Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Active learning assists chemical intuition identify a scalable conversion of chitin to 3-acetamido-5-acetylfuran†‡
Juliana G.
Pereira§
a,
João M. J. M.
Ravasco§
a,
Latimah
Bustillo
 a,
Inês S.
Marques
a,
Inês S.
Marques
 b,
Po-Yu
Kao
b,
Po-Yu
Kao
 c,
Po-Yi
Li
d,
Yen-Chu
Lin
ce,
Tiago
Rodrigues
c,
Po-Yi
Li
d,
Yen-Chu
Lin
ce,
Tiago
Rodrigues
 *a,
Vasco D. B.
Bonifácio
*a,
Vasco D. B.
Bonifácio
 fg,
Andreia F.
Peixoto
fg,
Andreia F.
Peixoto
 b,
Carlos A. M.
Afonso
*a and
Rafael F. A.
Gomes
b,
Carlos A. M.
Afonso
*a and
Rafael F. A.
Gomes
 *a
*a
aResearch Institute for Medicines (iMed.ULisboa), Faculty of Pharmacy, Universidade de Lisboa, Avenida Professor Gama Pinto, 1649-003, Lisbon, Portugal. E-mail: rafael.gomes@campus.ul.pt
bLAQV-REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências, Universidade do Porto, Rua do Campo Alegre s/n, 4169-007 Porto, Portugal
cInsilico Medicine Taiwan Ltd, Taipei 110208, Taiwan
dDepartment of Computer Science, University of Southern California, Salvatori Computer Science Center, 941 Bloom Walk, Los Angeles, CA 90089, USA
eDepartment of Pharmacy, National Yang Ming Chiao Tung University, Taipei 112304, Taiwan
fiBB-Institute for Bioengineering and Biosciences and i4HB-Institute for Health and Bioeconomy, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1049-001 Lisboa, Portugal
gBioengineering Department, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1049-001 Lisboa, Portugal
First published on 3rd January 2025
Abstract
The shift towards a more sustainable chemical and pharma industry led to considerable efforts on discoverying biorenewable synthons, amongst other approaches. Whereas lignocellulosic biomass has thrived as a source of furan building blocks, chitin has struggled in competing despite its abundance and being a source of sustainable nitrogen. This may be due to the difficulties in large scale production of chitin-derived furans. Here, we leverage active learning for the optimization of a multi-parameter reaction, namely the formation of 3-acetamido-5-acetylfuran. This active learning approach was able to outperform a trial-and-error optimization based on chemical intuition, yielding the desired N-rich furan in up to 70% yield from N-acetylglucosamine and in 10.5 mg g−1 directly from dry shrimp shells. The reaction was scalable up to a 4.5 mmol scale, bypasses the use of undesirable toxic, high boiling point solvents and allows the reuse of the reaction media, supporting the utility of machine learning to advance green chemistry and the valorization of biomasses.
Green foundation1. This study delivers an improved and greener method to yield a sustainable synthon (3A5AF), allowing further exploration of this biomass derivative. In addition, the method further corroborates the potential of machine learning for the upgrading of biomass.2. By using machine learning we were able to obtain the highest reported yield of 3A5AF directly from chitin, being able to reuse the reaction media and improving on some green metrics such as PMI. 3. This work can be further elevated by a process intensification endeavor, in particular continuous flow chemistry. Also, by further expanding the toolkit for modification of 3A5AF. |
Introduction
Nitrogen-containing compounds play an important role in the pharmaceutical industry; 80% of approved drugs have at least one nitrogen atom, the majority of which are obtained from ammonia produced by the Haber–Bosch process.1 This industrial reaction is responsible for more than 1% of the world's annual energy consumption and uses around 50% of the hydrogen supply.1,2 In this regard, the growing demand for sustainable development requires innovative solutions for the synthesis of N-containing value-added chemicals. Extensive research has been conducted on the synthesis and valorisation of biomass-derived intermediates prepared from lignocellulosic biomass primarily composed of carbon and oxygen atoms.3–7 In contrast, chitin, the most abundant oceanic biomass resource and the second most abundant polymer on Earth (after cellulose) has received less attention.8–11 Chitin is an important biologically-fixed nitrogen source with an annual production of up to 1011 tonnes. This biopolymer is composed of N-acetyl-D-glucosamine (NAG) residues, is readily available, non-toxic, and has the potential to serve as a platform for nitrogen-containing chemicals independent of the Haber-Bosh process. 