues).22 No intramolecular methylene bridges within these peptides were observed upon methanal 1 treatment. Pre-treatment of glycine with methanal 1 led to rapid formation the iminium adduct. This adduct reacted with the side-chains of several different residues within peptides to form methylene-bridged adducts, but crucially Cys 3 residues were unreactive. Li et al.,1 noted that peptides containing Lys 2 and Tyr or Arg were generally quite easy to cross-link using methanal 1, whilst cross-linking of Lys 2 and Cys 3 residues was relatively difficult to achieve. They also noted that methylene bridges between Cys 3 and Lys 2 were labile and could be easily hydrolysed.1
Formation of methylene bridges has also been described between Nα-Boc-amino acids and short peptides containing Lys 2, Trp, Cys 3, and His and deoxynucleosides and deoxynucleotides in the presence of methanal 1, although cross-linking between Lys 2 and DNA bases was reversible.21 Deoxythymidine was unreactive, but methylene-bridged adducts to other DNA nucleotides were formed. Carcinogenicity of methanal 1 is suggested to arise by cross-linking of Lys-containing histones with DNA.
Oxygen-bridged species
There has been considerable debate about the identity of the bridging atom.10,16,18,23 Wensien et al., 20212 noted that formation of the NOS bridge took place under oxidative conditions, with oxygen as the putative oxidizing agent. Moreover, the NOS bridge could be reduced using DTT or TCEP.Reanalysis10 of the original report of this novel covalent bond8 suggests the electron density observed in the crystal structure is more consistent with the presence of an oxygen than a –CH2– group, as does reinterpretation10 of the mass spectrometric data. A subsequent high-resolution X-ray structure of Neisseria gonorrhoeae transaldolase2 was also consistent with the presence of a bridging oxygen atom rather than a –CH2– group. Careful analysis also showed that the observed NOS bridge was unlikely to have resulted from radiation damage during X-ray diffraction experiments.2,11 Indeed it is generally accepted that formation of the NOS bridge requires oxidizing conditions, while synchrotron radiation causes reduction.9,12 A subsequent analysis of the same enzyme by sulfur K-edge X-ray absorption spectroscopy (XAS) conclusively showed the presence of a NOS bridge in solution.20
Analysis of crystal structures12–15 in the Acharya group were unable to definitively differentiate between an oxygen or methylene bridge, as these crystals did not diffract beyond 1.0 Å resolution, which is required to unequivocally determine the bridging atom.19 However, it was noted that crystals were obtained in non-reducing conditions.13 The bridging atom is also determinable at lower resolutions (1.1–2.0 Å), however, occupancy and local order impact the reliability of atom identification.11 There are examples where NOS linkages can be detected by mass spectrometry,24 although in general it is challenging to detect such a small change (∼14 Da compared to reduced enzyme) against background noise and extremely difficult to differentiate a NOS linkage from a methylene bridge (∼12 Da) because of the very small difference in mass between these linkages.
NOS bridges have been detected in a large number of other proteins (Table 1) of various types.11,24,25 A notable example of this is the formation of a NOS bridge within the active site of a mutant New Delhi metallo-β-lactamase-1 (NDM-1) which is Zn2+-deficient. Formation of this NOS bridge is thought to reduce thermal stability of the mutant enzyme compared to the wild-type enzyme by almost 10 °C.26 The significance of this observation is uncertain since the formation of the NOS bridge is incompatible with the presence of a metal-binding His residue. A novel SONOS extended covalent bond comprising of two Cys residues, a Lys residue and bridging oxygen atoms was also detected in several proteins11 including the SARS-CoV-2 virus main protease (Mpro) and related polyprotein sequences.27 The residues involved (Cys-22, Cys-44, and Lys-61) are conserved in the main proteases from a wide variety of coronaviruses, including Bat virus, Pangolin virus, and Middle East respiratory syndrome (MERS) virus.27
| Protein | NOS or SONOS | Organism | PDB codes |
|---|---|---|---|
| Arginine decarboxylase28 | NOS | Paramecium bursaria chlorella virus 1 | 2nv9 |
| DNA polymerase29 | NOS | Bacillus phage phi29 | 2py5 |
| Nuclear egress protein 2, nuclear egress protein 130 | NOS | Human cytomegalovirus (strain ad169) | 6t3x |
| Fibre protein31,32 | NOS | Human adenovirus type 19 and 37 | 1uxb, 2wgu |
| Fibre protein33 | NOS | Human adenovirus type 26 | 6qu8 |
| RNA-directed RNA polymerase34 | NOS | Hepatitis C virus subtype 1a | 3qgi |
| Main protease (MPRO) a.k.a. 3C-like proteinase27,35–39 | SONOS | Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) | 7zb6, 7zb7,7zb8, 7uua, 7uub, 7uuc, 7uud, 7uue, 6xmk, 7d1m, 7jr4, 6y2f, 3snd |
| Aldehyde dehydrogenase | NOS | Listeria monocytogenes | 3k9d |
| Carbohydrate binding family 6/xylanase40 | NOS | Clostridium thermocellum | 2y8k |
| Botulinum neurotoxin serotype A (subtypes A1, A2, A5, and A6)13,14,41,42 | NOS | Clostridium botulinum | 3fuo, 5mk6, 7z5t, 7z5s, 8alp, 6twp |
| 3-Deoxy-D-arabinoheptulosonate-7-phosphate synthase43,44 | NOS | Mycobacterium tuberculosis | 5e5g, 3rzi |
| 3-Deoxy-manno-octulosonate cytidylyltransferase | NOS | Acinetobacter baumannii | 4fcu |
| Dihydrodipicolinate synthase | NOS | Pseudomonas aeruginosa | 3na8 |
| dTDP-4-dehydro-6-deoxyglucose 3-epimerase45 | NOS | Streptomyces bikiniensis | 4hn1 |
| DUF4468 domain-containing protein | NOS | Parabacteroides distasonis | 4jhy |
| Glutaryl-CoA dehydrogenase46 | NOS | Burkholderia pseudomallei | 3gqt |
| Putative hydantoin racemase47 | NOS | Klebsiella pneumoniae subsp. Pneumoniae | 3qvl |
| Interferon-activable protein 20448 | NOS | Fischerella ambigua utex 1903 | 5y72 |
| Isochorismatase family protein | NOS | Desulfovibrio vulgaris | 3hu5 |
| KDPG (2-keto-3-deoxy-6-phosphogluconate) aldolase (putative)49 | NOS | Oleispira antarctica | 3vcr |
| LPG2147 (MavC)50 | NOS | Legionella pneumophila | 6ulh |
| Listeriolysin regulatory protein51 | NOS | Listeria monocytogenes egd-e | 6eut |
| Maleylpyruvate hydrolase52 | NOS | Sphingobium sp. (strain nbrc 103272/syk-6) | 6jvv |
| Metal binding protein RUMGNA_00854 | NOS | Ruminococcus gnavus | 3u7z |
