Open Access Article
Open Access ArticleUnique assembly of carbonylpyridinium and chromene reveals mitochondrial thiol starvation under ferroptosis and novel ferroptosis inducer†
Kaiqing
Ma
 a,
He
Yang
a,
Tianruo
Shen
c,
Yongkang
Yue
a,
Lingling
Zhao
a,
Xiaogang
Liu
a,
He
Yang
a,
Tianruo
Shen
c,
Yongkang
Yue
a,
Lingling
Zhao
a,
Xiaogang
Liu
 c,
Fangjun
Huo
c,
Fangjun
Huo
 b and
Caixia
Yin
b and
Caixia
Yin
 *a
*a
aKey Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education, Institute of Molecular Science, Shanxi University, Taiyuan 030006, PR China. E-mail: yincx@sxu.edu.cn
bResearch Institute of Applied Chemistry, Shanxi University, Taiyuan 030006, PR China
cUniversity of Technology and Design, 487372, Singapore
First published on 2nd March 2022
Abstract
To reveal the delicate function of mitochondria, spatiotemporally precise detection tools remain highly desirable. However, current probes with positively charged warheads for targeting mitochondria diffuse out of the mitochondria as the potential of the mitochondrial membrane changes, which directly influences the accuracy of the detection. Herein, we assembled carbonylpyridinium and chromene to afford the probe CM-Mit. Following the ultrafast response to thiol and the dissociation of carbonylpyridinium, the formation of o-quinone methide from CM-Mit was proposed to label proteins, thus avoiding diffusion out of the mitochondria. Therefore, the accurate spatiotemporal detection of thiol in mitochondria was realized. With this excellent probe, ferroptosis inducers were proved to stimulate thiol starvation in mitochondria for the first time in cancer cells. Moreover, CM-Mit was used to screen a compound library developed in-house and the stemona alkaloid analog SA-11 was shown to induce ferroptosis in various cancer cell lines, including a drug-resistant one.
Introduction
Mitochondrial thiol systems are central to protecting mitochondria against oxidative damage and are assumed to be the key means through which potential redox signals are transmitted, modulated and sensed.1,2 Activity-based sensing probes have been considered as desirable and indispensable tools for studying dynamic processes in the organelle.3 Thus, various mitochondrial-targeted fluorescent probes have been developed to understand the roles and functions of the analytes in mitochondria.4 Due to strong negative membrane potential, cations with strong lipophilicity tend to penetrate the membrane and accumulate inside the mitochondrial matrix.5 However, current positively charged warheads for targeting mitochondria, such as TPP (triphenylphosphine) and alkylpyridinium cations, can affect the membrane potential, triggering membrane rupture and disturbance of the microenvironment.6 Moreover, the probes with positive charge for targeting mitochondria can diffuse out of mitochondrial as the mitochondrial membrane potential changes, which could influence the accuracy of the detection. Notably, Zhang and co-workers reported their elegant mitochondria-targetable probe for accurate detection of Cys without interference from other sites.7 However, the fluorescence was re-distributed in the cell with the extension of the incubation time following the response to the analytes. Despite the solid-state organic fluorophores (SSOFs) being successfully constructed to avoid the diffusion out of mitochondria,8 the aqueous dispersibility and low biocompatibility of the SSOFs are currently unsatisfactory.9We recently took note of the thiol–chromene “click” nucleophilic reaction and developed a sensing probe for thiol with prominent selectivity, which enables us to monitor thiol fluctuations in various physiological and pathological processes (Fig. 1a and b)10–14. Ferroptosis is a unique type of non-apoptotic-regulated cell death culminating in overwhelming lipid peroxidation downstream of metabolic dysfunctions15,16 and holds promise as a novel therapeutic approach.17,18 Recent research indicated that cysteine deprivation could induce the ferroptosis of pancreatic ductal adenocarcinoma. However, the mitochondrial membrane potential (MMP) fluctuates in ferroptosis, which influences the accuracy of current mitochondrial-targeting probes.19 Therefore, this novel strategy for accurate visualization of thiol-mediated oxidative stress in mitochondria remains highly desirable but challenging. Moreover, the developed sensing probe could be further used for rapid screening of efficient ferroptosis-inducing compounds.20,21 It is noteworthy that an acrylate-based fluorescent probe was developed for monitoring intracellular cysteine in ferroptosis.22 However, the relatively low reaction rate and no organelle-targeting ability limited its use for further research on the role of thiol in ferroptosis.
