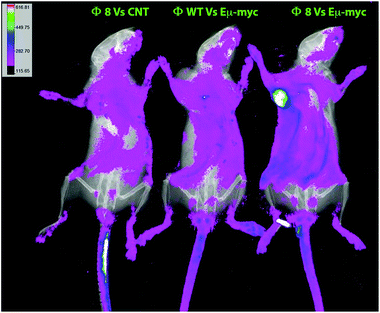A novel phage display based platform for exosome diversity characterization†
Domenico
Maisano‡
 a,
Selena
Mimmi‡
a,
Vincenzo
Dattilo
b,
Fabiola
Marino
a,
Massimo
Gentile
c,
Eleonora
Vecchio
a,
Giuseppe
Fiume
a,
Nancy
Nisticò
a,
Annamaria
Aloisio
a,
Maria Penelope
de Santo
d,
Giovanni
Desiderio
d,
Vincenzo
Musolino
e,
Saverio
Nucera
e,
Francesca
Sbrana
f,
Sebastiano
Andò
*g,
Simone
Ferrero
h,
Andrea
Morandi
i,
Francesco
Bertoni
a,
Selena
Mimmi‡
a,
Vincenzo
Dattilo
b,
Fabiola
Marino
a,
Massimo
Gentile
c,
Eleonora
Vecchio
a,
Giuseppe
Fiume
a,
Nancy
Nisticò
a,
Annamaria
Aloisio
a,
Maria Penelope
de Santo
d,
Giovanni
Desiderio
d,
Vincenzo
Musolino
e,
Saverio
Nucera
e,
Francesca
Sbrana
f,
Sebastiano
Andò
*g,
Simone
Ferrero
h,
Andrea
Morandi
i,
Francesco
Bertoni
 j,
Ileana
Quinto§
a and
Enrico
Iaccino§
*a
j,
Ileana
Quinto§
a and
Enrico
Iaccino§
*a
aDepartment of Experimental and Clinical Medicine, University “Magna Graecia of Catanzaro”, Catanzaro, Italy. E-mail: iaccino@unicz.it
bGenetics Units, IRCCS Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy
cHematology Unit, Annunziata Hospital, Cosenza, Italy
dCNR/Nanotec, Physics Department, University of Calabria, Rende, CS, Italy
eIRC-FSH Department of Health Sciences, University “Magna Græcia” of Catanzaro, Catanzaro, Italy
fSchaefer SEE srl, Via Luigi Einaudi 23, Rovigo, RO, Italy
gHealth Center, University of Calabria, Rende, CS, Italy. E-mail: sebastiano.ando@unical.it
hDivision of Hematology, Department of Molecular Biotechnologies and Health Sciences, University of Torino, Turin, Italy
iDepartment of Experimental and Clinical Biomedical Sciences, University of Florence, Florence, Italy
jIOR, Institute of Oncology Research, Faculty of Biomedical Sciences, USI, Bellinzona, Switzerland
First published on 18th January 2022
Abstract
We present an innovative approach allowing the identification, isolation, and molecular characterization of disease-related exosomes based on their different antigenic reactivities. The designed strategy could be immediately translated into any disease in which exosomes are involved. The identification of specific markers and their subsequent association with exosome subtypes, together with the possibility to engineer target-guided exosome-like particles, could represent the key for the effective adoption of exosomes in clinical practice.
The common approach in oncology diagnosis is currently represented by tissue biopsy, which over time has managed to earn the title of “gold standard” for microscopic and macroscopic examination of tumor tissues together with radio imaging-based investigations.1 Even though tissue biopsies represent a truthful tool in cancer detection and, despite the successes that this technique has accumulated over the years it remains very invasive and risky for the patient.2 In addition to invasiveness, which in some cases can cause negative implications from a clinical point of view,3,4 one of the major problems, and above all relating to small tumor masses, lies precisely in the impossibility, for tissue biopsy, to provide a wide assessment of tumor heterogeneity.5
A large contribution in the analysis of tumor heterogeneity, mostly considering the neoplasm in its continuous molecular evolution, is currently provided by the liquid biopsy.6 In fact, taking into consideration the circulating tumor components in blood vessels and body fluids – known as the tumor circulome7 – liquid biopsy aims to provide significant information about tumor dynamism, managing in many cases to anticipate the still-in-use prognostic and diagnostic approaches,8 always in association with the common investigation techniques in oncology.
