 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ultra-selective, ultra-clean 1D rotating-frame Overhauser effect spectroscopy†
Emma L.
Gates
 a,
Marshall J.
Smith
a,
Marshall J.
Smith
 a,
Jonathan P.
Bradley
a,
Jonathan P.
Bradley
 b,
Myron
Johnson
b,
Myron
Johnson
 b,
Göran
Widmalm
b,
Göran
Widmalm
 c,
Mathias
Nilsson
c,
Mathias
Nilsson
 a,
Gareth A.
Morris
a,
Gareth A.
Morris
 a,
Ralph W.
Adams
a,
Ralph W.
Adams
 *a and
Laura
Castañar
*a and
Laura
Castañar
 *ad
*ad
aDepartment of Chemistry, University of Manchester, Oxford Road, Manchester, M13 9PL, UK. E-mail: laura.castanaracedo@manchester.ac.uk; ralph.adams@manchester.ac.uk
bJohnson Matthey, Johnson Matthey Technology Centre, Sonning Common, Blounts Court Road, Reading RG4 9NH, UK
cDepartment of Organic Chemistry, Arrhenius Laboratory, Stockholm University, S-106 91 Stockholm, Sweden
dDepartment of Organic Chemistry, Faculty of Chemical Science, University Complutense of Madrid, Ciudad Universitaria, 28040 Madrid, Spain. E-mail: lcastana@ucm.es
First published on 3rd April 2023
Abstract
An ultra-selective 1D NMR experiment – GEMSTONE-ROESY – enables clear, unambiguous assignment of ROE signals in the not uncommon situation that traditional selective methods fail. Its usefulness is demonstrated in the analysis of the natural products cyclosporin and lacto-N-difucohexaose I, providing detailed insight into the structures and conformations of these molecules.
Solution-state nuclear magnetic resonance (NMR) spectroscopy is a versatile and powerful tool, which provides invaluable data about molecular structure and dynamics of molecules in solution. The structural information gained by NMR is unmatched by other solution-state analytical methods. Classically, 2D NMR methods1 are often used for structure elucidation of small molecules, providing a full profile of bond connections or through-space interactions of a molecule. However, 2D methods can be very time-consuming. Alternatively, direct selection of key signals can in many cases provide the required information for structural analysis more efficiently. In such instances, 1D selective experiments2 are fast alternative approaches, taking a fraction of the time of their 2D counterparts. One of the main barriers to the use of 1D selective methods is their inability to excite a single signal selectively when multiplets overlap. Signals can be resolved and identified using pure shift NMR approaches,3–6 but attempts at frequency-selective excitation of a single multiplet from an overlapped region using conventional 1D selective experiments give ambiguous results. Recent developments have led to the production of an ultra-selective method, GEMSTONE (gradient-enhanced multiplet-selective targeted-observation NMR experiment)7 which provides selectivity similar to that of the previously state-of-the-art ultra-selective method, the CSSF (chemical shift selective filter),8 but in a single transient. Unlike CSSF, GEMSTONE retains the full time advantage of selective 1D over 2D experiments. GEMSTONE has recently been demonstrated with common structure elucidation methods such as NOESY7,9 (nuclear Overhauser effect spectroscopy) and TOCSY10,11 (total correlation spectroscopy), providing detailed through-space and through-bond structural information, respectively.
The NOE (nuclear Overhauser effect) is the transfer of magnetisation via cross-relaxation between nuclei that share a dipolar interaction. Such through-space interactions give important insights into molecular conformation and configuration, which can greatly aid structure elucidation.12 NOEs can be either positive or negative depending on the magnetic field strength used and the dynamic properties of the species observed.9,13 A clear issue arises when these positive and negative contributions cancel. This is typically seen for molecules with a molecular weight in the region of 1000–2000 g mol−1, depending on the solvent viscosity and the magnetic field strength of the NMR spectrometer. The ROESY (rotating-frame Overhauser effect spectroscopy) experiment was developed to circumvent this issue.14 While NOEs can be positive or negative (or zero), ROEs are always positive. Here a new 1D GEMSTONE-ROESY method is therefore proposed as an ultra-selective method for observing through-space interactions where NOESY experiments fail to provide useful data.