3-Acetamido-5-acetylfuran (3A5AF), a furan obtained upon dehydration of NAG, is a particularly promising bio-renewable building block preserving the nitrogen atom present in chitin.12–14 Similarly to other high value biomass furanic synthons, such as furfurals, the 3A5AF synthetic utility has been showcased in several examples depicted in Scheme 1A.These range from simple acid-promoted condensation with ketones yielding dihydrodifuropyridines,15 to the complex synthesis of 3-acetamido-cyclopentenone via the Piancatelli rearrangement of 4-acetamidofurfuryl alcohol, derived from 3A5AF reduction.16 The oxidative ring expansion of the alcohol derived from the asymmetric reduction of 3A5AF yielded a 2-amino sugar named L-rednose.17 In addition, our group described a Diels–Alder reaction involving 3A5AF as a diene, which endorsed the formation of new molecular entities.18–20 These were further aromatized yielding the first examples of bio-based anilines from chitinous biomass. Additionally, the anticancer alkaloid proximicin A was successfully synthesized using the chitin-derived platform chemical 3A5AF as starting material.21
These examples highlight the importance of the novel furanic platform, which resulted in the emergence of several methodologies for the dehydration of NAG and chitin.
From the initial discovery of 3A5AF in 1984, where pyrolysis of the sugar yielded 2% of the desired furan,14 research has focused on increasing the yield of the product mostly by reacting NAG at high temperatures/MW irradiation in the presence of either acidic promoters or ionic liquids (IL) in polar aprotic solvents such as NMP, DMA or DMF.12,22–29 Despite recent methodologies reporting the furan in up to 67% yield, these conditions fail to deliver 3A5AF when scaling up, require complex ILs and use undesirable solvents.
The direct conversion of chitin into 3A5AF via depolymerization and dehydration reactions was also described by Chen and co-workers, yielding the furan molecule in 28% using [BMim]Cl in DMF at 180 °C for 10 minutes.30,31 This result supports the viability of obtaining the desired furan directly from the polymer, thus avoiding the time and reagent-consuming depolymerization to NAG.
Overall, we identified challenges in the preparation of 3A5AF to be (i) the use of MW, which limits scalability; (ii) the use of complex ILs which hinder their use in industrial applications; (iii) the use of toxic and high boiling point solvents such as NMP, DMA and DMF, which compromises both sustainability metrics and purification procedures; and finally (iv) poor scalability, potentially due to poor mass and heat transfer in >1 mmol scale, which cause either low conversions or runaway reactions.
Given the complexity of the reaction, which presents many tuneable variables, expert intuition can be inefficient in identifying optimal synthetic conditions.32 In recent years, there has been progress in the use of machine learning (ML) tools in chemistry, including in the optimization of reaction conditions, following active learning strategies.33–35 ML algorithms have the ability to identify hidden patterns in reaction conditions that may lead to desirable reaction outcomes. However, the use of ML tools is usually limited by the lack of quality training data.
The importance of the nitrogen-rich furan synthon together with its challenging preparation and numerous reaction variables makes its preparation a suitable candidate for ML-assisted optimization. Herein, we report a synthetic methodology identified by a learning algorithm that allows accessing 3A5AF from N-acetylglucosamine and chitin in high yield. Our results support the power of active learning to sustainably accelerate chemical research and valorize biomass-derived building blocks.