Exosomes are a subtype of extracellular vesicles (EV), 30–150 nm in diameter, with an essential role as mediators of both physiological and pathological processes, including cancer,9 neurodegenerative diseases,10 and cardiovascular disease.11 Together with their cargo of proteins, nucleic acids, and lipids, exosomes are secreted through endosome-mediated exocytosis by all cells and widely released in the blood, urine, cerebrospinal fluid, and other body fluids.9,12,13 Due to these features, exosomes are potentially capable of capturing the different biological properties associated with pathological alterations and could be used to design innovative biomarkers for longitudinal measures during disease progression, particularly relevant for those diseases in which early detection could significantly impact disease prognosis. Indeed, representing a molecular footprint of the cell of origin, exosomes are considered a promising tool to design reliable liquid biopsy options for non-invasive monitoring of disease evolution, recurrence, and response to therapy.14,15
In the last decade, a wide array of efficient protocols for exosome purification have been published but none of them allows the association of a specific marker with an exosome subtype and the exosome subtype with a particular function and/or group of functions.16–18 Indeed, the current standards for exosome characterization are largely based on the adoption of a set of markers that, similarly to those used in the identification of stem cells, can heavily differ among different tumor types and genetic backgrounds.19
Here, we describe an innovative method that is not based on a priori definition of exosome-associated markers by applying a phage display approach to a preclinical model of hematological malignancy, used as a proof of principle. Phage display is a powerful tool that allows the identification and the isolation of peptides with high affinity and specificity by a target-guided selective pressure.20 During the last few years, we successfully validated the screening of random peptide libraries (RPLs) as a method to identify phage-derived peptide binders for the idiotypic determinants of the immunoglobulin B cell receptors (IgBCRs).21–23 Using a multiple myeloma preclinical model, we demonstrated that tumor B cell-derived exosomes express the IgBCR of their parental malignant B-cells, thus constituting a personal “barcode” of tumor clones, which can be subsequently targeted by the idiotypic specific binders.8
Unlike our previous works in which we used the IgBCRs as bait for the screening, to verify the suitability of this technology even in the absence of an unequivocal exosome marker, in this work we adopted Eμ-myc transgenic mice lacking immunoglobulin expression.24
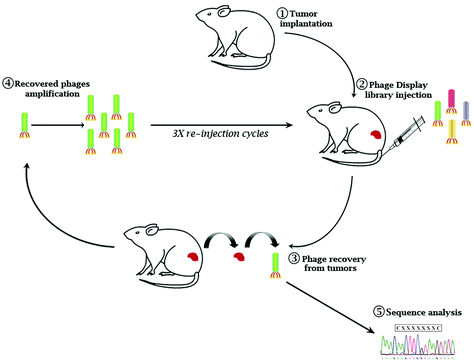
Eμ-myc B-lymphoma cells were isolated from the tumor mass of Eμ-myc tumor-bearing mice and subcutaneously re-inoculated into wild-type syngeneic mice (ESI Fig. 1A†). After 4 cycles of reinoculation, we established an Eμ-myc B-lymphoma xenograft model with homogeneous phenotypic characteristics in terms of the survival rate and time of tumor onset (ESI Fig. 1B and C†). A constrained random phage display library (CX7C) was intravenously injected into Eμ-myc tumor-bearing mice. After 1 hour of circulation, the mice were perfused to remove nonspecific binders. Tumor tissues were collected, phages from the tumor mass were amplified and used for the following cycles of selection (Fig. 1). After three biopanning cycles, 20 independent phages were randomly chosen to retrieve the selected epitopes’ amino acid sequence (ESI Table 1†). To verify the tumor specificity of the clones, phages were conjugated with fluorescein isothiocyanate (FITC) as a fluorophore and used in the cytofluorimetric analysis (ESI Fig. 2†). Based on the percentage of clonal identity and the high-affinity binding to target tumor cells, phage clone 8 (CNAGHLSQC) was chosen for further characterization studies.
