Rhoifolin loaded in PLGA nanoparticles alleviates oxidative stress and inflammation in vitro and in vivo†
Eveen
Al-Shalabi
,
Samah
Abusulieh
,
Alaa M.
Hammad
and
Suhair
Sunoqrot
 *
*
Department of Pharmacy, Faculty of Pharmacy, Al-Zaytoonah University of Jordan, P. O. Box 130, Amman 11733, Jordan. E-mail: suhair.sunoqrot@zuj.edu.jo; Fax: +962-6-4291432; Tel: +962-6-4291511, ext. 197
First published on 29th July 2022
Abstract
Rhoifolin (ROF) is a bioactive plant flavonoid with potent antioxidant and anti-inflammatory activity. However, no delivery system has yet been developed for ROF to overcome its biopharmaceutical limitations. The purpose of this study was to design a ROF-loaded polymeric nanocarrier as a potential anti-inflammatory nanomedicine. ROF was isolated from Jordanian Teucrium polium L. and entrapped into poly(lactide-co-glycolide) nanoparticles (PLGA NPs), followed by tannic acid-mediated surface modification with poly(ethylene glycol) (PEG). The optimal ROF NPs were highly monodisperse with an average diameter of 204 nm, a zeta potential of −28 mV, an entrapment efficiency of 45%, and drug loading of 9% w/w. The NPs exhibited excellent colloidal stability during storage and in the presence of serum and achieved sustained drug release for up to 96 h at physiologic (7.4) and acidic pH (5.0). In vitro cell-free antioxidant assays confirmed the potent radical scavenging activity of free ROF and ROF NPs. Moreover, ROF NPs were superior to free ROF in relieving oxidative stress in stimulated RAW 264.7 murine macrophages, which was attributed to enhanced cellular uptake of the NPs as confirmed by confocal microscopy and fluorimetry. In vivo anti-inflammatory activity was evaluated in a formalin-induced rat paw edema model. The results showed that ROF NPs were superior to free ROF in mitigating the histopathological changes in the inflamed paw tissues. Moreover, the NPs were equally potent to free ROF and the nonsteroidal anti-inflammatory drug diclofenac in terms of inhibiting the increase in paw thickness, normalizing nitric oxide levels, and modulating the gene expression of pro-inflammatory cytokines in the inflamed paw tissues. Our findings present a promising nanocarrier platform that can enhance the solubility and control the release of ROF, which will facilitate its administration in the treatment of inflammatory diseases.
Introduction
Inflammation is the underlying pathological process of most human diseases and plays an essential role in host defense against pathogens.1 The cardinal signs of inflammation include heat, swelling, redness, pain, and loss of function, which are mediated by the extravasation of leukocytes, vasodilation, and excessive release of inflammatory mediators.2,3 The acute phase of inflammation is characterized by vascular changes along with the accumulation of fluids, neutrophils, chemokines and cytokines, followed by tissue repair. Persistent acute inflammation leads to a progressive alteration in the type of cells and released mediators, resulting in chronic inflammation. If left uncontrolled, chronic inflammation may lead to a variety of diseases such as rheumatoid arthritis (RA), cancer, asthma, inflammatory bowel disease (IBD), Parkinson's disease, and auto-immune disorders.4 Corticosteroids and nonsteroidal anti-inflammatory drugs (NSAIDs) are the primary treatments for inflammatory disorders. However, studies have shown that their long-term use causes many adverse effects, such as cardiovascular complications, infections, diabetes, osteoporosis, and gastrointestinal ulcers and bleeding.5–9 For this reason, it is highly desirable to develop alternative drug candidates for the treatment of inflammation.Over the last few decades, there has been a growing interest in isolating and analyzing the bioactive constituents of medicinal plants to elucidate their health benefits, including their potential role in immunomodulation and inflammation.10–13 Plant polyphenols are among the leading candidates as anti-inflammatory agents due to their free-radical scavenging properties and their ability to modify the expression of pro-inflammatory mediators.14,15 Rhoifolin (ROF, apigenin 7-O-β-neohesperidoside) (Fig. 1) is a trisubstituted flavone and a naturally occurring glycoside of apigenin. It is found in several dietary sources and edible plants such as grapefruit, bitter orange, lemon, grapes, tomatoes, and bananas.16 Preclinical studies have shown that ROF possesses a variety of significant biological activities including anti-inflammatory,17,18 anti-arthritic,19 and anticancer effects.20 However, clinical use of plant polyphenols such as ROF has been limited due to many challenges such as poor water solubility, low stability, poor bioavailability and permeability, and fast release.21 Therefore, designing an appropriate drug delivery system is necessary to conquer those limitations.