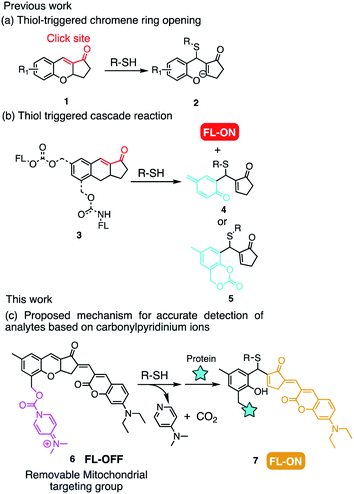 | ||
| Fig. 1 Previous work: (a) thiol-mediated nucleophilic attack on unsaturated α,β-ketone and (b) self-immolation to release the fluorophore. (c) Design strategy of this work. | ||
With these considerations in mind, we envisioned the possibility of utilizing the thiol–chromene “click” nucleophilic reaction to provide the probe with a high signal to background ratio (Fig. 1c). Besides, the formation of o-quinone methide after recognizing the analytes could label the proteins in the mitochondria, which may lead to mitochondrial targeting with high accuracy.23 Although alkylpyridinium ions has extensively been employed for mitochondrial targeting,8,24,25 there have been no reports concerning carbonylpyridinium as a mitochondrial-targeting group in the probe. Carbonylpyridinium cation is also assumed to accelerate the recognition reaction due to its excellent leaving ability.26
In this work, a novel fluorophore was constructed based on the chromene skeleton, which was integrated with the carbonylpyridinium ions to afford the ultrafast selective probe CM-Mit for accurate detection of thiol in mitochondria. The formation of the o-quinone methide enables it to label the proteins in the mitochondria, thus avoiding diffusion out of the mitochondria. With the probe CM-Mit, the ferroptosis inducer erastin and RSL3 were demonstrated for the first time to trigger thiol starvation in mitochondria. The screening approach was developed for discovering the potential thiol starvation modulator in mitochondria as a ferroptosis inducer. Notably, a stemona alkaloid analog (SA-11) with an α-methylene-γ-lactone moiety was first proved to induce thiol starvation and ferroptosis in various cancer cells, including drug-resistant ones. Overall, this work provides a robust and reliable tool for ferroptosis research.
Results and discussion
Probe design and synthesis
In our previous study, the chromene structure (8) was synthesized from (2-hydroxy-5-methyl-1,3-phenylene)dimethanol in two steps.10 To realize the condensation of compounds 8 and 9, mild conditions need to be identified. When catalytic amounts of acetic acid and piperidine were added with toluene as the solvent, the reaction was quite messy, with only a small amount of product 10 obtained (Scheme S1†). Using just piperidine as the catalyst with molecular sieves and ethanol as the solvent gave a better result. The crude product of the Knoevenagel condensation was submitted for the subsequent esterification and compound 11 was obtained with 32% yield for two steps, followed by reacting with DMAP to afford the probe CM-Mit with moderate yield.Spectral properties
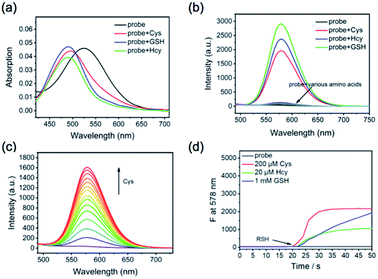
The capability of CM-Mit to detect thiols was evaluated using UV-vis absorption and fluorescence spectra (Fig. 2a and b). The selectivity of CM-Mit for thiols was investigated in DMSO/PBS (5 μM, pH 7.4, v/v, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9). The probe responded to various other amino acids, such as Ala, Arg, Asn, Asp, Cys, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Tyr, Val, Cys, and Hcy, and GSH was measured. Upon the addition of these analytes, only thiols caused significant changes in fluorescence emission at 578 nm with excitation at 475 nm. However, no obvious changes were observed for other amino acids (0.2 mM), anions and cations (Fig. S1†). Those results displayed that probe CM-Mit could detect thiols specifically. The addition of Cys at different pH values was also tested (Fig. S2†). The results indicated that probe CM-Mit showed almost no fluorescence emission in acidic and alkaline solutions.