| Methyl transferase53 | NOS | Burkholderia glumae | 5je5 |
| New Delhi metallo-β-lactamase 1 (NDM-1) mutant24 | NOS | Klebsiella pneumoniae | 7ct2 |
| Nitroreductase family protein | NOS | Clostridium novyi | 3g14 |
| Oxaloacetate decarboxylase 254 | NOS | Vibrio cholerae | 2nx9 |
| 3-Oxoacyl-[acyl-carrier-protein] synthase 255 | NOS | Escherichia coli | 3ho9 |
| Penicillin-binding protein 456 | NOS | Staphylococcus aureus | 6dz8 |
| Periplasmic divalent cation tolerance protein57 | NOS | Thermotoga maritima | 1vhf |
| Pyruvate-formate lyase-activating enzyme | NOS | Bacteroides vulgatus ATCC 8482 | 3can |
| Regulatory protein58 | NOS | Salmonella typhimurium (strain 14028s/sgsc) | 6ie9 |
| Ribonucleotide reductase R259 | NOS | Escherichia coli | 1mxr |
| L-Ribose isomerase60 | NOS | Acinetobacter | 4q0p, 4q0q, 4q0s |
| SH2 domain-containing protein61 | NOS | Legionella longbeachae serogroup 1 | 6e8h |
| Sucrose hydrolase62 | NOS | Xanthomonas axonopodis pv. Glycines | 3cze, 3czg |
| Sucrose hydrolase63 | NOS | Xanthomonas campestris pv. Campestris | 2wpg |
| Sulfur transferase DsrE | NOS | Allochromatium vinosum | 2hy5 |
| Transaldolase2 | NOS | Neisseria gonorrhoeae | 6zx4, 6zwj, 6zwh, 7b0l |
| DD-Transpeptidase64,65 | NOS | Streptomyces sp. | 1es2, 1es5, 1esi, 1skf |
| UPF0254 protein MJ125166 | NOS | Methanocaldococcus jannaschii | 3wva, 3wvb |
| S-Ureidoglycine aminohydrolase cupin domain-containing protein67 | NOS | Thermotoga maritima | 1o5u |
| Calexcitin68 | NOS | Doryteuthis pealeii | 4ndb |
| Calexcitin69 | NOS | Loligo pealeii | 2ccm |
| Farnesyl diphosphate synthase | NOS | Trypanosoma cruzi | 6sdp |
| Fluorescent protein Dronpa70 | NOS | Echinophyllia sp. Sc22 | 2iov |
| Histidinol-phosphate phosphatase (MtHPP)8 | NOS (originally interpreted as a methylene bridge) | Medicago truncatula | 5eqa |
| F-actin-capping protein71 | NOS | Gallus gallus | 3aa0, 3aa1, 3aa6, 3aa7 |
| Focal adhesion kinase 172 | NOS | Gallus gallus | 6cb0 |
| Glycolipid transfer protein73 | NOS | Bos taurus | 1tfj |
| Homeobox protein Hox-A974 | NOS | Mus musculus | 1puf |
| IfI20475 | NOS | Mus musculus domesticus | 5yzp |
| Kelch-like ech-associated protein 176 | NOS | Mus musculus | 6qmj |
| Profilin-277 | NOS | Mus musculus | 2v8f |
| Tubby protein78 | NOS | Mus musculus | 1i7e |
| cAMP and cAMP-inhibited cGMP 3′,5′-cyclic phosphodiesterase79 | NOS | Rattus norvegicus | 3qpo |
| Galectin-1 | SONOS | Rattus norvegicus | 4ga9 |
| Glutathione S-transferase A1-1 hybrid80 | NOS | Homo sapiens/Rattus norvegicus | 5ld0 |
| Rabphilin-3A81 | NOS | Rattus norvegicus | 4np9 |
| S-Adenosylmethionine synthase isoform type-282 | NOS | Homo sapiens | 6faj, 6fbp, 6g6r |
| Barrier-to-autointegration factor83 | NOS | Homo sapiens | 6usi |
| Butyrophilin subfamily 3 member A184 | NOS | Homo sapiens | 6ism |
| Casein kinase I isoform δ85 | NOS | Homo sapiens | 6f1w |
| Casein kinase I isoform γ-3 | NOS | Homo sapiens | 2izu |
| Cysteine desulfurase, mitochondrial | NOS and SONOS | Homo sapiens | 6uxe, 6w1d, 6wi2, 6wih |
| Density-regulated protein86 | NOS | Homo sapiens | 6vpq |
| Diphosphoinositol polyphosphate phosphohydrolase 187 | NOS | Homo sapiens | 6pck, 6pcl |
| Dual specificity protein kinase CLK188,89 | NOS | Homo sapiens | 6i5h, 6i5i, 6ft9 |
| Early activation antigen CD6990 | NOS | Homo sapiens | 1e8i |
| Gem-associated protein 591 | NOS | Homo sapiens | 5gxh, 5gxi, 5tha |
| Glutathione S-transferase A1-1 hybrid80 | NOS | Homo sapiens/Rattus norvegicus | 5ld0 |
| Histone-lysine N-methyltransferase SUV420H292 | NOS | Homo sapiens | 3rq4 |
| JAK2 protein tyrosine kinase93,94 | NOS | Homo sapiens | 4fvq, 5ut5 |
| Leucine carboxyl methyltransferase 1 | NOS | Homo sapiens | 3iei |
| OGG1, DNA hydrolase95,96 | NOS | Homo sapiens | 1m3q, 2xhi |
| PHD finger protein 297 | NOS | Homo sapiens | 3ptr, 3pu3, 3pu8, 3pua |
| Protein-lysine methyltransferase METTL21C | NOS | Homo sapiens | 4mtl |
| Protein-tyrosine kinase 698 | NOS | Homo sapiens | 6cz3 |
| Selenophosphate synthetase 199 | NOS | Homo sapiens | 3fd5 |
| Sprt-like domain-containing protein spartan100 | NOS | Homo sapiens | 6mdw, 6mdx |
| TBC1 domain family member 7 | NOS | Homo sapiens | 3qwl |
| Ubiquitin-conjugating enzyme E2101 | NOS | Homo sapiens | 1j7d |
| Ubiquitin-conjugating enzyme E2102 | NOS | Homo sapiens | 3e46 |
| Ubiquitin-conjugating enzyme E2 D2103 | NOS | Homo sapiens | 4v3l |
| Ubiquitin-conjugating enzyme E2104 | NOS | Homo sapiens | 6qhk |
A variety of different types of intramolecular and intermolecular bridges have been reported11 (Fig. 2, 6 to 10). The vast majority of these bridges are intramolecular, and occur between residues which are ten or less amino acids apart in the protein primary sequence.11 Intermolecular bridges are less frequently observed.11
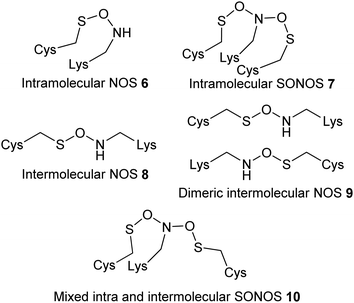 | ||
| Fig. 2 Different types of covalent bonds observed in proteins.11 | ||
Mechanism of NOS bridge formation
The mechanism of NOS and SONOS bridge (Fig. 2) formation has been extensively investigated.2,27,105 It is generally accepted that formation is an oxidative process. The oxidizing species has not been conclusively identified, but likely to be molecular oxygen2,17 or a reactive oxygen species.11,105 Formation of bridges in the presence of molecular oxygen and hydrogen peroxide has been observed.2 Bridge formation is also reversed in the presence of reducing agents in many cases,2 although there are examples where bridges are not reduced in the presence of 1 mM DTT but were in the presence of 5 mM DTT.27Various mechanisms for formation of the NOS bridge (Scheme 2) have been proposed,2,17,27,105 including those involving singlet and triplet oxygen, and various heterolytic and radical mechanisms. Theoretically, oxidation of the Lys 2 or the Cys 3 could take place as the first step. However, it is generally recognised that it is difficult for amines to be oxidized in aqueous solution because they are often protonated.10 Hence oxidation of the Cys 3 sulfur atom is generally considered as the most likely first step.2,27,105 Both heterolytic and radical reactions have been considered as potential mechanisms. Initial calculations favoured a heterolytic mechanism generating ionic species.2 However, subsequent analysis105 suggested that this type of mechanism would favour formation of the sulfinic acid rather than a NOS bridge. On the other hand, a radical reaction gives rise to the desired NOS linkage.