Nanocarriers have achieved considerable steps forward in bringing anti-inflammatory phytochemicals closer to therapeutic applications.22 Polymeric nanoparticle (NP) drug delivery systems provide a versatile paradigm for targeting inflammation because of their biocompatibility, low toxicity, and biodegradability.23 In addition, they can enhance drug absorption because of their nanometer size and ability to permeate across biological barriers. They can protect drugs from premature enzymatic degradation, increase the stability and solubility of hydrophobic drugs, and offer sustained drug release and prolonged circulation times, which can increase patient compliance. Furthermore, they can be prepared using cost effective and scalable methods.24–26 Poly(lactide-co-glycolide) (PLGA) is among the commonly used polymers for NP fabrication because of its biocompatibility, biodegradability, stability, and possibility to modify surface properties. It is also approved by the US Food and Drug Administration (FDA) for human administration.27,28
Despite the potent role of ROF as an anti-inflammatory agent,17–19 like other molecules in its class, it suffers from poor aqueous solubility,29 which can greatly hinder its clinical application. At the same time, no delivery system has yet been developed for this promising drug candidate. Thus, we hypothesized that PLGA polymeric NPs could serve as a delivery platform for ROF to overcome its biopharmaceutical challenges in therapeutic applications such as inflammation. To validate our hypothesis, ROF was first isolated from Jordanian Teucrium polium (germander), an herbaceous medicinal plant known to be rich in flavonoids and abundant in the Mediterranean region.30–32 ROF was then entrapped in poly(ethylene glycol) (PEG)-modified PLGA NPs. The NPs were prepared, optimized, and characterized to determine their physicochemical properties. Then, the antioxidant and anti-inflammatory activities of the optimized NPs were evaluated in vitro in RAW 264.7 murine macrophages and in vivo in a formalin-induced rat paw edema model.
Materials and Methods
Materials
ROF was isolated from Jordanian T. polium (germander) as described below. PLGA (lactide/glycolide ratio 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 5–10 kDa) and bicine were purchased from Acros Organics (USA). Poly(vinyl alcohol) (PVA, MW 13–23 kDa, 87–89% hydrolyzed), tannic acid (TA), Nile red (NR), acetic acid, and formalin (∼37% in water) were purchased from Sigma-Aldrich (USA). Diclofenac deanolate (DIC, Tratul® ampoules) was obtained from Gerot Pharmazeutika (Australia). Methoxy poly(ethylene glycol)-thiol (mPEG-SH, MW 5 kDa) was purchased from Laysan Bio (USA). Dimethyl sulfoxide (DMSO), dichloromethane (DCM), and potassium bromide (KBr) were obtained from Fisher Chemicals (Thermo Fisher Scientific, UK). Phosphate buffered saline solution (PBS, 10×, pH 7.4) was purchased from Fisher Bioreagents (USA). Tween 80 was procured from RCFL Limited (India). 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS diammonium salt chromogenic peroxidase substrate) was obtained from Abcam (UK). Potassium permanganate (KMnO4) was purchased from Riedel-de Haën (Germany). Ultrapure water was prepared using an EMD Millipore Direct-Q 5UV system (USA).
50, 5–10 kDa) and bicine were purchased from Acros Organics (USA). Poly(vinyl alcohol) (PVA, MW 13–23 kDa, 87–89% hydrolyzed), tannic acid (TA), Nile red (NR), acetic acid, and formalin (∼37% in water) were purchased from Sigma-Aldrich (USA). Diclofenac deanolate (DIC, Tratul® ampoules) was obtained from Gerot Pharmazeutika (Australia). Methoxy poly(ethylene glycol)-thiol (mPEG-SH, MW 5 kDa) was purchased from Laysan Bio (USA). Dimethyl sulfoxide (DMSO), dichloromethane (DCM), and potassium bromide (KBr) were obtained from Fisher Chemicals (Thermo Fisher Scientific, UK). Phosphate buffered saline solution (PBS, 10×, pH 7.4) was purchased from Fisher Bioreagents (USA). Tween 80 was procured from RCFL Limited (India). 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS diammonium salt chromogenic peroxidase substrate) was obtained from Abcam (UK). Potassium permanganate (KMnO4) was purchased from Riedel-de Haën (Germany). Ultrapure water was prepared using an EMD Millipore Direct-Q 5UV system (USA).