Results and discussion
To enable the use of a ML workflow previously reported by us,36 we first generated a reaction dataset. We started this task by exploring the dehydration reaction of NAG under standard conditions with various heterogeneous catalysts, including amberlyst (amberlyst 15w, 16w and 21), amberlite (IRA400, IRA410, IRA458, IRC86) and glucose-derived amorphous carbon (GC400) with DMA as solvent. However, the desired product was not obtained under these conditions. Based on our previous experience with HMF, we next queried the use of ILs as solvent.37 In fact, we observed that most ILs led to the formation of 3A5AF. Interestingly, tetraethylammonium chloride (TEAC) proved effective, leading to a remarkable 40% yield. This is consistent with the importance of chloride anion in the reaction mechanism. Different promoters were also screened, both homogeneous and heterogeneous38–40 Brønsted acids, leading to yields ranging from 27% to 51%. Phosphoric acid was the best homogeneous promoter, whereas SO3H-functionalized Montmorillonite K10 was the best heterogeneous acid, yielding 3A5AF in 51%. In general, no correlation between the acidity and the yield of 3A5AF, or between homogeneous or heterogeneous catalysts was observed. In the absence of acid, NAG dehydration occurred to some extent promoted by the high temperatures and the IL, albeit with a lower reaction yield. Other additives were also included in the reaction condition search space, in particular boric acid and boronic acids, given the reported formation of boronate complexes in the dehydration of NAG. However, these had little influence on the overall reaction yield. NaCl was also added as additive due to its reported enhancement of the product yield. Mechanistic studies reported in previous literature suggest that the boronic acid complexes with the diols, and the chloride anion helps the dehydration to form the furan ring.41Stabilizing reagents, such as antioxidants – e.g., Na2SO4, Na2S2O5 – and glycine which have been shown to avoid HMF decomposition were also screened.37,42–44
Concentration values in the range of 0.1–0.9 M was another parameter of importance to the dataset. A steep drop in reaction yield was observed at higher concentration values potentially due to humin formation.23 Finally, we studied the effect of water in the system either by adding water as co-solvent or by removing water through vacuum. However, both strategies had a detrimental effect on the formation of 3A5AF. Taken together, following this intuition-based, divide and conquer strategy, we were able to obtain 3A5AF in 54% yield. For detailed information of the dataset and the unique reaction conditions see the ESI.‡
Overall, we iteratively performed 149 unique reactions and 10 replicates that were purposefully used as noise in the training data, given the variability of reaction yield measurement. While those reactions explored only a small fraction of a vast reactivity space, they informed on which variables appeared most important toward improved reaction yields. With that information in hand, we built a search space combining >108 discrete reaction conditions and randomly sampled 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for downstream studies. Interestingly, the sampled reactions explore different regions of the search space in comparison to the training data, suggesting a potential bias in established practices and our previous reactions. Using this small dataset to train a ML algorithm can be challenging since it does not entirely represent the optimization task. For a successful application, the ML routine must transfer knowledge. Toward that end, we used our LabMate.ML routine.33,35 In short, the method uses self-tuning random forests and allows optimizing categorical (e.g., solvent) and real value (e.g., temperature) variables simultaneously by predicting reaction yields. The prioritized reactions will thus correspond to conditions that meet a pre-established objective function criterion, such as maximizing reaction yield. Following this active learning cycle, the method balances both exploitation and exploration of the reactivity space, taking the experimentally generated data to close a re-training loop and suggest a new set of experiments.
000 for downstream studies. Interestingly, the sampled reactions explore different regions of the search space in comparison to the training data, suggesting a potential bias in established practices and our previous reactions. Using this small dataset to train a ML algorithm can be challenging since it does not entirely represent the optimization task. For a successful application, the ML routine must transfer knowledge. Toward that end, we used our LabMate.ML routine.33,35 In short, the method uses self-tuning random forests and allows optimizing categorical (e.g., solvent) and real value (e.g., temperature) variables simultaneously by predicting reaction yields. The prioritized reactions will thus correspond to conditions that meet a pre-established objective function criterion, such as maximizing reaction yield. Following this active learning cycle, the method balances both exploitation and exploration of the reactivity space, taking the experimentally generated data to close a re-training loop and suggest a new set of experiments.
For the first iteration, LabMate.ML suggested three reactions that afforded 3A5AF in 24–53% yield (Fig. 1b and Table 8 from ESI‡). Those reaction conditions, together with the corresponding yields, allowed refinement of the model. An optimal yield (62%) was obtained in iteration 4 while using tetrapropylammonium chloride (TPAC), pTsOH, B(OH)3 and NaCl as additives at 169 °C for 12 minutes.
Moreover, scaling up the reaction from 0.1 to 1.1 mmol consistently yielded the desired product in 66% yield after extraction. To further show the applicability of the ML-suggested method, we performed a larger scale synthesis, starting with 4.5 mmol of NAG to obtain the isolated product in 68% yield. In contrast with the unoptimized methodology, no significant formation of side products and insoluble tars (i.e., humins) was observed.