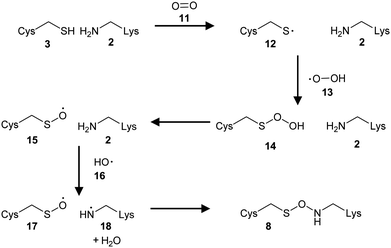 | ||
| Scheme 2 Formation of the NOS linkage in the presence of oxygen.105 | ||
Although it was initially suggested that a Glu residue promoted formation of the NOS bridge,2 it was subsequently noted that the environment in which the bridge exists is highly variable with different surrounding residues.11,25 It is now generally accepted that acid/base catalysis is not required for bridge formation in most proteins.11
It appears that formation of the SONOS bridge (Fig. 2, 7) in SARS-CoV-2 main protease, which contains two Cys residues bridged to a central Lys residue, is asymmetrical.105 Formation of a bridge between Cys-22 and Lys-61 is thermodynamically more favoured than is formation of the alternative NOS intermediate where Cys-44 reacts with Lys-61. A similar mechanism to NOS linkage formation has been proposed.105 However, the presence of two Cys residues mean that formation of a disulfide bond could potentially compete for SONOS formation.35 The Cys 3 to Lys 2 distance is around 2.6 to 2.7 Å in the NOS linkage,11 which compares to 2.18 Å and 2.05 Å for reversible and structural disulfide bonds, respectively.106 It thus appears that some proteins may interconvert between a NOS bridge, a disulfide bond, or the reduced form depending on conditions.2,12,14,17,35 A similar situation was observed with NOS linkages in BoNT structures.13
Although it is generally accepted that NOS and SONOS bridge formation is reversible under reducing conditions,2,27,105 there have been no reported mechanistic or computational studies on the reduction of NOS bridges to Cys and Lys residues. It is therefore not clear whether the reductive process is a direct reversal of NOS formation or if a different mechanism operates.17 It is noted that bridge formation is thermodynamically difficult but reduction is easy.17
Implications of NOS bridge formation in protein structures
Formation of NOS or SONOS bridges can have a wide variety of effect on the protein. For example, formation of the NOS bridge results in allosteric conformational changes.2,13 In the cell binding domain of BoNT serotype A2,12 slight changes in a β-hairpin were observed on going from the reduced to NOS bridged forms. In other cases it can result in subtle allosteric changes resulting in greater protein stability, as measured by resistance to heat-denaturation.2 In enzymes there is also the potential for modulation of catalytic activity. N. gonorrhoeae transaldolase2 has been shown to be substantially inactivated by NOS bridge formation. Catalytic activity of the wild-type enzyme was diminished by a factor of ∼50-fold (as judged by kcat/Km) in the oxidized form. On the other hand, the enzyme from N. meningitidis was ∼4.5-fold more active than the N. gonorrhoeae enzyme, and activity was diminished to a much lesser extent upon NOS bridge formation (∼3.5-fold). Reversal of this inactivation was achieved using reducing agents (DTT or TCEP).2 Similar observations were made with SARS-CoV-2 main protease (Mpro), which is slowly inactivated under non-reducing conditions.35 The enzyme treated with H2O2 at moderate concentrations is fully reactivated upon treatment with DTT. However, treatment of enzyme with high H2O2 concentrations results in irreversible inactivation. It is also notable that the enzyme undergoes a dimer to monomer transition upon treatment with H2O2, and results in reduced protein stability to thermal denaturation. How generally applicable these phenomena will be is difficult to tell, given that intracellular enzymes usually exist in reducing environments which disfavours NOS bridge formation. There has been discussion about NOS bridge formation as part of a generalized redox response,11,25 oxidative stress response,11 and there is some suggestion that NOS or SONOS bridges may also function as oxygen sensors. It is notable that BoNTs eventually end up in cell endosomes, which is an oxidizing environment,107 implying that NOS formation could be pathologically relevant.12–14Exploiting protein NOS linkages for drug design
The demonstration that protein NOS linkages can allosterically modulate the activity of enzymes2,35 opens a new avenue of drug discovery research.2,35 Allosteric drugs have a number of advantages, including the potential for fewer side-effects due to higher selectivity,108 acting with naturally occurring ligands rather than having their effects diminished by the ligand,108 and the ability to drug “undruggable” targets.109 Covalent drugs108,110 modifying the Cys 3 or Lys 2 residue would be particularly useful for targets where NOS bridge formation were activating, although there are currently no examples of this. On the other hand, examples where NOS bridge formation is inhibitory2 may be more difficult to target as this requires that bridge formation (oxidation) is enhanced. Allosteric drugs11 may lend themselves to this approach by enhancing the required protein conformation for NOS bridge formation. A challenge with this approach is the ability to reliably detect NOS bridge formation in the presence of the drug by crystallographic or other biophysical methods. There is also likely to be a complex interplay between NOS bridge formation, inhibitor potency, and cellular redox environment.A recent study on the SARS-CoV-2 main protease (Mpro)35 provides proof of concept (Scheme 3). Treatment of Mpro with the heterobifunctional cross-linker maleimidoacetic acid N-hydroxysuccinimide ester 19 inhibited the enzyme (IC50 = 18 ± 1.8 μM) and caused dimer to monomer conversion, consistent with SONOS bridge formation. Further analysis demonstrated that the derivatized Cys 3 and Lys 2 residues were not those involved in the SONOS linkage, and that Cys-145 (the catalytic cysteine) was derivatized. Nevertheless, this demonstrates that cross-linking of Cys 3 and Lys 2 residues can be achieved.