9). The probe responded to various other amino acids, such as Ala, Arg, Asn, Asp, Cys, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Tyr, Val, Cys, and Hcy, and GSH was measured. Upon the addition of these analytes, only thiols caused significant changes in fluorescence emission at 578 nm with excitation at 475 nm. However, no obvious changes were observed for other amino acids (0.2 mM), anions and cations (Fig. S1†). Those results displayed that probe CM-Mit could detect thiols specifically. The addition of Cys at different pH values was also tested (Fig. S2†). The results indicated that probe CM-Mit showed almost no fluorescence emission in acidic and alkaline solutions.
Based on the above result, we selected pH 7.4 as the measurement system in our following experiments in terms of the body pH value. The fluorescence spectrum of CM-Mit was monitored in the presence of CM-Mit (5 μM, DMSO/PBS, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) with increasing concentration of Cys (1–40 μM) (Fig. 2c). The kinetic study showed that the reaction was completed within 10 s when Cys (200 μM) was added, while the reactions for GSH (1 mM) and Hcy (20 μM) were completed in 200 and 30 s (Fig. 2d and S3†). As we expected, the introduction of the carbonylpyridinium ion significantly improved the reaction rate compared with the probe (Cys: 7 min, Hcy: 3 min, GSH: 7 min) in our previous work10 (Table S2†).
1, v/v) with increasing concentration of Cys (1–40 μM) (Fig. 2c). The kinetic study showed that the reaction was completed within 10 s when Cys (200 μM) was added, while the reactions for GSH (1 mM) and Hcy (20 μM) were completed in 200 and 30 s (Fig. 2d and S3†). As we expected, the introduction of the carbonylpyridinium ion significantly improved the reaction rate compared with the probe (Cys: 7 min, Hcy: 3 min, GSH: 7 min) in our previous work10 (Table S2†).
A significant increase in the fluorescence emission intensity at 575 nm was measured in DMSO/PBS (v/v, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, pH 7.4) after the addition of Cys (0–3.6 equiv.) into the mixture. Meanwhile, a linear relation was observed between the fluorescence intensity and the concentration of Cys (Fig. 3). Furthermore, the detection limit of CM-Mit for Cys based on the IUPAC definition (CDL = 3Sb/m) is 0.49 μM, which is comparable to that for NIR-HMPC reported by our group.13 The above results demonstrated that CM-Mit is an ultrafast fluorescent probe for thiol detection with excellent selectivity and sensitivity.
9, pH 7.4) after the addition of Cys (0–3.6 equiv.) into the mixture. Meanwhile, a linear relation was observed between the fluorescence intensity and the concentration of Cys (Fig. 3). Furthermore, the detection limit of CM-Mit for Cys based on the IUPAC definition (CDL = 3Sb/m) is 0.49 μM, which is comparable to that for NIR-HMPC reported by our group.13 The above results demonstrated that CM-Mit is an ultrafast fluorescent probe for thiol detection with excellent selectivity and sensitivity.