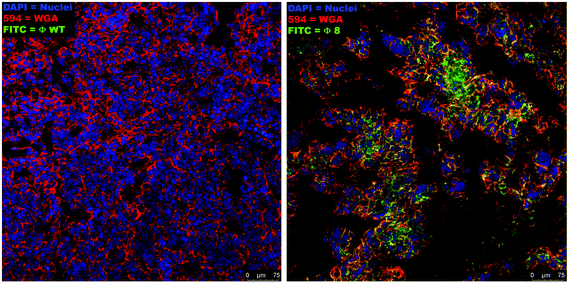
The binding of the selected phage (Φ8) to the target tumor cells was identified by confocal analysis of the sections of fresh tumor tissues stained with Φ8-FITC conjugated, demonstrating a clear overlap of the signal derived from Φ8-FITC (green) and that of wheat germ agglutinin staining (red) directed against glycoproteins of the cancer cells within the tumor mass (Fig. 2). Wild-type (WT) phage was used as a control and no fluorescence signal was recorded.
The Φ8-mediated tumor-specific targeting was also in vivo validated, phages were again used in combination with FITC for conjugating to primary amines on the phage proteins and used as a probe. Phage particles were intravenously administered, and after 2 hours of circulation, conversely to the controls, an evident accumulation of fluorescence in Eμ-myc subcutaneously tumor-bearing mice was seen in Φ8 FITC-conjugated recipients (Fig. 3).
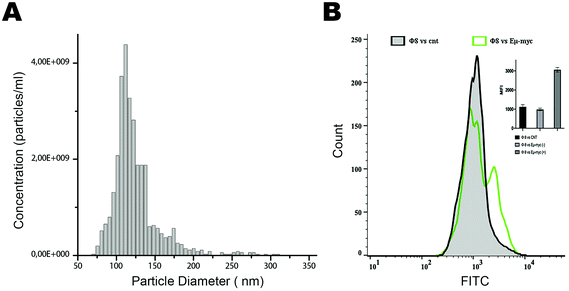
Based on these positive results, we aim to validate this approach for the identification and selection of cancer-released exosomes. After a preliminary exosome purification protocol based on size exclusion chromatography (Fig. 4A), we tested the ability of phage 8 in exosomes detection. In contrast to the exosomes purified from control mice, FACS analysis identified a distinct Φ8 FITC-conjugated positive exosome sub-population in the exosome preparations derived from the Eμ-myc-derived exosomes (Fig. 4B).
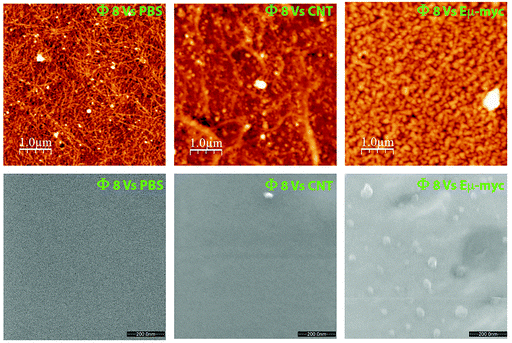
Encouraged by these pieces of evidence, and to validate the potential phage-mediated exosome trapping ability, we performed a prelaminar bacteriophage coating of atomically flat substrates followed by a short incubation with the target exosomes. Both atomic force microscopy (AFM) and environmental scanning electron microscopy (ESEM) showed an appreciable enrichment of Eμ-myc derived exosomes with respect to those incubated with tumor-free derived exosomes and to the substrates coated with the WT phage (Fig. 5).