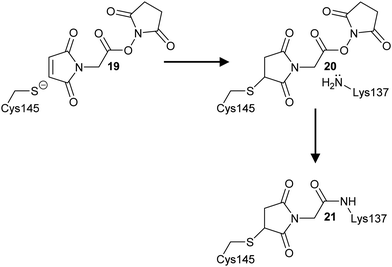 | ||
| Scheme 3 Cross-linking of Cys145 and Lys137 residues in SARS-CoV-2 main protease by maleimidoacetic acid N-hydroxysuccinimide 19 to form adducts 20 and 21. | ||
NOS intermediates in natural product biosynthesis
A recent paper on streptothricin 22 and 23 antibiotic biosynthesis by Wang et al., revealed a NOS linkage 24 between a catalytic Cys residue and a hydroxylamine small-molecule intermediate 25 was formed during enzyme catalysis (Scheme 4).111 This N-formimidoyl fortimicin A synthase enzyme requires flavin adenine dinucleotide to perform this redox reaction. Examination of the enzymes crystal structure111 confirmed the presence of a NOS linkage 25, with geometry and bond lengths like those observed between Cys 3 and Lys 2 residues. Decarboxylation of intermediate 26 to give the formimidoylated product 22 readily cleaves the NOS linkage. In contrast, formation of the N-iminoacetylated product 23 is less well defined since the proposed mechanism requires oxidation of the side-chain and concomitant reduction of the NOS linkage.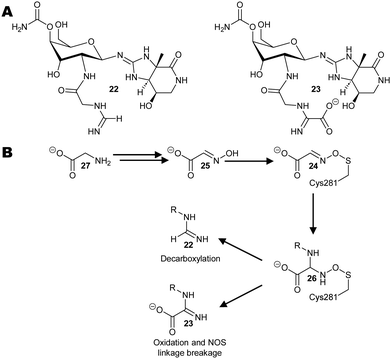 | ||
| Scheme 4 Biosynthesis of streptothricin antibiotics from glycine 27.111 (A) Structures of representative streptothricin antibiotics 22 and 23. (B) Putative reaction pathway via the NOS linked intermediate 24 observed by X-ray crystallography. | ||
Similar enzymes are found in many Streptomyces and other Gram-positive bacteria, and NOS intermediates may play a role in the biosynthesis of complex antibiotics which have similar structures to streptothricins or other natural products where a reactive intermediate needs to be tethered in the enzyme active site.
Conclusions
The discovery of the NOS linkage revealed a new protein post-translation modification which has been shown to be widely distributed amongst proteins.8,11,17,27,105 A mechanism for its formation has been proposed,2,27,105 as have a number of roles in reactive oxygen species biology.25,105 The NOS linkage and the branched SONOS linkage27 may be as widespread within proteins as disulfide bonds. There are still many questions to be answered, including if NOS or SONOS linkages regulate the activity of many different types of proteins,25 if they modulate protein turnover,9 whether the linkage interacts with small molecules such as glutathione or amines,9,25 and what role these linkages play in disease?The reduction of NOS and SONOS linkages in cells is completely unexplored. Although promiscuous molybdenum-containing enzymes (mitochondrial amidoxime reducing component; mARC) which can reduce N–O bonds have been characterized,112–115 it is unclear whether they can reduce NOS or SONOS linkages. All the currently known enzymes are all located within mitochondria. Reduction of N–O bonds has been demonstrated for a wide variety of small molecules. The location of these linkages within substrate proteins would be important. An internal linkage implies that reduction requires either quite large conformational changes or reversible protein unfolding to allow the reductase access. The mARC proteins require NADH and auxiliary proteins to catalyse their reduction, such as cytochrome b5 and cytochrome b5 reductase.113–115 Intriguingly, in plants the enzyme nitrite reductase (which catalyses the reduction of nitrite to nitric oxide) has also been shown to interact with mARC proteins, implying a potential link with cellular signalling.113,114
Abbreviations
| BoNT | Botulinum neurotoxins |
| dTDP | deoxythymidine diphosphate |
| DTT | dithiothreitol |
| mARC | mitochondrial amidoxime reducing component |
| NDM-1 | New Delhi metallo-β-lactamase-1 |
| NOS | nitrogen–oxygen–sulfur linkage |
| SARS-CoV-2 | severe-acute-respiratory-syndrome-related coronavirus |
| SONOS | sulfur–oxygen–nitrogen–oxygen–sulfur linkage |
| TCEP | tris(carboxyethyl) phosphine |
Author contributions
MDL wrote the first draft. All authors reviewed the draft, were involved in revision of the manuscript, and approved the final version.Data availability
No primary research results, software or code are included, and no new data were generated or analysed.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
No specific funding was obtained for this highlight.Notes and references
- B. Li, H. Tang, A. Turlik, Z. Wan, X. S. Xue, L. Li, X. X. Yang, J. Y. Li, G. He, K. N. Houk and G. Chen, Angew. Chem., Int. Ed., 2021, 60, 6646–6652 CrossRef CAS PubMed.
- M. Wensien, F. R. von Pappenheim, L. M. Funk, P. Kloskowski, U. Curth, U. Diederichsen, J. Uranga, J. Ye, P. Fang, K. T. Pan, H. Urlaub, R. A. Mata, V. Sautner and K. Tittmann, Nature, 2021, 593, 460–465 CrossRef CAS PubMed.
- M. Narayan, Protein J., 2021, 40, 134–139 CrossRef CAS PubMed.
- H. J. Kang, F. Coulibaly, F. Clow, T. Proft and E. N. Baker, Science, 2007, 318, 1625–1628 CrossRef CAS PubMed.
- O. Lockridge and L. M. Schopfer, Chem.–Biol. Interact., 2023, 376, 110460 CrossRef CAS PubMed.
- W. J. Gui, G. A. Davidson and Z. H. Zhuang, RSC Chem. Biol., 2021, 2, 450–467 RSC.
- Y. H. Zhou, Q. S. Xie, H. G. Wang and H. Sun, J. Pept. Sci., 2022, 28, e3367 CrossRef CAS PubMed.
- M. Ruszkowski and Z. Dauter, J. Biol. Chem., 2016, 291, 9960–9973 CrossRef CAS PubMed.
- M. Ruszkowski and Z. Dauter, Protein Sci., 2016, 25, 1734–1736 CrossRef CAS PubMed.
- J. M. Wang, Protein Sci., 2019, 28, 472–477 CrossRef CAS PubMed.
- F. Rabe von Pappenheim, M. Wensien, J. Ye, J. Uranga, I. Irisarri, J. de Vries, L. M. Funk, R. A. Mata and K. Tittmann, Nat. Chem. Biol., 2022, 18, 368–375 CrossRef CAS PubMed.