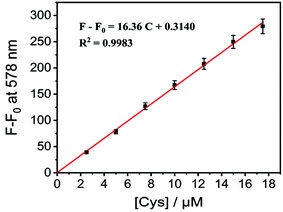 | ||
| Fig. 3 The working curve of CM-Mit (5 μM) in the presence of different concentrations of Cys. Each point represents the average and standard deviation of three independent repeated experiments. | ||
Proposed mechanism
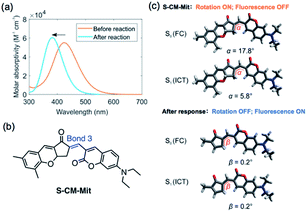
Originally, a photoinduced electron transfer (PET) mechanism was assumed to be responsible for the fluorescence quenching of CM-Mit, due to the existence of the carbonylpyridinium moiety. The S1 photoexcitation of CM-Mit mainly involves the transition from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO). Between these two orbitals, no other molecular orbitals from a quencher are available. HOMO−1 and LUMO+1 are 0.971 eV and 0.878 eV away from the HOMO and LUMO, respectively. These energy levels do not meet the requirement of PET27 (Fig. S5†). Further computational analysis showed that two factors contribute to the low quantum yield of the CM-Mit and subsequent fluorescence turn-on after recognizing the analytes (Fig. 3). A hypsochromic shift in the UV-vis absorption spectrum occurred upon the reaction of CM-Mit with biological thiols and the distinct difference in the absorbance could effectively contribute to the fluorescence intensification of the product.28 The second factor for modulating the fluorescence intensity is related to intramolecular rotations upon photoexcitation in probe CM-Mit. To reduce the computational load, we analyzed the intramolecular rotations in a simplified CM-Mit structure (S-CM-Mit) (Fig. 4b). In the most stable structure of S-CM-Mit, the dihedral angle along bond 3 is 17.8° in the ground state (the Franck–Condon state) but reduced to 5.8° in the ICT state in water, which could activate low-frequency vibrations and reduce the fluorescence intensity of S-CM-Mit, while the most stable molecular structure after S-CM-Mit recognizing thiol is planar in both the ground state and the ICT state (Fig. 4c).29 Our viscosity-dependence study supports the intramolecular rotations of CM-Mit (Fig. S6†). As the volume ratio of glycerol increased, the emission intensity of CM-Mit was enhanced by four times.The thiol–chromene “click” nucleophilic mechanism was proposed via the 1H NMR titration experiment (Fig. S4†). The signal of the original proton (H-3) at 5.32 ppm shifted low-field to 6.85. Meanwhile, the signal of the proton (H-4) on the double bond shifted from 7.25 to 5.53 ppm. These NMR data delivered direct proof for the thiol–chromene-based ring-opening reaction and the formation of quinone methide as the intermediate.
Imaging in cells and tumors
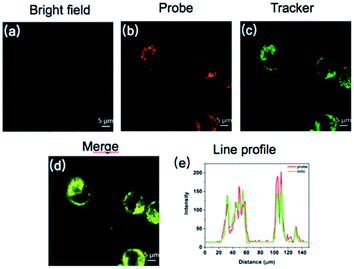
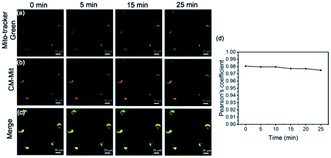
Since probe CM-Mit showed an increased fluorescence emission response to thiol in vitro and the detection limits of probe CM-Mit for thiol was micromolar, bioimaging detection of thiol-containing species in vivo may become possible. Before bioimaging assays, MTT assays were performed to check the cytotoxicity of CM-Mit against HeLa cells. As shown in Fig. S7,† the HeLa cells treated with various concentrations of CM-Mit (1–50 μM) for 5 and 10 h showed high cell viability, indicating that the agent had fairly low toxicity to living cells. Then, the ability of probe CM-Mit to detect thiol in living cells was evaluated (Fig. S8†). The results indicated that probe CM-Mit penetrated the cell membrane well and selectively monitored the level of thiols in the living cells. The feasibility for real-time visualization of endogenous thiols in HeLa tumor-bearing mice was also investigated. As shown in Fig. S9 and S10,† intratumor injection with CM-Mit resulted in a significant fluorescence signal in the tumor only 2 min after injection, which indicated that CM-Mit could be activated rapidly by endogenous thiol in the tumor.Subsequently, the colocalization experiment was conducted and the results showed that probe CM-Mit selectively localized in mitochondria (Fig. 5) and the Pearson's coefficient is around 0.98. This result indicated that probe CM-Mit could selectively detect thiols in mitochondria without interference from the cytoplasmic thiols. Moreover, the fluorescence reached a high level after incubation with the probe for only 2 min (Fig. S11†). Previous research into accurate detection of thiols in mitochondria indicated that the product of the probe and the analytes may escape from mitochondria with extended incubation time.7 To test whether the probe could accurately monitor the thiol level in mitochondria over a longer time, the Pearson's coefficient was utilized to evaluate. In the colocalization experiment, it is interesting to find that the Pearson's coefficient is around 0.97 even with another 25 min incubation time, which was probably due to the formation of the o-quinone methide after response to the thiol, followed by labeling the protein in the mitochondria.23 To examine this hypothesis, the cells were incubated with CM-Mit, followed by analyzing the lysate by SDS-PAGE. The result indicated that certain proteins were indeed stained compared to the lysate with no probe (Fig. S12e†). Moreover, the fluorescence improved significantly when the cells were treated with Cys and CM-Mit (Fig. S12f†), which was probably due to the formation of o-quinone methide promoted by the thiol. Overall, the above data confirmed that probe CM-Mit could firmly monitor the mitochondrial thiols (Fig. 6).