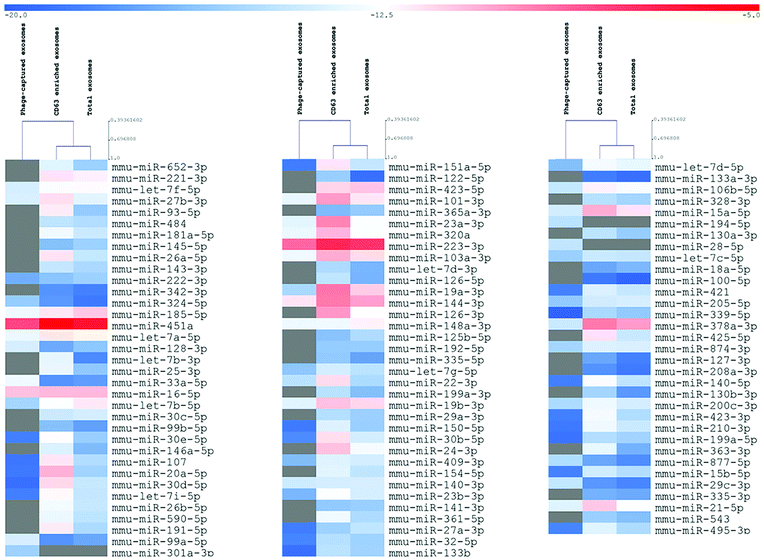
To verify whether the described approach could satisfy the need for reliable technologies for the molecular characterization of disease-related exosome subcategories, we lastly analyzed three differently sorted exosome subpopulations (total, CD63, Φ8) by qRT-PCR analysis of a predefined set of miRNAs that were reported to be associated with exosomes.
Even if not directly addressed to identify the expression levels of miRNAs, the analysis results highlighted the deregulation of crucial tumor progression, invasion, and proliferation players such as miR-15a,25 miR33a-5p,26 and miR-151a-5p.27 The unsupervised hierarchical clustering showed that the total and CD63 positive exosomes demonstrated substantial uniform expression levels of miRNAs, whereas exosomes sorted for their affinity to Φ8 showed an independent exosome-derived miRNA profile (Fig. 6).
Conclusions
We described a proof-of-principle approach of phage display technology as a tool for the identification and isolation of disease-related exosomes. In this regard, phage-based technology could be crucial in the era of personalized therapy. Indeed, the majority of neoantigens should arise from random, spontaneous mutations with little overlap between individual patients. Such a “private” antigen expression indicates the need for a personal biopanning procedure making the proposed model a potentially revolutionary approach. Adapting our validated approach to the enormous possibilities given by the use of patient-derived xenograft (PDX), the reported in vivo phage-display-based platform could represent the cornerstone for a comprehensive molecular characterization of disease-related exosomes. Capturing the biological implications of exosome production and release in different pathological contexts could be useful for further describing innovative biomarker nanotechnological platforms to be applied to diseases in which exosomes play a role, such as cancer and neurodegenerative disorders, and greatly impacting the new era of liquid biopsy.Author contributions
Conceptualization: EI, SM, DM, and IQ; methodology: DM, SM, FB, EV, GF, NN, AA, MPDS, GD, VM, MG, SN, and FS; investigation: DM, SM, VD, FB, EV, GF, NN, AA, MPDS, GD, VM, MG, SN, and FS; funding acquisition: EI, SM, DM, and IQ; supervision: EI, SM, and IQ; writing – the original draft: EI; and writing – review and editing: AM, SF, SA, FB, IQ, and VM.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the following grants: MIUR-PRIN 2017MHJJ55_002 to IQ; GILEAD Fellowship 2018 to EI; EU PON Salute ARS01_00568 to S.A; POR FES/FESR 2014-20-ATS ALCMEONE cup J18C17000610006 to IQ.Notes and references
- H. Liu, M. Ruan, H. Wang, H. Wang, X. Li and G. Song, Can fewer transperineal systematic biopsy cores have the same prostate cancer detection rate as of magnetic resonance imaging/ultrasound fusion biopsy?, Prostate Cancer Prostatic Dis., 2020, 23, 589–595 CrossRef PubMed.
- J. Marrugo-Ramírez, M. Mir and J. Samitier, Blood-based cancer biomarkers in liquid biopsy: A promising non-invasive alternative to tissue biopsy, Int. J. Mol. Sci., 2018, 19, 2877 CrossRef PubMed.