- K. S. Gregory and K. R. Acharya, Toxins, 2023, 15, 92 CrossRef CAS PubMed.
- K. S. Gregory, T. B. Mahadeva, S. M. Liu and K. R. Acharya, Toxins, 2022, 14, 356 CrossRef CAS PubMed.
- K. S. Gregory, A. R. Newell, O. O. Mojanaga, S. M. Liu and K. R. Acharya, Int. J. Mol. Sci., 2022, 23, 9620 CrossRef CAS PubMed.
- J. R. Davies, A. Britton, S. M. Liu and K. R. Acharya, FEBS Open Bio, 2020, 10, 1474–1481 CrossRef CAS PubMed.
- B. W. Matthews, Protein Sci., 2021, 30, 1491–1492 CrossRef CAS PubMed.
- D. Fass and S. N. Semenov, Nature, 2021, 593, 343–344 CrossRef CAS PubMed.
- M. Ruszkowski and Z. Dauter, Protein Sci., 2019, 28, 470 CrossRef CAS PubMed.
- P. Neumann and K. Tittmann, Curr. Opin. Struct. Biol., 2014, 29, 122–133 CrossRef CAS PubMed.
- A. Tamhankar, M. Wensien, S. A. V. Jannuzzi, S. Chatterjee, B. Lassalle-Kaiser, K. Tittmann and S. Debeer, J. Phys. Chem. Lett., 2024, 15, 4263–4267 CrossRef CAS PubMed.
- K. Lu, W. J. Ye, L. Zhou, L. B. Collins, X. Chen, A. Gold, L. M. Ball and J. A. Swenberg, J. Am. Chem. Soc., 2010, 132, 3388–3399 CrossRef CAS PubMed.
- B. Metz, G. F. A. Kersten, P. Hoogerhout, H. F. Brugghe, H. A. M. Timmermans, A. de Jong, H. Meiring, J. ten Hove, W. E. Hennink, D. J. A. Crommelin and W. Jiskoot, J. Biol. Chem., 2004, 279, 6235–6243 CrossRef CAS PubMed.
- J. M. Wang, Protein Sci., 2019, 28, 470–471 CrossRef PubMed.
- W. P. Kong, Y. W. Chen and K. Y. Wong, J. Struct. Biol., 2022, 214, 107922 CrossRef CAS PubMed.
- F. Rabe von Pappenheim and K. Tittmann, Trends Biochem. Sci., 2022, 47, 372–374 CrossRef CAS PubMed.
- Y.-H. Fung, W.-P. Kong, A. S. L. Leung, R. Du, P.-K. So, W.-L. Wong, Y.-C. Leung, Y. W. Chen, K.-Y. Wong and Y. Wong, Biochim. Biophys. Acta, Proteins Proteomics, 2022, 1870, 140833 CrossRef CAS PubMed.
- K. S. Yang, L. R. Blankenship, S. T. A. Kuo, Y. J. Sheng, P. W. Li, C. A. Fierke, D. H. Russell, X. Yan, S. Q. Xu and W. R. Liu, ACS Chem. Biol., 2023, 18, 449–455 CrossRef CAS PubMed.
- R. Shah, R. Akella, E. J. Goldsmith and M. A. Phillips, Biochemistry, 2007, 46, 2831–2841 CrossRef CAS PubMed.
- A. J. Berman, S. Kamtekar, J. L. Goodman, J. M. Lazaro, M. de Vega, L. Blanco, M. Salas and T. A. Steitz, EMBO J., 2007, 26, 3494–3505 CrossRef CAS PubMed.
- Y. A. Muller, S. Haege, S. Alkhashrom, T. Hoellriegl, S. Weigert, S. Dolles, K. Hof, S. A. Walzer, C. Egerer-Sieber, M. Conrad, S. Holst, J. Loesing, E. Sonntag, H. Sticht, J. Eichler and M. Marschall, J. Biol. Chem., 2020, 295, 3189–3201 CrossRef CAS PubMed.
- W. P. Burmeister, D. Guilligay, S. Cusack, G. Wadell and N. Arnberg, J. Virol., 2004, 78, 7727–7736 CrossRef CAS PubMed.
- S. Johansson, E. Nilsson, W. Qian, D. Guilligay, T. Crepin, S. Cusack, N. Arnberg and M. Elofsson, J. Med. Chem., 2009, 52, 3666–3678 CrossRef CAS PubMed.
- A. T. Baker, R. M. Mundy, J. A. Davies, P. J. Rizkallah and A. L. Parker, Sci. Adv., 2019, 5, eaax3567 CrossRef CAS PubMed.
- R. G. Gentles, S. Sheriff, B. R. Beno, C. Wan, K. Kish, M. Ding, X. Zheng, L. Chupak, M. A. Poss, M. R. Witmer, P. Morin, Y.-K. Wang, K. Rigat, J. Lemm, S. Voss, M. Liu, L. Pelosi, S. B. Roberts, M. Gao and J. F. Kadow, Bioorg. Med. Chem. Lett., 2011, 21, 2212–2215 CrossRef CAS PubMed.
- L. M. Funk, G. Poschmann, F. R. von Pappenheim, A. Chari, K. M. Stegmann, A. Dickmanns, M. Wensien, N. Eulig, E. Paknia, G. Heyne, E. Penka, A. R. Pearson, C. Berndt, T. Fritz, S. Bazzi, J. Uranga, R. A. Mata, M. Dobbelstein, R. Hilgenfeld, U. Curth and K. Tittmann, Nat. Commun., 2024, 15, 411 CrossRef CAS PubMed.
- A. D. Rathnayake, J. Zheng, Y. Kim, K. D. Perera, S. Mackin, D. K. Meyerholz, M. M. Kashipathy, K. P. Battaile, S. Lovell, S. Perlman, W. C. Groutas and K.-O. Chang, Sci. Transl. Med., 2020, 12, eabc5332 CrossRef CAS PubMed.
- L. Fu, F. Ye, Y. Feng, F. Yu, Q. Wang, Y. Wu, C. Zhao, H. Sun, B. Huang, P. Niu, H. Song, Y. Shi, X. Li, W. Tan, J. Qi and G. F. Gao, Nat. Commun., 2020, 11, 4417 CrossRef CAS PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed.
- L. Zhu, S. George, M. F. Schmidt, S. I. Al-Gharabli, J. Rademann and R. Hilgenfeld, Antiviral Res., 2011, 92, 204–212 CrossRef CAS PubMed.
- M. A. S. Correia, K. Mazumder, J. L. A. Bras, S. J. Firbank, Y. Zhu, R. J. Lewis, W. S. York, C. M. G. A. Fontes and H. J. Gilbert, J. Biol. Chem., 2011, 286, 22510–22520 CrossRef CAS PubMed.