Isolation of ROF from T. polium
Jordanian T. polium was collected, defatted, and extracted using ethanol as described in our previous publication.33 The crude ethanolic extract was dried under vacuum and then partitioned between chloroform and water (1.1 L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.1 L). The water layer was extracted with 2.5 L butanol to give 264 g of butanol extract and 120 g of water extract. The butanol extract (TB, 146 g) was adsorbed on 150 g of silica gel 60 and subjected to column chromatography (62 × 7.0 cm) using 800 g of the same adsorbent. The column was packed in chloroform and the polarity was gradually increased using methanol, till pure methanol was added. Five hundred milliliter fractions (a total of 110 fractions) were collected and grouped into six groups designated TBI–TBVI. Fraction TBVI was further treated with methanol, which led to precipitation of a yellow UV-active solid (2993 mg) that was identified as ROF. M. p.: 312 °C. HRMS (Bruker Daltonics Apex IV, 7.0 T Ultra Shield Plus) m/z: 601.15278 [M + Na]+. IR (KBr; Thermo-Nicolet Nexus 870 FT-IR spectrophotometer) υ max cm−1: 3275, 2904, 1654, 1602, 1586, 1495, 1344, 1292, 1180, 833. UV λmax (MeOH; Shimadzu UV-1800 spectrophotometer) nm: 333 (band I), 226 (band II); + NaOMe: 391 (band I), 229 (band II); + AlCl3: 387 (band I), 229 (band II); + HCl: 387 (band I), 229 (band II). 1H NMR (300 MHz, DMSO-d6) δ ppm: 3.12–5.44 (complex CH, CH2, CH3, diglucoside moiety), 6.42 (1H, d, J = 1.2 Hz, H-6), 6.76 (1H, d, J = 1.3 Hz, H-8), 6.83 (1H, s, H-3), 6.92 (2H, d, J = 8.6 Hz, H-3′,5′), 7.91 (2H, d, J = 8.6 Hz, H-2′,6′), 12.93 (C5–OH). 13C NMR (75 MHz, DMSO-d6) δ ppm: 18.3 (C-6′′′), 66.5 (C-6′′), 68.8 (C-5′′′), 70.0 (C-4′′), 70.8 (C-3′′′), 71.2 (C-2′′′), 72.5 (C-4′′′), 73.6 (C-2′′), 76.1 (C-5′′), 76.7 (C-3′′), 95.3 (C-8), 100 (C-6), 100.3 (C-1′′), 101.0 (C-1′′′), 103.6 (C-3), 105.9 (C-10), 116.6 (C-3′,5′), 121.5 (C-1′), 129.1 (C-2′,6′), 157.4 (C-9), 161.7 (C-4′), 161.9 (C-5), 163.3 (C-7), 164.8 (C-2), 182.5 (C-4). Spectral characterization of ROF is presented in the ESI.†
1.1 L). The water layer was extracted with 2.5 L butanol to give 264 g of butanol extract and 120 g of water extract. The butanol extract (TB, 146 g) was adsorbed on 150 g of silica gel 60 and subjected to column chromatography (62 × 7.0 cm) using 800 g of the same adsorbent. The column was packed in chloroform and the polarity was gradually increased using methanol, till pure methanol was added. Five hundred milliliter fractions (a total of 110 fractions) were collected and grouped into six groups designated TBI–TBVI. Fraction TBVI was further treated with methanol, which led to precipitation of a yellow UV-active solid (2993 mg) that was identified as ROF. M. p.: 312 °C. HRMS (Bruker Daltonics Apex IV, 7.0 T Ultra Shield Plus) m/z: 601.15278 [M + Na]+. IR (KBr; Thermo-Nicolet Nexus 870 FT-IR spectrophotometer) υ max cm−1: 3275, 2904, 1654, 1602, 1586, 1495, 1344, 1292, 1180, 833. UV λmax (MeOH; Shimadzu UV-1800 spectrophotometer) nm: 333 (band I), 226 (band II); + NaOMe: 391 (band I), 229 (band II); + AlCl3: 387 (band I), 229 (band II); + HCl: 387 (band I), 229 (band II). 1H NMR (300 MHz, DMSO-d6) δ ppm: 3.12–5.44 (complex CH, CH2, CH3, diglucoside moiety), 6.42 (1H, d, J = 1.2 Hz, H-6), 6.76 (1H, d, J = 1.3 Hz, H-8), 6.83 (1H, s, H-3), 6.92 (2H, d, J = 8.6 Hz, H-3′,5′), 7.91 (2H, d, J = 8.6 Hz, H-2′,6′), 12.93 (C5–OH). 13C NMR (75 MHz, DMSO-d6) δ ppm: 18.3 (C-6′′′), 66.5 (C-6′′), 68.8 (C-5′′′), 70.0 (C-4′′), 70.8 (C-3′′′), 71.2 (C-2′′′), 72.5 (C-4′′′), 73.6 (C-2′′), 76.1 (C-5′′), 76.7 (C-3′′), 95.3 (C-8), 100 (C-6), 100.3 (C-1′′), 101.0 (C-1′′′), 103.6 (C-3), 105.9 (C-10), 116.6 (C-3′,5′), 121.5 (C-1′), 129.1 (C-2′,6′), 157.4 (C-9), 161.7 (C-4′), 161.9 (C-5), 163.3 (C-7), 164.8 (C-2), 182.5 (C-4). Spectral characterization of ROF is presented in the ESI.†