This showcases the potential of ML that in 4 iterations was able to build upon the previous method and further fine tune the reaction conditions. When generating the initial dataset, chemical intuition required over 100 reactions to plateau at ∼50% yield. Overall comparing to the previously reported methodologies,12,22–29 this procedure is easily scalable due to the homogeneity of the reaction, does not use complex reagents and avoids the use of undesirable solvents, such as DMF.
Additionally, the use of TPAC as solvent allows the reuse of the reaction media, further improving the green metrics of our methodology. To this end, successive dehydration reactions were performed, allowing the IL to be reused for up to eight cycles without significant loss of 3A5AF yield (Table 1). TPAC was recovered from the aqueous phase during the extraction procedure in each cycle. Thus, the reutilization of the reaction medium makes our process even more favourable in terms of green chemistry, with low environmental impact. Importantly, the yield of the reaction ranges around ∼65% in most of our experiments, with yields going down to 56% and up to 72%. This may be due to heat transfer challenges that are more noticeable in higher scales, that can be further optimized by adjusting reaction set-ups.
Having successfully developed a simple, efficient, reusable, and scalable method for the transformation of NAG into 3A5AF, we envisioned the synthesis of 3A5AF directly from the biopolymer chitin. We carried out this study by milling chitin to break its crystalline structure (hydrogen bond networks), allowing it to react more easily.45 In particular, we grinded 2 g of commercial chitin in a planetary ball mill, using a ZrO2 reactor and 100 ZrO2 balls at 650 rpm for 1 h 30 min (with rotation inversion cycles of 15 minutes and 5 s pause between inversion cycles). This procedure afforded a 10-fold reduction in chitin particle size, breaking its crystalline structure and increasing its surface area (Fig. 2).
Furthermore, FTIR analysis shows no changes in the spectra, retaining the bands corresponding to the hydrogen bond O–H and N–H stretching at 3440 cm−1 and ∼3000 cm−1 correspondingly. Importantly, no changes were observed to the strong bands at 1600, corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of the amide, highlighting that no chemical modification of the chitin occurred.
O stretching of the amide, highlighting that no chemical modification of the chitin occurred.
To our delight, 3A5AF was isolated with 37% yield (compared with 20% reported in the literature) directly from chitin. This result demonstrates that, in addition to NAG dehydration, our system promotes the hydrolysis of chitin glycosidic bonds. Additionally, we reused the TPAC employing chitin as starting material for up to 3 cycles (Table 2) and similar behaviour to the monomeric starting material was observed.
Our final experiment was an integrated approach starting from the shrimp shells to the desired final furan as depicted in Fig. 3. To this end, we extracted chitin from the shrimp shells using naturally occurring citric acid – less toxic than the commonly used acids in extraction of chitin46 – followed by ball milling and our dehydration method. This yielded the 3A5AF in 10.5 mg g−1 of shrimp shell, further highlighting the potential of this methodology for the valorization of waste biomass.47 The extracted chitin was prepared and characterized, and the results are presented in the ESI.‡
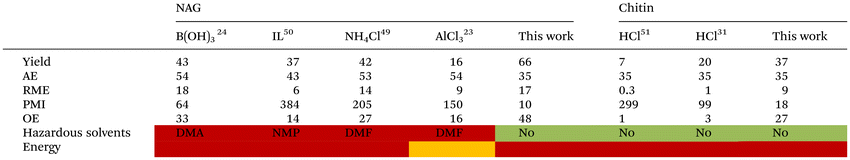
Finally, we performed a quantitative and qualitative green metrics analysis for the formation of the furan from NAG and chitin of some representative methods from the literature and ours according to the CHEM21 toolkit (Table 3).48 These methods encompass initial reports using B(OH)3 and NaCl,24 as well as more recent methods that employ AlCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 or simple NH4Cl.49 Most methodologies from NAG provide similar “atom economy (AE)”, as well as “reaction mass efficiency (RME)”, with the use of NH4Cl and IL50 affording lower RMEs. However, concerning “process mass intensity” (PMI) our method that allows the reuse of the IL, with only ca. 20% losses, affords significantly lower PMIs for the reaction. It is note highlighting that these PMIs do not account for isolation solvents due to the lack of information available on previous literature. Concerning chitin, previous methods31,51 afford lower RMEs, whereas our method affords better PMIs for the same reason as NAG. Importantly, both our method and the previous ones using chitin do not use hazardous solvents (Table 3).
23 or simple NH4Cl.49 Most methodologies from NAG provide similar “atom economy (AE)”, as well as “reaction mass efficiency (RME)”, with the use of NH4Cl and IL50 affording lower RMEs. However, concerning “process mass intensity” (PMI) our method that allows the reuse of the IL, with only ca. 20% losses, affords significantly lower PMIs for the reaction. It is note highlighting that these PMIs do not account for isolation solvents due to the lack of information available on previous literature. Concerning chitin, previous methods31,51 afford lower RMEs, whereas our method affords better PMIs for the same reason as NAG. Importantly, both our method and the previous ones using chitin do not use hazardous solvents (Table 3).
Conclusions
In summary, we here report the use of active learning to steer the valorization of biomasses and leverage the discovery of a novel and efficient system for converting NAG into 3A5AF. The chitin-derived 3A5AF was isolated from NAG in 66% yield (conditions: pTsOH, B(OH)3, NaCl in TPAC at 169 °C for 12 minutes) and the reaction media was reused for up to 8 cycles without significant loss efficiency. Moreover, the reaction was efficiently scaled up to 4.52 mmol. The one-pot reaction of chitin into 3A5AF, resulted in 37% yield of the required product. This is ca. 2-fold higher than the previously best described literature method. Further, the ML-designed method proved robust and generalizable as it led to 3A5AF in 10.5 mg g−1 of shrimp shell, highlighting the potential for the implementation of the methodology in a “real world” scenario. The reaction media can be reused for up to 8 cycles without significant loss of productivity. We envision that the disclosed method for obtaining 3A5AF directly from chitin will expand the utility of nitrogen rich synthons in materials science and medicinal chemistry and anticipate that ML can play a decisive role uncovering new roles for biomasses and promoting sustainable chemistry.Data availability
The data supporting this article have been included as part of the ESI.‡Conflicts of interest
T. R. is co-founder and shareholder of TargTex S.A., Vernus BioLabs, a consultant to the pharmaceutical and biotechnology industry and a member of the Acceleration Consortium, University of Toronto.Acknowledgements
The authors acknowledge Fundação para a Ciência e Tecnologia (FCT) for financial support (PTDC/QUI-QOR/32008/2017, PTDC/MECONC/29327/2017, UIDB/04138/2020, 2022.08851.PTDC, EXPL/BII-BIO/0436/2021, UIDB/50006/2020, 2020.01614.CEECIND/CP1596/CT0007). The project leading to this application has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 951996. The NMR spectrometers are part of the National NMR Network (PTNMR) are partially supported by Infrastructure Project No 022161 (co-financed by FEDER through COMPETE 2020, POCI and PORL and FCT through PIDDAC). The authors acknowledge Prof. Cristina Silva Pereira from Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, for kindly allowing the use of the cryomill equipment and guide us in the grinding process.References
- K. H. R. Rouwenhorst, P. M. Krzywda, N. E. Benes, G. Mul and L. Lefferts, in Techno-Economic Challenges of Green Ammonia as an Energy Vector, Elsevier, 2021, 41–83, DOI:10.1016/B978-0-12-820560-0.00004-7.
- X. Zhang, E. A. Davidson, D. L. Mauzerall, T. D. Searchinger, P. Dumas and Y. Shen, Nature, 2015, 528, 51–59 CrossRef CAS PubMed.
- K. I. Galkin and V. P. Ananikov, ChemSusChem, 2019, 12, 2976–2982 CrossRef CAS PubMed.
- N. Li and M. H. Zong, ACS Catal., 2022, 12, 10080–10114 CrossRef CAS.
- L. T. Mika, E. Cséfalvay and Á. Németh, Chem. Rev., 2018, 118, 505–613 CrossRef CAS PubMed.
- C. Espro, E. Paone, F. Mauriello, R. Gotti, E. Uliassi, M. L. Bolognesi, D. Rodríguez-Padrón and R. Luque, Chem. Soc. Rev., 2021, 50, 11191–11207 RSC.
- C. Xu, E. Paone, D. Rodríguez-Padrón, R. Luque and F. Mauriello, Chem. Soc. Rev., 2020, 49, 4273–4306 RSC.