- Z. Fu, C. Chen, J. T. Barbieri, J.-J. P. Kim and M. R. Baldwin, Biochemistry, 2009, 48, 5631–5641 CrossRef CAS PubMed.
- J. R. Davies, G. S. Hackett, S. M. Liu and K. R. Acharya, Peer J, 2018, 6, e4552 CrossRef PubMed.
- S. Reichau, N. J. Blackmore, W. Jiao and E. J. Parker, PLoS One, 2016, 11, e0152723 CrossRef PubMed.
- W. Jiao, R. D. Hutton, P. J. Cross, G. B. Jameson and E. J. Parker, J. Mol. Biol., 2012, 415, 716–726 CrossRef CAS PubMed.
- R. L. Kubiak, R. K. Phillips, M. W. Zmudka, M. R. Ahn, E. M. Maka, G. L. Pyeatt, S. J. Roggensack and H. M. Holden, Biochemistry, 2012, 51, 9375–9383 CrossRef CAS PubMed.
- D. W. Begley, D. R. Davies, R. C. Hartley, S. N. Hewitt, A. L. Rychel, P. J. Myler, W. C. Van Voorhis, B. L. Staker and L. J. Stewart, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2011, 67, 1060–1069 CrossRef CAS PubMed.
- J. B. French, D. B. Neau and S. E. Ealick, J. Mol. Biol., 2011, 410, 447–460 CrossRef CAS PubMed.
- C. P. Wong, T. Awakawa, Y. Nakashima, T. Mori, Q. Zhu, X. Liu and I. Abe, Angew. Chem., Int. Ed., 2018, 57, 560–563 CrossRef CAS PubMed.
- M. Kube, T. N. Chernikova, Y. Al-Ramahi, A. Beloqui, N. Lopez-Cortez, M.-E. Guazzaroni, H. J. Heipieper, S. Klages, O. R. Kotsyurbenko, I. Langer, T. Y. Nechitaylo, H. Luensdorf, M. Fernandez, S. Juarez, S. Ciordia, A. Singer, O. Kagan, O. Egorova, P. A. Petit, P. Stogios, Y. Kim, A. Tchigvintsev, R. Flick, R. Denaro, M. Genovese, J. P. Albar, O. N. Reva, M. Martinez-Gomariz, H. Tran, M. Ferrer, A. Savchenko, A. F. Yakunin, M. M. Yakimov, O. V. Golyshina, R. Reinhardt and P. N. Golyshin, Nat. Commun., 2013, 4, 2156 CrossRef PubMed.
- K. Puvar, S. Iyer, J. Fu, S. Kenny, K. I. N. Teron, Z.-Q. Luo, P. S. Brzovic, R. E. Klevit and C. Das, Nat. Commun., 2020, 11, 2365 CrossRef CAS PubMed.
- M. Kulen, M. Lindgren, S. Hansen, A. G. Cairns, C. Grundstrom, A. Begum, I. van der Lingen, K. Brannstrom, M. Hall, U. H. Sauer, J. Johansson, A. E. Sauer-Eriksson and F. Almqvist, J. Med. Chem., 2018, 61, 4165–4175 CrossRef CAS PubMed.
- H. Hong, H. Seo and K.-J. Kim, Biochem. Biophys. Res. Commun., 2019, 514, 765–771 CrossRef CAS PubMed.
- M. K. Fenwick, B. Philmus, T. P. Begley and S. E. Ealick, Biochemistry, 2016, 55, 2748–2759 CrossRef CAS PubMed.
- R. Studer, P. Dahinden, W.-W. Wang, Y. Auchli, X.-D. Li and P. Dimroth, J. Mol. Biol., 2007, 367, 547–557 CrossRef CAS PubMed.
- S. B. Singh, J. G. Ondeyka, K. B. Herath, C. Zhang, H. Jayasuriya, D. L. Zink, G. Parthasarathy, J. W. Becker, J. Wang and S. M. Soisson, Bioorg. Med. Chem. Lett., 2009, 19, 4756–4759 CrossRef CAS PubMed.
- R. Maya-Martine, J. A. N. Alexander, C. F. Otten, I. Ayala, D. Vollmer, J. Gray, C. M. Bougault, A. Burt, C. Laguri, M. Fonvielles, M. Arthur, N. C. J. Strynadka, W. Vollmer and J.-P. Simorre, Front. Microbiol., 2019, 9, 3223 CrossRef PubMed.
- J. Badger, J. M. Sauder, J. M. Adams, S. Antonysamy, K. Bain, M. G. Bergseid, S. G. Buchanan, M. D. Buchanan, Y. Batiyenko, J. A. Christopher, S. Emtage, A. Eroshkina, I. Feil, E. B. Furlong, K. S. Gajiwala, X. Gao, D. He, J. Hendle, A. Huber, K. Hoda, P. Kearins, C. Kissinger, B. Laubert, H. A. Lewis, J. Lin, K. Loomis, D. Lorimer, G. Louie, M. Maletic, C. D. Marsh, I. Miller, J. Molinari, H. J. Muller-Dieckmann, J. M. Newman, B. W. Noland, B. Pagarigan, F. Park, T. S. Peat, K. W. Post, S. Radojicic, A. Ramos, R. Romero, M. E. Rutter, W. E. Sanderson, K. D. Schwinn, J. Tresser, J. Winhoven, T. A. Wright, L. Wu, J. Xu and T. J. R. Harris, Proteins: Struct., Funct., Bioinf., 2005, 60, 787–796 CrossRef CAS PubMed.
- S. Yamasaki, R. Nakashima, K. Sakurai, S. Baucheron, E. Giraud, B. Doublet, A. Cloeckaert and K. Nishino, Sci. Rep., 2019, 9, 177 CrossRef PubMed.
- M. Högbom, M. Galander, M. Andersson, M. Kolberg, W. Hofbauer, G. Lassmann, P. Nordlund and F. Lendzian, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3209–3214 CrossRef PubMed.
- H. Yoshida, A. Yoshihara, M. Teraoka, Y. Terami, G. Takata, K. Izumori and S. Kamitori, FEBS J., 2014, 281, 3150–3164 CrossRef CAS PubMed.
- T. Kaneko, P. J. Stogios, X. Ruan, C. Voss, E. Evdokimova, T. Skarina, A. Chung, X. Liu, L. Li, A. Savchenko, A. W. Ensminger and S. S. C. Li, Nat. Commun., 2018, 9, 4549 CrossRef PubMed.
- M. Kim, II, H.-S. Kim, J. Jung and S. Rhee, J. Mol. Biol., 2008, 380, 636–647 CrossRef PubMed.
- E. Champion, M. Remaud-Simeon, L. K. Skov, J. S. Kastrup, M. Gajhede and O. Mirza, Acta Crystallogr., Sect. D: Struct. Biol., 2009, 65, 1309–1314 CrossRef CAS PubMed.