Cyclosporin, an oral immunosuppressant used to prevent organ transplant rejection, falls into the 1000–2000 g mol−1 molecular weight range and generally shows poor NOEs.15 Unexpectedly, cyclosporin is orally active despite failing to meet the usual requirements for bioavailability;16 it is a “beyond the rule-of-five” compound. A key reason for its oral activity is the protean nature of cyclosporin, which adopts different conformations in different chemical environments.17 Analysis of 3D conformation is therefore vital to understanding how it behaves in different contexts, in addition to facilitating structure elucidation. The need for ultra-selective excitation methods such as GEMSTONE-ROESY is apparent in the extensive multiplet overlap present in several regions of the 1H NMR spectrum of cyclosporin (Fig. 1a). Very weak NOEs are observed (see ESI,† Fig. S3) highlighting the need for ROESY experiments if through-space interactions are to be measured.
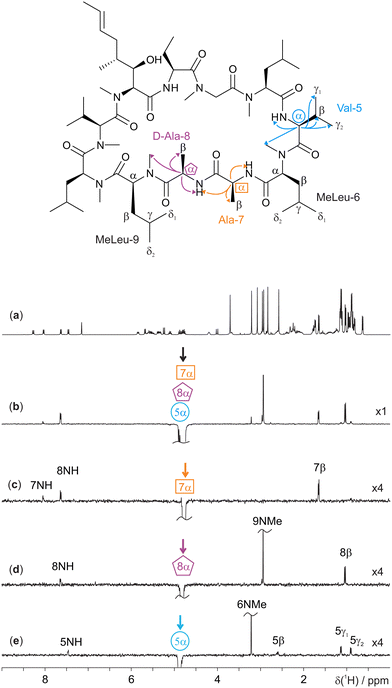 | ||
| Fig. 1 400 MHz 1H NMR spectra of 70 mM cyclosporin in C6D6. Molecular structure with relevant assignments18 is shown above the spectra, with arrows representing the 1H-1H ROEs observed; for simplicity, individual protons are not shown. (a) Conventional 1H 1D spectrum, (b) 1D selective EASY-ROESY centred at 4.82 ppm using a 20 Hz selective refocusing pulse, and (c–e) GEMSTONE-ROESY spectra exciting at 4.78 (Ala-7α), 4.82 (D-Ala-8α), and 4.88 (Val-5α) ppm, respectively. The GEMSTONE adiabatic pulse duration was set as 100 ms and a selective refocusing pulse with a 200 Hz bandwidth was used. All ROESY spectra were acquired using a 200 ms spin-lock mixing period. Further experimental details are given in the ESI.† Spectra (c–e) are scaled by a factor of 4 to match the intensity of the spectrum in panel b. | ||
GEMSTONE ensures that only on-resonance signals are retained, while all off-resonance signals are dephased.7 It therefore requires that the exact chemical shift of the signal of interest be known. Pure shift methods3–6 offer the ability to measure chemical shifts without multiplet structure; a single peak at the chemical shift is observed for each chemical environment. In this case, a PSYCHE (pure shift yielded by chirp excitation) pure shift 1H NMR spectrum was acquired (see ESI,† Fig. S2).19,20 The three overlapping multiplets in the spectral region 4.7–4.9 ppm (Fig. 1a) correspond to the alanine-7α (4.78 ppm), D-alanine-8α (4.82 ppm) and valine-5α (4.88 ppm) signals,18 with chemical shifts which are seen to be approximately 20 Hz apart in the pure shift spectrum. As expected, a conventional 1D selective EASY-ROESY21,22 spectrum (Fig. 1b) struggles to distinguish between the three resonances. All three overlapped signals are excited, to different extents, resulting in ROE peaks that cannot be assigned unequivocally to a given proton. Ilgen et al. recently published a pure shift 2D EASY-ROESY method to resolve the observed ROE signals better.22 Here, an alternative, faster solution is presented in the form of ultra-selective 1D excitation. This retains the advantage of short experiment time, and excites one chemical shift at a time, allowing ROEs to be assigned directly. This application of the ultra-selective GEMSTONE-ROESY method is illustrated in Fig. 1c–e, where the previously inseparable signals have been individually selected. In contrast to Fig. 1b, Fig. 1c–e show ROEs originating in magnetisation transfer from a single resonance. Enabling the selection of individual α-protons allows the signals of each amino acid residue to be identified, along with those of adjacent residues.