- J. Dai, F. Li and X. Fu, ChemSusChem, 2020, 13, 6498–6508 CrossRef CAS PubMed.
- X. Shi, X. Ye, H. Zhong, T. Wang and F. Jin, Mol. Catal., 2021, 505, 111517 CrossRef CAS.
- H. Kobayashi, T. Sagawa and A. Fukuoka, Chem. Commun., 2023, 59, 6301–6313 RSC.
- M. Bisht, I. P. E. Macário, M. C. Neves, J. L. Pereira, S. Pandey, R. D. Rogers, J. A. P. Coutinho and S. P. M. Ventura, ACS Sustainable Chem. Eng., 2021, 9, 16073–16081 CrossRef CAS.
- M. W. Drover, K. W. Omari, J. N. Murphy and F. M. Kerton, RSC Adv., 2012, 2, 4642–4644 RSC.
- Z. Guo, C. Chen, J. Zhao, X. Guo, L. Jia, P. Liu, C. M. Pedersen, X. Hou, Y. Qiao and Y. Wang, J. Mol. Liq., 2022, 365, 120219 CrossRef CAS.
- R. A. Franich, S. J. Goodin and A. L. Wilkins, J. Anal. Appl. Pyrolysis, 1984, 7, 91–100 CrossRef CAS.
- T. T. Pham, X. Chen, N. Yan and J. Sperry, Monatsh. Chem., 2018, 149, 857–861 CrossRef CAS.
- T. T. Pham, X. Chen, T. Söhnel, N. Yan and J. Sperry, Green Chem., 2020, 22, 1978–1984 RSC.
- T. T. Pham, G. Gözaydın, T. Söhnel, N. Yan and J. Sperry, Eur. J. Org. Chem., 2019, 1355–1360 CrossRef CAS.
- J. G. Pereira, J. M. J. M. Ravasco, J. R. Vale, F. Queda and R. F. A. Gomes, Green Chem., 2022, 24, 7131–7136 RSC.
- C. Souza Santos, R. R. Mattioli, J. S. Baptista, V. H. Menezes da Silva, D. L. Browne and J. C. Pastre, Green Chem., 2023, 25, 5059–5067 RSC.
- C. H. M. van der Loo, J. P. Kaniraj, T. Wang, J. O. P. Broekman, M. L. G. Borst, K. Pouwer, A. Heeres, P. J. Deuss and A. J. Minnaard, Org. Biomol. Chem., 2023, 21, 8372–8378 RSC.
- A. D. Sadiq, X. Chen, N. Yan and J. Sperry, ChemSusChem, 2018, 11, 532–535 CrossRef CAS PubMed.
- K. Yamazaki, N. Hiyoshi and A. Yamaguchi, ChemistryOpen, 2023, 12, e202300148 CrossRef CAS PubMed.
- D. Padovan, H. Kobayashi and A. Fukuoka, ChemSusChem, 2020, 13, 3594–3598 CrossRef CAS PubMed.
- K. W. Omari, L. Dodot and F. M. Kerton, ChemSusChem, 2012, 5, 1767–1772 CrossRef CAS PubMed.
- X. Ji, J. Kou, G. Gözaydın and X. Chen, Appl. Catal., B, 2024, 342, 123379 CrossRef CAS.
- Y. Du, H. Zang, Y. Feng, K. Wang, Y. Lv and Z. Liu, J. Mol. Liq., 2022, 347, 117970 CrossRef CAS.
- H. Zang, J. Lou, S. Jiao, H. Li, Y. Du and J. Wang, J. Mol. Liq., 2021, 330, 115667 CrossRef CAS.
- H. Zang, Y. Feng, M. Zhang, K. Wang, Y. Du, Y. Lv, Z. Qin and Y. Xiao, Carbohydr. Res., 2022, 522, 108679 CrossRef CAS PubMed.
- H. Zang, Y. Feng, J. Lou, K. Wang, C. Wu, Z. Liu and X. Zhu, J. Mol. Liq., 2022, 366, 120281 CrossRef CAS.
- K. Chen, C. Wu, C. Wang, A. Zhang, F. Cao and P. Ouyang, Mol. Catal., 2021, 516, 112001 CrossRef CAS.
- X. Chen, Y. Gao, L. Wang, H. Chen and N. Yan, ChemPlusChem, 2015, 80, 1565–1572 CrossRef CAS PubMed.
- G. S. Halford, R. Baker, J. E. McCredden and J. D. Bain, Psychol. Sci., 2005, 16, 70–76 CrossRef PubMed.
- A. F. Almeida, F. A. P. Ataíde, R. M. S. Loureiro, R. Moreira and T. Rodrigues, J. Org. Chem., 2021, 86, 14192–14198 CrossRef CAS PubMed.
- B. J. Shields, J. Stevens, J. Li, M. Parasram, F. Damani, J. I. Martinez Alvarado, J. M. Janey, R. P. Adams and A. G. Doyle, Nature, 2021, 590, 89–96 CrossRef CAS PubMed.
- D. Reker, E. A. Hoyt, G. J. L. Bernardes and T. Rodrigues, Cell Rep. Phys. Sci., 2020, 1, 100247 CrossRef.
- A. F. Almeida, F. A. P. Ataíde, R. M. S. Loureiro, R. Moreira and T. Rodrigues, J. Org. Chem., 2021, 86, 14192–14198 CrossRef CAS PubMed.
- R. F. A. Gomes, Y. N. Mitrev, S. P. Simeonov and C. A. M. Afonso, ChemSusChem, 2018, 11, 1612–1616 CrossRef CAS PubMed.
- P. A. Russo, M. M. Antunes, P. Neves, P. V. Wiper, E. Fazio, F. Neri, F. Barreca, L. Mafra, M. Pillinger, N. Pinna and A. A. Valente, J. Mater. Chem. A, 2014, 2, 11813–11824 RSC.
- O. H. P. Cuervo, S. P. Simeonov, A. F. Peixoto, M. D. Popova, H. I. Lazarova, G. P. Romanelli, J. J. Martínez, C. Freire and C. A. M. Afonso, Energy Technol., 2019, 7, 1–9 Search PubMed.
- A. F. Peixoto, R. Ramos, M. M. Moreira, O. S. G. P. Soares, L. S. Ribeiro, M. F. R. Pereira, C. Delerue-Matos and C. Freire, Fuel, 2021, 303, 121227 CrossRef CAS.
- N. Bossons and R. F. A. Gomes, Curr. Opin. Green Sustain. Chem., 2024, 49, 100961 CrossRef.
- H. Liu, C. Hua, C. Song, S. Dai, H. Wang, W. Zhu and H. Li, Inorg. Chim. Acta, 2015, 428, 32–36 CrossRef CAS.
- K. Sun, L. Zhang, Y. Shao, Q. Li, H. Fan, G. Gao, S. Zhang, Q. Liu, Y. Wang and X. Hu, J. Chem. Technol. Biotechnol., 2019, 94, 3676–3686 CrossRef CAS.
- P. Y. Nikolov and V. A. Yaylayan, J. Agric. Food Chem., 2011, 59, 10104–10113 CrossRef CAS PubMed.
- X. Chen, Y. Gao, L. Wang, H. Chen and N. Yan, ChemPlusChem, 2015, 80, 1565–1572 CrossRef CAS PubMed.
- J. Pohling, D. Dave, Y. Liu, W. Murphy and S. Trenholma, Green Chem., 2022, 24, 1141–1151 RSC.
- N. Bossons and R. F. A. Gomes, Curr. Opin. Green Sustainable Chem., 2024, 49, 100961 CrossRef.
- C. R. McElroy, A. Constantinou, L. C. Jones, L. Summerton and J. H. Clark, Green Chem., 2015, 17, 3111–3121 RSC.
- C. Wang, C. Wu, A. Zhang, K. Chen, F. Cao and P. Ouyang, ChemistrySelect, 2022, 7, e202104574 CrossRef CAS.
- H. Zang, Y. Feng, M. Zhang, K. Wang, Y. Du, Y. Lv, Z. Qin and Y. Xiao, Carbohydr. Res., 2022, 522, 108679 CrossRef CAS PubMed.
- X. Chen, Y. Liu, F. M. Kerton and N. Yan, RSC Adv., 2015, 5, 20073–20080 RSC.
Footnotes |
| † Code used in this study can be accessed at https://hub.docker.com/r/tcorodrigues/labmate.ml. |
| ‡ Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc04280h |
| § These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2025 |