- N. Rhazi, P. Charlier, D. Dehareng, D. Engher, M. Vermeire, J. M. Frère, M. Nguyen-Distèche and E. Fonzé, Biochemistry, 2003, 42, 2895–2906 CrossRef CAS PubMed.
- E. Fonzé, M. Vermeire, M. Nguyen-Distèche, R. Brasseur and P. Charlier, J. Biol. Chem., 1999, 274, 21853–21860 CrossRef PubMed.
- T. Fujishiro, J. Kahnt, U. Ermler and S. Shima, Nat. Commun., 2015, 6, 6895 CrossRef CAS PubMed.
- D. McMullan, R. Schwarzenbacher, L. Jaroszewski, F. von Delft, H. E. Klock, J. Vincent, K. Quijano, P. Abdubek, E. Ambing, T. Biorac, L. S. Brinen, J. M. Canaves, X. P. Dai, A. M. Deacon, M. DiDonato, M. A. Elsliger, S. Eshaghi, R. Floyd, A. Godzik, C. Grittini, S. K. Grzechnik, E. Hampton, C. Karlak, E. Koesema, A. Kreusch, P. Kuhn, I. Levin, T. M. McPhillips, M. D. Miller, A. Morse, K. Moy, O. Y. Jie, R. Page, R. Reyes, F. Rezezadeh, A. Robb, E. Sims, G. Spraggon, R. C. Stevens, H. van den Bedem, J. Velasquez, X. H. Wang, B. West, G. Wolf, Q. P. Xu, K. O. Hodgson, J. Wooley, S. A. Lesley and I. A. Wilson, Proteins: Struct., Funct., Bioinf., 2004, 56, 615–618 CrossRef CAS PubMed.
- P. T. Erskine, A. Fokas, C. Muriithi, H. Rehman, L. A. Yates, A. Bowyer, I. S. Findlow, R. Hagan, J. M. Werner, A. J. Miles, B. A. Wallace, S. A. Wells, S. P. Wood and J. B. Cooper, Acta Crystallogr., Sect. D: Struct. Biol., 2015, 71, 615–631 CrossRef CAS PubMed.
- P. T. Erskine, G. D. E. Beaven, R. Hagan, I. S. Findlow, J. M. Werner, S. P. Wood, J. Vernon, K. P. Giese, G. Fox and J. B. Cooper, J. Mol. Biol., 2006, 357, 1536–1547 CrossRef CAS PubMed.
- A. C. Stiel, S. Trowitzsch, G. Weber, M. Andresen, C. Eggeling, S. W. Hell, S. Jakobs and M. C. Wahl, Biochem. J., 2007, 402, 35–42 CrossRef CAS PubMed.
- S. Takeda, S. Minakata, R. Koike, I. Kawahata, A. Narita, M. Kitazawa, M. Ota, T. Yamakuni, Y. Maeda and Y. Nitanai, PLoS Biol., 2010, 8, e1000416 CrossRef PubMed.
- T. Marlowe, A. Dementiev, S. Figel, A. Rivera, M. Flavin and W. Cance, BMC Mol. Cell Biol., 2019, 20, 10 CrossRef PubMed.
- T. T. Airenne, H. Kidron, Y. Nymalm, M. N. G. West, P. Mattjus and T. A. Salminen, J. Mol. Biol., 2006, 355, 224–236 CrossRef CAS PubMed.
- N. A. LaRonde-LeBlanc and C. Wolberger, Genes Dev., 2003, 17, 2060–2072 CrossRef CAS PubMed.
- X. Fan, J. Jiang, D. Zhao, F. Chen, H. Ma, P. Smith, L. Unterholzner, T. S. Xiao and T. Jin, Nucleic Acids Res., 2021, 49, 2959–2972 CrossRef CAS PubMed.
- T. D. Heightman, J. F. Callahan, E. Chiarparin, J. E. Coyle, C. Griffiths-Jones, A. S. Lakdawala, R. McMenamin, P. N. Mortenson, D. Norton, T. M. Peakman, S. J. Rich, C. Richardson, W. L. Rumsey, Y. Sanchez, G. Saxty, H. M. G. Willems, L. Wolfe, III, A. J. A. Woolford, Z. Wu, H. Yan, J. K. Kerns and T. G. Davis, J. Med. Chem., 2019, 62, 4683–4702 CrossRef CAS PubMed.
- P. Kursula, I. Kursula, M. Massimi, Y.-H. Song, J. Downer, W. A. Stanley, W. Witke and M. Wilmanns, J. Mol. Biol., 2008, 375, 270–290 CrossRef CAS PubMed.
- S. Santagata, T. J. Boggon, C. L. Baird, C. A. Gomez, J. Zhao, W. S. Shan, D. G. Myszka and L. Shapiro, Science, 2001, 292, 2041–2050 CrossRef CAS PubMed.
- C. J. Helal, Z. Kang, X. Hou, J. Pandit, T. A. Chappie, J. M. Humphrey, E. S. Marr, K. F. Fennell, L. K. Chenard, C. Fox, C. J. Schmidt, R. D. Williams, D. S. Chapin, J. Siuciak, L. Lebel, F. Menniti, J. Cianfrogna, K. R. Fonseca, F. R. Nelson, R. O'Connor, M. MacDougall, L. McDowell and S. Liras, J. Med. Chem., 2011, 54, 4536–4547 CrossRef CAS PubMed.
- I. Axarli, A. W. Muleta, E. G. Chronopoulou, A. C. Papageorgiou and N. E. Labrou, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 3416–3428 CrossRef CAS PubMed.
- J. Guillen, C. Ferrer-Orta, M. Buxaderas, D. Perez-Sanchez, M. Guerrero-Valero, G. Luengo-Gil, J. Pous, P. Guerra, J. C. Gomez-Fernandez, N. Verdaguer and S. Corbalan-Garcia, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 20503–20508 CrossRef CAS PubMed.
- J. Panmanee, J. Bradley-Clarke, J. M. Mato, P. M. O'Neill, S. V. Antonyuk and S. S. Hasnain, FEBS J., 2019, 286, 2135–2154 CrossRef CAS PubMed.
- S. Agarwal, M. Smith, I. De La Rosa, K. A. Verba, P. Swartz, M. Segura-Totten and C. Mattos, Acta Crystallogr., Sect. D: Struct. Biol., 2020, 76, 1001–1014 CrossRef CAS PubMed.
- Y. Yang, L. Li, L. Yuan, X. Zhou, J. Duan, H. Xiao, N. Cai, S. Han, X. Ma, W. Liu, C.-C. Chen, L. Wang, X. Li, J. Chen, N. Kang, J. Chen, Z. Shen, S. R. Malwal, W. Liu, Y. Shi, E. Oldfield, R.-T. Guo and Y. Zhang, Immunity, 2019, 50, 1043–1053 CrossRef CAS PubMed.
- A. Luxenburger, D. Schmidt, C. Ianes, C. Pichlo, M. Krueger, T. von Drathen, E. Brunstein, G. J. Gainsford, U. Baumann, U. Knippschild and C. Peifer, Molecules, 2019, 24, 873 CrossRef PubMed.