CM-Mit-based fluorescence assays for discovery of potential ferroptosis inducers
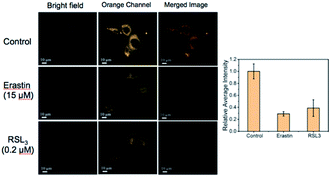
We further attempted to assess the potential use of probe CM-Mit to visualize the thiol profile of ferroptosis. We applied two ferroptosis inducers, including erastin and RSL3, which are respectively selective inhibitors of system Xc− and glutathione peroxidase 4 (GPX4). Cells incubated with probe CM-Mit displayed bright fluorescence in the orange channel, whereas pre-treatment with erastin and RSL3 significantly decreased the fluorescence intensity (Fig. 7), indicating that probe CM-Mit could specifically monitor mitochondrial thiols of ferroptosis and the ferroptosis inducer could trigger thiol starvation in mitochondria.Stemona alkaloids possess diverse structures and interesting biological activities.30,31 In our previous work,32–35 we synthesized several stemona alkaloids and their derivatives. To identify stemona alkaloids that induce ferroptosis, we performed CM-Mit-based fluorescence assays to detect the effects of these stemona alkaloids on mitochondrial thiol levels. The live HeLa cells were pretreated with 15 μM of different compounds as well as the positive control erastin, followed by imaging and analysis of the endogenous thiol levels in mitochondria with the probe CM-Mit. The structure of the stemona alkaloids and their derivatives is shown in Table S1.† The results indicated that SA-11 induced a significant decrease in the fluorescence signal, which suggested that SA-11 significantly reduced the mitochondrial thiol level (Fig. 8). Previously, SA-11 was proved to overcome the resistance to 5-fluorouracil in colorectal cancer cells.36 However, the exact underlying mechanism is still unknown. Given that SA-11 triggered thiol starvation in mitochondria, we propose that SA-11 could induce ferroptosis of cancer cells to overcome drug resistance.
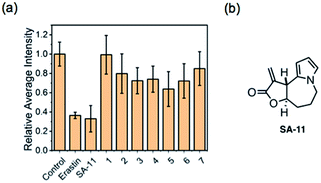 | ||
| Fig. 8 Screening of stemona alkaloids and their derivatives for controlling the thiol level. (a) HeLa cells were pretreated with ferroptosis inducer erastin (5 μM), SA-11 and its analogs (15 μM) in a 5% carbon dioxide incubator at 37 °C for 24 h, and the cells were incubated with 10 μM CM-Mit for 15 min. The mean intensity from Fig. S9,† where the control group is defined as 1.0. Data are represented as mean ± SD (n = 3); (b) the structure of SA-11. | ||
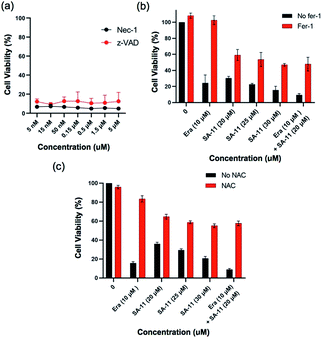
To examine this hypothesis, ES-2 cells were first treated with SA-11 (30 μmol) and the apoptosis inhibitor z-VAD or the necroptosis inhibitor necrostatin-1 (Nec-1) (Fig. 9a). The cytotoxicity of SA-11 was not abrogated by z-VAD and Nec-1, indicating that SA-11 had no effect on apoptosis and necroptosis. When ES-2 cells were treated with various concentrations of SA-11, the ES-2 cells were inhibited in a dose-dependent manner (Fig. 9b). In contrast, when ES-2 cells were incubated with SA-11 and the ferroptosis inhibitor ferrostatin-1 (fer-1), the cell death was significantly rescued. When ES-2 cells were treated with SA-11 combined with erastin, the cell viability was further reduced compared to the effect of erastin alone, which suggested that SA-11 and erastin could collaboratively induce cell death. Meanwhile, the lethality induced by SA-11 was also suppressed by N-acetyl cysteine (NAC), which also served as an antioxidant (Fig. 9c). A further experiment was conducted to monitor the effect of SA-11 in HCT-116 and the drug-resistant HCT-116p53−/−. The results indicated that both cell lines treated with different concentrations of NAC could be rescued from the death induced by SA-11 (Fig. 10). These results clearly indicate that SA-11 triggers ferroptotic cell death in cancer cells.