- M. J. Resnick, D. J. Lee, L. Magerfleisch, K. N. Vanarsdalen, J. E. Tomaszewski, A. J. Wein, S. B. Malkowicz and T. J. Guzzo, Repeat prostate biopsy and the incremental risk of clinically insignificant prostate cancer, Urology, 2011, 77, 548–552 CrossRef PubMed.
- K. Shyamala, H. C. Girish and S. Murgod, Risk of tumor cell seeding through biopsy and aspiration cytology, J. Int. Soc. Prev. Community Dent., 2014, 4, 5–11 CrossRef CAS PubMed.
- H. M. Huang and H. X. Li, Tumor heterogeneity and the potential role of liquid biopsy in bladder cancer, Cancer Commun., 2021, 41, 91–108 CrossRef PubMed.
- M. Russano, A. Napolitano, G. Ribelli, M. Iuliani, S. Simonetti, F. Citarella, F. Pantano, E. Dell'Aquila, C. Anesi, N. Silvestris, A. Argentiero, A. G. Solimando, B. Vincenzi, G. Tonini and D. Santini, Liquid biopsy and tumor heterogeneity in metastatic solid tumors: the potentiality of blood samples, J. Exp. Clin. Cancer Res., 2020, 39, 95 CrossRef CAS PubMed.
- J. Wu, S. Hu, L. Zhang, L. Xin, C. Sun, L. Wang, K. Ding and B. Wang, Tumor circulome in the liquid biopsies for cancer diagnosis and prognosis, Theranostics, 2020, 10, 4544–4556 CrossRef CAS PubMed.
- E. Iaccino, S. Mimmi, V. Dattilo, F. Marino, P. Candeloro, A. Di Loria, D. Marimpietri, A. Pisano, F. Albano, E. Vecchio, S. Ceglia, G. Golino, A. Lupia, G. Fiume, I. Quinto and G. Scala, Monitoring multiple myeloma by idiotype specific peptide binders of tumor- derived exosomes, Mol. Cancer, 2000, 16, 159 CrossRef PubMed.
- R. Kalluri, The biology and function of exosomes in cancer, J. Clin. Invest., 2016, 26, 1208–1215 CrossRef PubMed.
- Y. Men, J. Yelick, S. Jin, Y. Tian, M. S. R. Chiang, H. Higashimori, E. Brown, R. Jarvis and Y. Yang, Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS, Nat. Commun., 2019, 10, 4136 CrossRef PubMed.
- A. Ibrahim and E. Marbán, Exosomes: Fundamental Biology and Roles in Cardiovascular Physiology, Annu. Rev. Physiol., 2016, 78, 67–83 CrossRef CAS PubMed.
- R. Kalluri and V. S. LeBleu, The biology, function, and biomedical applications of exosomes, Science, 2020, 367, eaau6977 CrossRef CAS PubMed.
- M. Mathieu, L. Martin-Jaular, G. Lavieu and C. Théry, Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication, Nat. Cell Biol., 2019, 21, 9–17 CrossRef CAS PubMed.
- T. B. Steinbichler, J. Dudás, H. Riechelmann and I. I. Skvortsova, The role of exosomes in cancer metastasis, Semin. Cancer Biol., 2017, 44, 170–181 CrossRef CAS PubMed.
- G. De Rubis, S. Rajeev Krishnan and M. Bebawy, Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis, Trends Pharmacol. Sci., 2019, 40, 172–186 CrossRef CAS PubMed.
- M. Tkach, J. Kowal and C. Théry, Why the need and how to approach the functional diversity of extracellular vesicles, Philos. Trans. R. Soc. London, Ser. B, 2018, 373, 20160479 CrossRef PubMed.
- R. Crescitelli, C. Lässer and J. Lötvall, Isolation and characterization of extracellular vesicle subpopulations from tissues, Nat. Protoc., 2021, 16(3), 1548–1580 CrossRef CAS PubMed.