Preparation of ROF-loaded PEGylated PLGA NPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 40 min at 4 °C and washed with water to remove trace DMSO, PVA, and unentrapped drug. The supernatant was collected, and the NP pellet was resuspended in ultrapure water under sonication (Elmasonic S 40-H, Germany) and stored at 4 °C. For the preparation of NR-labeled PLGA NPs, ROF was replaced with 0.5 mg NR and the NPs were prepared using 50 mg PLGA as described above.
000g for 40 min at 4 °C and washed with water to remove trace DMSO, PVA, and unentrapped drug. The supernatant was collected, and the NP pellet was resuspended in ultrapure water under sonication (Elmasonic S 40-H, Germany) and stored at 4 °C. For the preparation of NR-labeled PLGA NPs, ROF was replaced with 0.5 mg NR and the NPs were prepared using 50 mg PLGA as described above.
PEGylation of ROF-loaded PLGA NPs
The optimal NP formulation was chosen based on drug loading efficiency and coated with TA as an adhesive layer to facilitate PEGylation as previously reported.35 Briefly, NPs were suspended in 2 mL bicine buffer (0.1 M, pH 8) containing 1 mg mL−1 of TA and incubated for 30 min under sonication (Elmasonic S 40-H), then for 90 min under vigorous stirring. The NP dispersion was then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 40 min at 4 °C and washed with water to remove excess bicine and TA. The supernatant was discarded, and the NP pellets were collected and called PLGA@TA NPs. For PEGylation, PLGA@TA NPs were conjugated with mPEG-SH by relying on the reactivity of oxidized TA toward nucleophiles as previously described.36 Briefly, NPs were resuspended in 2 mL bicine buffer (0.1 M, pH 8) containing 50 mg mPEG-SH and incubated for 30 min under sonication (Elmasonic S 40-H) and then for 90 min under vigorous stirring. The NP dispersion was then centrifuged at 10
000g for 40 min at 4 °C and washed with water to remove excess bicine and TA. The supernatant was discarded, and the NP pellets were collected and called PLGA@TA NPs. For PEGylation, PLGA@TA NPs were conjugated with mPEG-SH by relying on the reactivity of oxidized TA toward nucleophiles as previously described.36 Briefly, NPs were resuspended in 2 mL bicine buffer (0.1 M, pH 8) containing 50 mg mPEG-SH and incubated for 30 min under sonication (Elmasonic S 40-H) and then for 90 min under vigorous stirring. The NP dispersion was then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 40 min at 4 °C and washed with water to remove excess bicine and mPEG-SH. The supernatant was discarded, and the NP pellets were collected, resuspended in ultrapure water under sonication (Elmasonic S 40-H), stored at 4 °C, and called PLGA@PEG NPs (ROF NPs). NR-labeled PLGA NPs were also primed with TA and PEGylated as described above for ROF NPs.
000g for 40 min at 4 °C and washed with water to remove excess bicine and mPEG-SH. The supernatant was discarded, and the NP pellets were collected, resuspended in ultrapure water under sonication (Elmasonic S 40-H), stored at 4 °C, and called PLGA@PEG NPs (ROF NPs). NR-labeled PLGA NPs were also primed with TA and PEGylated as described above for ROF NPs.
Particle size and zeta potential analysis
The particle size and polydispersity index (PDI) of the prepared NPs were measured by dynamic light scattering (DLS). Freshly prepared NPs were diluted appropriately with ultrapure water and the measurements were acquired using a Nicomp® Nano Z3000 particle sizing system (Entegris, USA). Zeta potential was measured via electrophoretic light scattering using the same instrument in the zeta potential mode. Measurements were reported as the mean ± standard deviation (SD) from at least three different NP batches.Entrapment efficiency (EE) and drug loading (DL) determination
The entrapment efficiency of ROF in the various batches of PLGA NPs was determined indirectly by analyzing the supernatant from the first centrifugation step by UV-Vis (UV-1800 spectrophotometer, Shimadzu, Japan). A calibration curve of ROF absorbance at 340 nm versus concentration in DMSO was constructed to determine the amount of unentrapped drug in the supernatant. EE and DL were calculated according to eqn (1) and (2), respectively, as follows: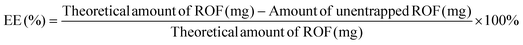 | (1) |
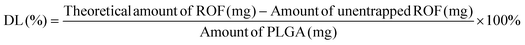 | (2) |
The amount of NR entrapped in NR-labeled NPs was calculated as described above for ROF, except that the UV absorbance of the supernatant was measured at 552 nm.
Fourier transform-infrared (FTIR) spectroscopy
ROF NPs were lyophilized using a freeze dryer system (FreeZone 4.5 L benchtop freeze dryer, Labconco, USA) at −50 °C and 0.5 mbar and prepared as KBr pellets. FTIR spectra of free ROF and ROF NPs were recorded using a Shimadzu IR Affinity-1 spectrometer (Japan) with data scanning from 4500 to 650 cm−1.X-ray photoelectron spectroscopy (XPS)
XPS was carried out to verify surface modification of ROF NPs. Freshly prepared PLGA, PLGA@TA, and PLGA@PEG NPs (ROF NPs) were each resuspended in 100 μL ultrapure water and drop-casted on pre-cleaned TiO2-coated Si wafers. The wafers were dried under vacuum and analyzed using a Kratos Axis SUPRA instrument fitted with a monochromated Al Kα X-ray source (1486.7 eV). All data was recorded at 150 W and a spot size of 700 × 300 μm. Survey scans and high-resolution scans were recorded at 160 eV and 20 eV pass energies, respectively. Electronic charge neutralization was achieved using a magnetic immersion lens. Data was analyzed using CasaXPS v2.3.20PR1.0 and the spectra were calibrated by setting the C 1s peak at 284.8 eV.Transmission electron microscopy (TEM)
ROF NPs were observed using TEM (Morgagni 268D, FEI, Netherlands) at an accelerating voltage of 60 kV. A drop of the NP dispersion was placed on a Formvar-coated copper grid (300 mesh, Electron Microscopy Sciences, USA) for 1 min, and the excess was wicked with filter paper. The grid was kept at room temperature overnight to air dry prior to imaging.Stability of ROF NPs
Storage stability of ROF NPs was evaluated by tracking the change in particle size up to 4 months of storage at 4 °C. Stability was also evaluated by incubating freshly prepared NPs in Dulbecco's Modified Eagle's Medium (DMEM, Euroclone, Italy) supplemented with 10% fetal bovine serum (FBS; Capricorn, Germany) at 37 °C under shaking at 100 rpm (Biosan ES-20 Orbital Shaker-Incubator, Latvia), withdrawing samples every 24 h up to 96 h to measure the size. Fold increase in size relative to fresh NPs was calculated from the ratio of particle size after storage or incubation to the particle size of freshly prepared NPs. Each experiment was performed in triplicate.In vitro release of ROF from loaded NPs
ROF release was conducted by the centrifugation method.37 Pellets of ROF NPs were suspended in 2 mL of phosphate buffered saline (PBS, pH 7.4) or acetate buffer (pH 5.0) containing 0.5% w/v Tween 80 to solubilize the released ROF. Experiments were performed in triplicate in 2 mL microtubes. The tubes were placed in a shaking incubator (Biosan ES-20 Orbital Shaker-Incubator) operating at 37 °C and 100 rpm. At predetermined time intervals (1, 2, 4, 6, 24, 48, 72, and 96 h), NP dispersions were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min to separate the released ROF from the NPs. After centrifugation, 500 μL of the supernatant was withdrawn and replaced with an equal amount of fresh PBS. The precipitated pellets in each tube were redispersed and incubated until the next sampling point. The amount of released ROF in the supernatant was determined by UV-Vis at 340 nm, based on a calibration curve of ROF in the release medium. The percentage of cumulative release was calculated from the ratio of the cumulative amount of ROF released at each time interval to the total amount of ROF entrapped in the NPs. Cumulative release (%) of ROF was then plotted against time (h) to obtain the release profile.