The new GEMSTONE-ROESY pulse sequence is shown in Fig. 2. The ‘EASY-ROESY’ mixing element has been used here, to attenuate TOCSY interference, along with zero-quantum coherence (ZQC) suppression elements.23 The ultra-selective nature of the experiment originates from the GEMSTONE element.7 The combination of dual swept-frequency pulses with simultaneous pulsed field gradients (PFGs), labelled G1, causes all signals within the sample to become spatially encoded. Only on-resonance signals survive, all off-resonance signals are dephased. The selectivity of the experiment is inversely proportional to the total duration of the swept-frequency pulses. However, sensitivity decreases with increased duration, due in part to relaxation, so a compromise must be made between selectivity and sensitivity. The GEMSTONE-ROESY sequence uses additional PFGs to minimise subtraction artefacts, as previously described in the literature,22,24,25 leaving ultra-clean spectra. Incomplete suppression of unwanted signals can lead to the presence of subtraction artefacts in the spectra, hampering the identification of low intensity ROEs (see ESI,† Fig. S4). A sensitivity penalty is paid when PFGs are used to select the desired coherences, as a single coherence transfer pathway (CTP) is selected and there are diffusion and convection losses, but this is a small price to pay for the greatly improved spectral quality. Small phase and intensity anomalies are occasionally observed due to the final hard spin echo; details are provided in Fig. S6 (ESI†).
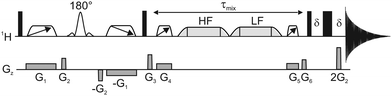 | ||
| Fig. 2 Schematic representation of the 1D GEMSTONE-ROESY pulse sequence. Narrow and wide rectangles represent hard 90° and 180° radiofrequency pulses, respectively. The open trapezoids with directional arrows are adiabatic 180° pulses used for GEMSTONE selection and ZQC suppression. The labelled 180° pulse shape denotes a band-selective refocusing pulse. The spin-lock element is shown by light grey trapezoids, held first at a high-field (HF) and then at a low-field (LF) offset. The mixing period, τmix, incorporates the spin-lock and the ZQC suppression elements. Pulsed field gradients are represented by grey boxes in the Gz channel. The delay δ is just sufficient for the gradient pulse and recovery delay. Further details of the pulse sequence are given in the ESI.† | ||
A further example of the utility of the new GEMSTONE-ROESY method is shown in a structural study of lacto-N-difucohexaose I (LNDFH I), a hexasaccharide found in human breast milk.26,27 LNDFH I exemplifies the complexity of carbohydrate structures by being branched, having different anomeric configurations of the constituent sugar residues that are linked at various positions, and having some of the sugar residues being of the same kind (fucose and galactose are present twice in this case) thereby increasing the chance of 1H NMR spectral overlap of resonances. TOCSY experiments can be employed to determine the bond connectivities within individual monosaccharide units. However, to identify neighbouring monosaccharides and gain insight into the 3D configuration and conformation, determination of through-space interactions is vital. Fig. 3c and d show 1D GEMSTONE-ROESY spectra for the methyl groups of the two fucosyl residues.