- Y. Chen, Q. Zhu, L. Cheng, Y. Wang, M. Li, Q. Yang, L. Hu, D. Lou, J. Li, X. Dong, L. P. Lee and F. Liu, Exosome detection via the ultrafast isolation system: EXODUS, Nat. Methods, 2021, 18, 212–218 CrossRef CAS PubMed.
- I. Manna, E. Iaccino, V. Dattilo, S. Barone, E. Vecchio, S. Mimmi, E. Filippelli, G. Demonte, S. Polidoro, A. Granata, S. Scannapieco, I. Quinto, P. Valentino and A. Quattrone, Exosome-associated miRNA profile as a prognostic tool for therapy response monitoring in multiple sclerosis patients, FASEB J., 2018, 32, 4241–4246 CrossRef CAS PubMed.
- S. Mimmi, D. Maisano, I. Quinto and E. Iaccino, Phage Display: An Overview in Context to Drug Discovery, Trends Pharmacol. Sci., 2019, 40, 87–91 CrossRef CAS PubMed.
- C. Palmieri, C. Falcone, E. Iaccino, F. M. Tuccillo, M. Gaspari, F. Trimboli, A. De Laurentiis, L. Luberto, M. Pontoriero, A. Pisano, E. Vecchio, O. Fierro, M. R. Panico, M. Larobina, S. Gargiulo, N. Costa, F. Dal Piaz, M. Schiavone, C. Arra, A. Giudice, G. Palma, A. Barbieri, I. Quinto and G. Scala, In vivo targeting and growth inhibition of the A20 murine B-cell lymphoma by an idiotype- specific peptide binder, Blood, 2010, 116, 226–238 CrossRef CAS PubMed.
- S. Mimmi, E. Vecchio, E. Iaccino, M. Rossi, A. Lupia, F. Albano, F. Chiurazzi, G. Fiume, A. Pisano, S. Ceglia, M. Pontoriero, G. Golino, P. Tassone, I. Quinto, G. Scala and C. Palmieri, Evidence of shared epitopic reactivity among independent B-cell clones in chronic lymphocytic leukemia patients, Leukemia, 2016, 30, 2419–2422 CrossRef CAS PubMed.
- S. Mimmi, D. Maisano, N. Nisticò, E. Vecchio, F. Chiurazzi, K. Ferrara, M. Iannalfo, A. D'Ambrosio, G. Fiume, E. Iaccino and I. Quinto, Detection of chronic lymphocytic leukemia subpopulations in peripheral blood by phage ligands of tumor immunoglobulin B cell receptors, Leukemia, 2021, 35, 610–614 CrossRef CAS PubMed.
- A. W. Harris, C. A. Pinkert, M. Crawford, W. Y. Langdon, R. L. Brinster and J. M. Adams, The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells, J. Exp. Med., 1988, 167, 353–371 CrossRef CAS PubMed.
- S. B. Tuncer, D. Akdeniz, B. Celik, S. Kilic, O. Sukruoglu, M. Avsar, L. Ozer, M. Ekenel, S. Ozel and H. Yazici, The expression levels of miRNA-15a and miRNA-16-1 in circulating tumor cells of patients with diffuse large B-cell lymphoma, Mol. Biol. Rep., 2019, 46, 975–980 CrossRef CAS PubMed.
- M. Sasaki, T. Ishikawa, M. Ishiguro, S. Okazaki, S. Yamauchi, A. Kikuchi, T. Matsuyama, K. Kawada, M. Tokunaga, H. Uetake and Y. Kinugasa, The effectiveness of plasma miR-33a-5p as a predictive biomarker for the efficacy of colorectal cancer chemotherapy, Oncol. Lett., 2021, 21, 489 CrossRef CAS PubMed.
- P. Roth, A. Keller, J. D. Hoheisel, P. Codo, A. S. Bauer, C. Backes, P. Leidinger, E. Meese, E. Thiel, A. Korfel and M. Weller, Differentially regulated miRNAs as prognostic biomarkers in the blood of primary CNS lymphoma patients, Eur. J. Cancer, 2015, 51, 382–390 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr06804k |
| ‡ These authors shared first authorship. |
| § These authors shared senior authorship. |
| This journal is © The Royal Society of Chemistry 2022 |