- I. B. Lomakin, S. Swastik De, J. Wang, A. N. Borkar and T. A. Steitz, Comput. Struct. Biotechnol. J., 2020, 18, 696–704 CrossRef CAS PubMed.
- D. E. Dollins, W. Bai, P. C. Fridy, J. C. Otto, J. L. Neubauer, S. G. Gattis, K. P. M. Mehta and J. D. York, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 9356–9364 CrossRef PubMed.
- V. Nemec, M. Hylsova, L. Maier, J. Flegel, S. Sievers, S. Ziegler, M. Schroeder, B.-T. Berger, A. Chaikuad, B. Valcikova, S. Uldrijan, S. Drapela, K. Soucek, H. Waldmann, S. Knapp and K. Paruch, Angew. Chem., Int. Ed., 2019, 58, 1062–1066 CrossRef CAS PubMed.
- A. Walter, A. Chaikuad, R. Helmer, N. Loaec, L. Preu, I. Ott, S. Knapp, L. Meijer and C. Kunick, PLoS One, 2018, 13, e0196761 CrossRef PubMed.
- A. S. Llera, F. Viedma, F. Sánchez-Madrid and J. Tormo, J. Biol. Chem., 2001, 276, 7312–7319 CrossRef CAS PubMed.
- C. Xu, H. Ishikawa, K. Izumikawa, L. Li, H. He, Y. Nobe, Y. Yamauchi, H. M. Shahjee, X.-H. Wu, Y.-T. Yu, T. Isobe, N. Takahashi and J. Min, Genes Dev., 2016, 30, 2376–2390 CrossRef CAS PubMed.
- H. Wu, A. Siarheyeva, H. Zeng, R. Lam, A. Dong, X.-H. Wu, Y. Li, M. Schapira, M. Vedadi and J. Min, FEBS Lett., 2013, 587, 3859–3868 CrossRef CAS PubMed.
- R. M. Bandaranayake, D. Ungureanu, Y. Shan, D. E. Shaw, O. Silvennoinen and S. R. Hubbard, Nat. Struct. Mol. Biol., 2012, 19, 754–759 CrossRef CAS PubMed.
- A. S. Newton, L. Deiana, D. E. Puleo, J. A. Cisneros, K. J. Cutrona, J. Schlessinger and W. L. Jorgensen, ACS Med. Chem. Lett., 2017, 8, 614–617 CrossRef CAS PubMed.
- S. J. Chung and G. L. Verdine, Chem. Biol., 2004, 11, 1643–1649 CrossRef CAS PubMed.
- B. Dalhus, M. Forsbring, I. H. Helle, E. S. Vik, R. J. Forstrom, P. H. Backe, I. Alseth and M. Bjoras, Structure, 2011, 19, 117–127 CrossRef CAS PubMed.
- J. R. Horton, A. K. Upadhyay, H. Hashimoto, X. Zhang and X. Cheng, J. Mol. Biol., 2011, 406, 1–8 CrossRef CAS PubMed.
- L. Qiu, K. Levine, K. S. Gajiwala, C. N. Cronin, A. Nagata, E. Johnson, M. Krause, J. Tatlock, R. Kania, T. Foley and S. Sun, PLoS One, 2018, 13, e0198374 CrossRef PubMed.
- K.-T. Wang, J. Wang, L.-F. Li and X.-D. Su, J. Mol. Biol., 2009, 390, 747–759 CrossRef CAS PubMed.
- F. Li, J. E. Raczynska, Z. Chen and H. Yu, Cell Rep., 2019, 26, 3336–3346 CrossRef CAS PubMed.
- T. F. Moraes, R. A. Edwards, S. McKenna, L. Pastushok, W. Xiao, J. N. M. Glover and M. J. Ellison, Nat. Struct. Biol., 2001, 8, 669–673 CrossRef CAS PubMed.
- R. C. Wilson, R. C. Hughes, J. W. Flatt, E. J. Meehan, J. D. Ng and P. D. Twigg, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2009, 65, 440–444 CrossRef CAS PubMed.
- L. Buetow, M. Gabrielsen, N. G. Anthony, H. Dou, A. Patel, H. Aitkenhead, G. J. Sibbet, B. O. Smith and D. T. Huang, Mol. Cell, 2015, 58, 297–310 CrossRef CAS PubMed.
- A. K. L. Liess, A. Kucerova, K. Schweimer, L. Yu, T. I. Roumeliotis, M. Diebold, O. Dybkov, C. Sotriffer, H. Urlaub, J. S. Choudhary, J. Mansfeld and S. Lorenz, Structure, 2019, 27, 1195–1210 CrossRef CAS PubMed.
- J. Ye, S. Bazzi, T. Fritz, K. Tittmann, R. A. Mata and J. Uranga, Angew. Chem., Int. Ed., 2023, 62, e202304163 CrossRef CAS PubMed.
- M. A. Sun, Y. J. Wang, Q. Zhang, Y. J. Xia, W. Ge and D. J. Guo, BMC Genom., 2017, 18, 279 CrossRef PubMed.
- C. D. Austin, X. H. Wen, L. Gazzard, C. Nelson, R. H. Scheller and S. J. Scales, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17987–17992 CrossRef CAS PubMed.
- R. Nussinov and C. J. Tsai, The design of covalent allosteric drugs, in Annual Review of Pharmacology and Toxicology, ed. P. A. Insel, 2015, pp. 249–267 Search PubMed.
- W. V. Tee and I. N. Berezovsky, Curr. Opin. Struct. Biol., 2024, 84, 102758 CrossRef CAS PubMed.
- L. Hillebrand, X. J. Liang, R. A. M. Serafim and M. Gehringer, J. Med. Chem., 2024, 67, 7668–7758 CrossRef CAS PubMed.
- Y.-L. Wang, C.-Y. Chang, N.-S. Hsu, I. W. Lo, K.-H. Lin, C.-L. Chen, C.-F. Chang, Z.-C. Wang, Y. Ogasawara, T. Dairi, C. Maruyama, Y. Hamano and T.-L. Li, Nat. Commun., 2023, 14, 2528 CrossRef CAS PubMed.
- B. Clement and M. A. Struwe, Molecules, 2023, 28, 4713 CrossRef CAS PubMed.
- M. Tejada-Jimenez, A. Chamizo-Ampudia, V. Calatrava, A. Galvan, E. Fernandez and A. Llamas, Molecules, 2018, 23, 3287 CrossRef PubMed.
- A. Llamas, A. Chamizo-Ampudia, M. Tejada-Jimenez, A. Galvan and E. Fernandez, BioFactors, 2017, 43, 486–494 CrossRef CAS PubMed.
- G. Ott, A. Havemeyer and B. Clement, JBIC, J. Biol. Inorg. Chem., 2015, 20, 265–275 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |