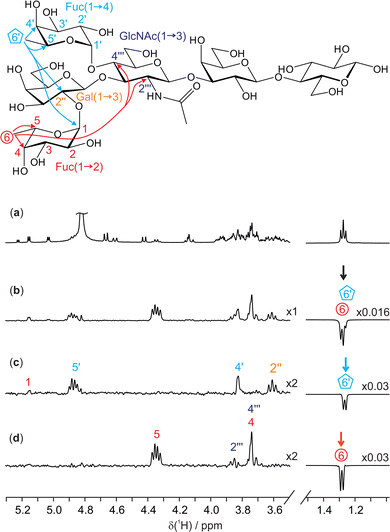 | ||
| Fig. 3 400 MHz 1H NMR spectra of 25 mM lacto-N-difucohexaose I (LNDFH I) in D2O. Molecular structure with relevant assignments28 is shown above the spectra, with arrows representing the 1H–1H ROEs observed; for simplicity, individual protons are not shown. (a) Conventional 1D 1H spectrum; (b) 1D selective EASY-ROESY using a 10 Hz selective refocusing pulse, centred at 1.28 ppm, and (c and d) GEMSTONE-ROESY spectra exciting the individual methyl doublets for H6′ (1.27 ppm) and H6 (1.28 ppm), respectively. The GEMSTONE adiabatic pulse duration was set between 120 and 130 ms and a selective refocusing pulse with a 200 Hz bandwidth was used. All ROESY spectra were acquired using a 200 ms mixing period. Further experimental details are given in the ESI.† Spectra (c and d) are scaled by a factor of 2 to match the intensity of the spectrum in panel b. | ||
The 1H NMR spectrum (Fig. 3a) shows an apparent triplet at 1.27 ppm, but this is actually the sum of two doublets: the pure shift spectrum (see ESI,† Fig. S7) shows two singlets with a chemical shift separation of 6.1 Hz (12 ppb). The two signals originate from the methyl groups H6 and H6′ of the 1,2- and 1,4-linked fucosyl residues in LNDFH I, respectively. As seen in Fig. 3b, a conventional 1D selective experiment cannot single out just one of the two doublets, making an ultra-selective technique such as GEMSTONE necessary. In contrast, Fig. 3c and d show GEMSTONE-ROESY spectra in which the individual methyl groups have been cleanly selected. From Fig. 3c, the ROE correlation between H6′ and the signal at 3.62 ppm, H2′′ of the 1,3-linked galactose, provides evidence for the close proximity of these monosaccharide units. Similar conclusions can be drawn from Fig. 3d, where an ROE is observed between H6 and the signals at 3.74 and 3.85 ppm, assigned to H4′′′ and H2′′′ of N-acetylglucosamine, respectively. Both H6′ and H6 show a large ROE to their respective close neighbours H5′ and H5, 4.88 and 4.35 ppm, respectively. Significant ROEs are also observed to H4′ and H4 (at 3.84 and 3.74 ppm, respectively), indicating that protons 4 and 5 are on the same side of the pyranose ring in each case, giving insight into the configuration and conformation of these sugar residues. Small ROEs are also observed between the two fucosyl residues (H6′ to H1 in LNDFH I). ROEs show that the 1,2-linked fucosyl and N-acetylglucosamine residues are relatively close in space, providing valuable insight into the 3D conformation of the hexasaccharide.
The new ultra-selective, ultra-clean 1D GEMSTONE-ROESY experiment has been shown to provide unambiguous ROE data even in spectral regions of severe multiplet overlap. GEMSTONE-ROESY offers a simple alternative where traditional 1D selective experiments have insufficient selectivity, leading to unclear and often uninterpretable data. Due to the ultra-clean nature of the spectra obtained, quantitative extraction of inter-proton distances should be possible.29 GEMSTONE-ROESY complements the recently published GEMSTONE-NOESY experiment, for use as an alternative where nuclear Overhauser effects are small or zero.
Conceptualisation of the project: EG, LC and RA. Data curation and NMR analysis: EG. Discussion of the results: EG, LC, RA, GAM, MN, MJS, GW. Manuscript draft: EG. Supervision: LC, RA, JPB, MJ. Manuscript review and editing: EG, LC, RA, JPB, MJ, GAM, MN, MJS, GW. Funding acquisition: LC, JPB. All authors have given approval for the final version of the manuscript.
This work was funded by an iCASE award from Johnson Matthey and the Engineering and Physical Sciences Research Council (grant numbers EP/V519613/1 2509660 and EP/R018790/1), by The University of Manchester (Dame Kathleen Ollerenshaw Fellowship to LC), and in part by the Swedish Research Council (grant number 2017-03703 to GW) and the Comunidad de Madrid (grant number 2022-T1/BMD-24030 to LC). For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising. All experimental data for all the spectra shown, pulse program codes and experimental parameters are freely available at https://doi.org/10.48420/21905049.
Conflicts of interest
There are no conflicts to declare.Notes and references
- W. P. Aue, E. Bartholdi and R. R. Ernst, J. Chem. Phys., 1976, 64, 2229–2246 CrossRef CAS.
- R. Freeman, Chem. Rev., 1991, 91, 1397–1412 CrossRef CAS.
- L. Castañar and T. Parella, Magn. Reson. Chem., 2015, 53, 399–426 CrossRef PubMed.
- R. W. Adams, eMagRes, 2014, 3, 295–309 CAS.
- K. Zangger, Prog. Nucl. Magn. Reson. Spectrosc., 2015, 86–87, 1–20 CrossRef CAS PubMed.
- L. Castañar, Magn. Reson. Chem., 2017, 55, 47–53 CrossRef PubMed.
- P. Kiraly, N. Kern, M. P. Plesniak, M. Nilsson, D. J. Procter, G. A. Morris and R. W. Adams, Angew. Chem., Int. Ed., 2021, 60, 666–669 CrossRef CAS PubMed.
- L. D. Hall and T. J. Norwood, J. Magn. Reson., 1988, 78, 582–587 CAS.
- I. Solomon, Phys. Rev., 1955, 99, 559–565 CrossRef CAS.
- P. Kiraly, M. Nilsson, G. A. Morris and R. W. Adams, Chem. Commun., 2021, 57, 2368–2371 RSC.
- L. Braunschweiler and R. R. Ernst, J. Magn. Reson., 1983, 53, 521–528 CAS.
- D. Neuhaus, Nuclear Overhauser Effect, in eMagRes, ed. R. K. Harris and R. L. Wasylishen, 2011, https://doi.org/10.1002/9780470034590.emrstm0350.pub2 Search PubMed.
- A. Kalk and H. J. C. Berendsen, J. Magn. Reson., 1976, 24, 343–366 CAS.
- A. A. Bothner-By, R. L. Stephens, J. Lee, C. D. Warren and R. W. Jeanloz, J. Am. Chem. Soc., 1984, 106, 811–813 CrossRef CAS.
- R. M. Wenger, Angew. Chem., Int. Ed. Engl., 1985, 24, 77–85 CrossRef.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 1997, 23, 3–25 CrossRef CAS.
- N. El Tayar, A. E. Mark, P. Vallat, R. M. Brunne, B. Testa and W. F. van Gunsteren, J. Med. Chem., 1993, 36, 3757–3764 CrossRef CAS PubMed.
- H. Kessler, H.-R. Looslib and H. Oschkinat, Helv. Chim. Acta, 1985, 68, 661–681 CrossRef CAS.
- M. Foroozandeh, R. W. Adams, N. J. Meharry, D. Jeannerat, M. Nilsson and G. A. Morris, Angew. Chem., Int. Ed., 2014, 53, 6990–6992 CrossRef CAS PubMed.
- M. Foroozandeh, G. A. Morris and M. Nilsson, Eur. J. Chem., 2018, 24, 13988–14000 CrossRef CAS PubMed.
- C. M. Thiele, K. Petzold and J. Schleucher, Eur. J. Chem., 2009, 15, 585–588 CrossRef CAS PubMed.
- J. Ilgen, J. Nowag, L. Kaltschnee, V. Schmidts and C. M. Thiele, J. Magn. Reson., 2021, 324, 106900 CrossRef CAS PubMed.
- S. Boros and G. Batta, Magn. Reson. Chem., 2016, 54, 947–952 CrossRef CAS PubMed.
- J. Furrer, J. Nat. Prod., 2009, 72, 1437–1441 CrossRef CAS PubMed.
- P. Adella, T. Parella, F. Sánchez-Ferrando and A. Virgili, J. Magn. Reson., 1995, 108, 77–80 CrossRef.
- S. Austin, C. A. De Castro, N. Sprenger, A. Binia, M. Affolter, C. L. Garcia-Rodenas, L. Beauport, J. F. Tolsa and C. J. F. Fumeaux, Nutrients, 2019, 11, 1282 CrossRef CAS PubMed.
- C. B. Lebrilla, J. Liu, G. Widmalm and J. H. Prestegard, Essentials of Glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 4th edn, 2022, pp. 32–42 Search PubMed.
- Q. Shi, J. Yan, B. Jiang, X. Chi, J. Wang, X. Liang and X. Ai, Carbohydr. Polym., 2021, 267, 118218 CrossRef CAS PubMed.
- C. P. Butts, C. R. Jones, E. C. Towers, J. L. Flynn, L. Appleby and N. J. Barron, Org. Biomol. Chem., 2011, 9, 177–184 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental details, further experimental results. See DOI: https://doi.org/10.1039/d3cc00550j |
| This journal is © The Royal Society of Chemistry 2023 |