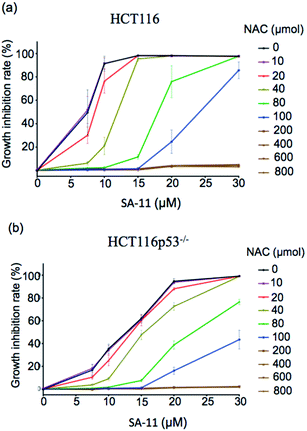 | ||
| Fig. 10 Effect of NAC on viability of (a) HCT-116 and (b) HCT116P53−/− after incubation with different concentration of SA-11. Data are represented as mean ± SD (n = 3). | ||
Conclusion
In this work, the novel probe CM-Mit was constructed for accurate detection of thiol in mitochondria based on the chromene skeleton and carbonylpyridinium structure. Extensive computational studies indicated that the hypsochromic shift after the response to analytes and the intramolecular rotations between the ground state and the ICT state contribute to the fluorescence turn-on. CM-Mit displays excellent mitochondrial-targeting ability and could accurately detect the thiols in mitochondria without interference from other sites. Moreover, the formation of the o-quinone methide after the response to thiols could label the proteins, which enables it to localize in mitochondria with the extension of incubation time. With the probe CM-Mit, the ferroptosis inducers erastin and RSL3 were demonstrated for the first time to trigger thiol starvation in mitochondria. Additionally, the screening approach was successfully developed for discovering potential thiol-level modulators as ferroptosis inducers. The stemona alkaloid derivative SA-11 with an α-methylene-γ-lactone moiety was found to trigger thiol starvation in mitochondria and demonstrated to induce the ferroptosis of cancer cells, including drug-resistant ones. This work not only provides carbonylpyridinium as a robust mitochondrial-targeting group for mitochondrial function research but also illustrates a versatile strategy for the discovery of small molecules as ferroptosis inducers.Author contributions
K. M. and C. Y. conceived the project. K. M., H. Y. and L. Z. synthesized the dyes and conducted photophysical characterization and analysis. H. Y. and L. Z. performed the fluorescence imaging experiments. T. R. and X. L. conducted the theoretical calculations. F. H. characterized the structures. All authors participated in writing the manuscript. We thank Xingkang Wu for the discussion about the biological assays, and Zhihong Wei for the help in computational analysis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Natural Science Foundation of China (No. 22077076, 22074084, and 21878180), the Shanxi Province “1331 Project” Key Innovation Team Construction Plan Cultivation Team (2018-CT-1), the 2018 Xiangyuan County Solid Waste Comprehensive Utilization Science and Technology Project (2018XYSDJS-05), the Shanxi Collaborative Innovation Center of High Value-added Utilization of Coal-related Wastes (2015-10-B3), the Key R&D Program of Shanxi Province (No. 201803D421059 and 201903D421069), the Shanxi Province Science Foundation (No. 201901D111012 and 201901D111015), Key R&D and Transformation Plan of Qinghai Province (No. 2020-GX-101), and Scientific Instrument Center of Shanxi University (201512).Notes and references
- M. P. Murphy, Antioxid. Redox Signaling, 2012, 16, 476 CrossRef CAS PubMed.
- L. M. Booty, J. M. Gawel, F. Cvetko, S. T. Caldwell, A. R. Hall, J. F. Mulvey, A. M. James, E. C. Hinchy, T. A. Prime, S. Arndt, C. Beninca, T. P. Bright, M. R. Clatworthy, J. R. Ferdinand, H. A. Prag, A. Logan, J. Prudent, T. Krieg, R. C. Hartley and M. P. Murphy, Cell Chem. Biol., 2019, 26, 449 CrossRef CAS PubMed.
- K. J. Bruemmer, S. W. M. Crossley and C. J. Chang, Angew. Chem., Int. Ed., 2020, 59, 13734 CrossRef CAS PubMed.
- W. Xu, Z. Zeng, J. H. Jiang, Y. T. Chang and L. Yuan, Angew. Chem., Int. Ed., 2016, 55, 13658 CrossRef CAS PubMed.
- M. P. Murphy, Biochim. Biophys. Acta, Bioenerg., 2008, 1777, 1028 CrossRef CAS PubMed.
- G. Kroemer, L. Galluzzi and C. Brenner, Physiol. Rev., 2007, 87, 99 CrossRef CAS PubMed.
- T. B. Ren, Q. L. Zhang, D. Su, X. X. Zhang, L. Yuan and X. B. Zhang, Chem. Sci., 2018, 9, 5461 RSC.
- Z. Li, T.-B. Ren, X.-X. Zhang, S. Xu, X.-Y. Gong, Y. Yang, G. Ke, L. Yuan and X.-B. Zhang, Anal. Chem., 2021, 93, 2235 CrossRef CAS PubMed.
- K. Li, T.-B. Ren, S. Huan, L. Yuan and X.-B. Zhang, J. Am. Chem. Soc., 2021, 143, 21143 CrossRef CAS PubMed.
- K. Ma, L. Zhao, Y. Yue, F. Huo, J. Chao and C. Yin, Anal. Chem., 2020, 92, 15936 CrossRef CAS PubMed.
- F.-J. Huo, Y.-Q. Sun, J. Su, J.-B. Chao, H.-J. Zhi and C.-X. Yin, Org. Lett., 2009, 11, 4918 CrossRef CAS PubMed.
- K. Ma, L. Zhao, Y. Yue and C. Yin, Chem. Phys. Rev., 2022, 3, 011302 CrossRef.
- Y. Yang, T. Zhou, M. Jin, K. Zhou, D. Liu, X. Li, F. Huo, W. Li and C. Yin, J. Am. Chem. Soc., 2020, 142, 1614 CrossRef CAS PubMed.
- Y. Yue, C. Yin, F. Huo, J. Chao and Y. Zhang, Sens. Actuators, B, 2016, 223, 496 CrossRef CAS.
- H. Bayir, T. S. Anthonymuthu, Y. Y. Tyurina, S. J. Patel, A. A. Amoscato, A. M. Lamade, Q. Yang, G. K. Vladimirov, C. C. Philpott and V. E. Kagan, Cell Chem. Biol., 2020, 27, 387 CrossRef CAS PubMed.
- S. J. Dixon, K. M. Lemberg, M. R. Lamprecht, R. Skouta, E. M. Zaitsev, C. E. Gleason, D. N. Patel, A. J. Bauer, A. M. Cantley, W. S. Yang, B. Morrison 3rd and B. R. Stockwell, Cell, 2012, 149, 1060 CrossRef CAS PubMed.
- V. S. Viswanathan, M. J. Ryan, H. D. Dhruv, S. Gill, O. M. Eichhoff, B. Seashore-Ludlow, S. D. Kaffenberger, J. K. Eaton, K. Shimada, A. J. Aguirre, S. R. Viswanathan, S. Chattopadhyay, P. Tamayo, W. S. Yang, M. G. Rees, S. Chen, Z. V. Boskovic, S. Javaid, C. Huang, X. Wu, Y. Y. Tseng, E. M. Roider, D. Gao, J. M. Cleary, B. M. Wolpin, J. P. Mesirov, D. A. Haber, J. A. Engelman, J. S. Boehm, J. D. Kotz, C. S. Hon, Y. Chen, W. C. Hahn, M. P. Levesque, J. G. Doench, M. E. Berens, A. F. Shamji, P. A. Clemons, B. R. Stockwell and S. L. Schreiber, Nature, 2017, 547, 453 CrossRef CAS PubMed.
- M. A. Badgley, D. M. Kremer, H. C. Maurer, K. E. DelGiorno, H.-J. Lee, V. Purohit, I. R. Sagalovskiy, A. Ma, J. Kapilian and C. E. J. S. Firl, Science, 2020, 368, 85 CrossRef CAS PubMed.
- M. Gao, J. Yi, J. Zhu, A. M. Minikes, P. Monian, C. B. Thompson and X. Jiang, Mol. Cell, 2019, 73, 354 CrossRef CAS PubMed.
- C. Shao, J. Yuan, Y. Liu, Y. Qin, X. Wang, J. Gu, G. Chen, B. Zhang, H. K. Liu, J. Zhao, H. L. Zhu and Y. Qian, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10155 CrossRef CAS PubMed.
- T. Hirayama, M. Niwa, S. Hirosawa and H. Nagasawa, ACS Sens., 2020, 5, 2950 CrossRef CAS PubMed.
- Z. Li, Y. Li, Y. Yang, Z. Gong, H. Zhu and Y. Qian, Anal. Chim. Acta, 2020, 1125, 66 CrossRef CAS PubMed.
- H. Iwashita, E. Castillo, M. S. Messina, R. A. Swanson and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2018513118 CrossRef CAS PubMed.
- Y. Zhang, Q. Wang, Z. Zhu, W. Zhao, C. Yan, Z. Liu, M. Liu, X. Zhao, H. Tian and W.-H. Zhu, CCS Chem., 2021, 3, 1800 CrossRef.
- H. Chen, J. Wang, X. Feng, M. Zhu, S. Hoffmann, A. Hsu, K. Qian, D. Huang, F. Zhao and W. Liu, Chem. Sci., 2019, 10, 7946 RSC.
- H. Yan, F. Huo, Y. Yue, J. Chao and C. Yin, J. Am. Chem. Soc., 2021, 143, 318 CrossRef CAS PubMed.
- W. Chi, J. Chen, W. Liu, C. Wang, Q. Qi, Q. Qiao, T. M. Tan, K. Xiong, X. Liu, K. Kang, Y.-T. Chang, Z. Xu and X. Liu, J. Am. Chem. Soc., 2020, 142, 6777 CrossRef CAS PubMed.
- W. Chi, L. Huang, C. Wang, D. Tan, Z. Xu and X. Liu, Mater. Chem. Front., 2021, 5, 7012 RSC.
- Q. Peng, Y. Yi, Z. Shuai and J. Shao, J. Am. Chem. Soc., 2007, 129, 9333 CrossRef CAS PubMed.
- H. Greger, Phytochem. Rev., 2019, 18, 463 CrossRef CAS.
- X. Wu and K. Ma, Prog. Chem., 2020, 32, 752 Search PubMed.
- K. Ma, X. Yin and M. Dai, Angew. Chem., Int. Ed., 2018, 57, 15209 CrossRef CAS PubMed.
- K. Ma, H.-B. Ren, J.-B. Chao and X.-M. Qin, J. Asian Nat. Prod. Res., 2019, 22, 655 CrossRef PubMed.
- K. Ma, H. Ren, X. Wu, J. Chao and X. Qin, Chin. J. Org. Chem., 2019, 39, 2094 CrossRef CAS.
- X. Yin, K. Ma, Y. Dong and M. Dai, Org. Lett., 2020, 22, 5001 CrossRef CAS PubMed.
- K. Ma, M. Zhang, X. Wu, P. Yang and C. Yin, Bioorg. Med. Chem., 2021, 30, 115929 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2sc00328g |
| This journal is © The Royal Society of Chemistry 2022 |