000g for 30 min to separate the released ROF from the NPs. After centrifugation, 500 μL of the supernatant was withdrawn and replaced with an equal amount of fresh PBS. The precipitated pellets in each tube were redispersed and incubated until the next sampling point. The amount of released ROF in the supernatant was determined by UV-Vis at 340 nm, based on a calibration curve of ROF in the release medium. The percentage of cumulative release was calculated from the ratio of the cumulative amount of ROF released at each time interval to the total amount of ROF entrapped in the NPs. Cumulative release (%) of ROF was then plotted against time (h) to obtain the release profile.
In vitro antioxidant activity of ROF NPs
Antioxidant activity of ROF and ROF NPs was determined using the ABTS assay following a previously described method.38 Briefly, the ABTS radical was produced by reacting the ABTS salt (3.6 mM in water) with KMnO4 solution (2.4 mM in water) in the dark for 18 h under stirring at room temperature. The produced blue-green solution was diluted ten times with ultrapure water to give an absorbance of 0.7 at 734 nm. Different amounts (1.25–200 μg) of ROF as free drug or an equivalent amount of ROF NPs were dissolved in 200 μL DMSO (the NPs were incubated overnight in DMSO to ensure complete breakdown and release of ROF). After adding 1 mL of diluted ABTS solution to 200 μL of each sample, samples were incubated at room temperature for 30 min in the dark, and the absorbance (Abs.) was measured at 734 nm. Solvent blanks (1 mL ABTS + 200 μL DMSO) were also run in each assay. The experiment was performed in triplicate. Scavenging activity (%) was calculated according to eqn (3):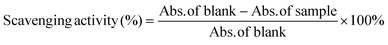 | (3) |
Antioxidant activity of ROF NPs in RAW 264.7 murine macrophages
Cell viability assay
When the cells reached 80% confluence, they were removed from the flask using a cell scraper and the cell suspension was counted. Cells were seeded in 96-well plates at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and allowed to attach overnight. Then, cells (n = 5) were incubated with various concentrations of ROF and ROF NPs (0–300 μM) diluted in complete culture medium for 24 h. After 24 h, the media was completely removed and replaced with 100 μL of fresh culture medium containing 0.5 mg mL−1 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, USA). Cells were incubated for another 2 h (37 °C, 5% CO2), after which the media was carefully removed and replaced with 100 μL of DMSO to dissolve the formazan crystals. The absorbance of the plates was measured at 540 nm using a Synergy HTX multi-mode microplate reader (Biotek, USA). Cell viability was determined relative to untreated cells.
000 cells per well and allowed to attach overnight. Then, cells (n = 5) were incubated with various concentrations of ROF and ROF NPs (0–300 μM) diluted in complete culture medium for 24 h. After 24 h, the media was completely removed and replaced with 100 μL of fresh culture medium containing 0.5 mg mL−1 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, USA). Cells were incubated for another 2 h (37 °C, 5% CO2), after which the media was carefully removed and replaced with 100 μL of DMSO to dissolve the formazan crystals. The absorbance of the plates was measured at 540 nm using a Synergy HTX multi-mode microplate reader (Biotek, USA). Cell viability was determined relative to untreated cells.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and allowed to attach overnight. Then, cells were treated with ROF and ROF NPs (0–40 μM) for 24 h. After 24 h, the wells were gently washed with PBS, followed by adding 500 μL of PBS containing 20 μM DCFDA and 50 μM tert-butyl hydroperoxide (tBHP) to stimulate the cells to produce ROS.40 After incubation with DCFDA/tBHP for 45 min, the fluorescence of each well was read using a Synergy HTX multi-mode microplate reader at 460 nm excitation and 528 nm emission wavelengths. Antioxidant activity was expressed as the fold increase in DCF fluorescence relative to untreated/unstimulated cells.
000 cells per well and allowed to attach overnight. Then, cells were treated with ROF and ROF NPs (0–40 μM) for 24 h. After 24 h, the wells were gently washed with PBS, followed by adding 500 μL of PBS containing 20 μM DCFDA and 50 μM tert-butyl hydroperoxide (tBHP) to stimulate the cells to produce ROS.40 After incubation with DCFDA/tBHP for 45 min, the fluorescence of each well was read using a Synergy HTX multi-mode microplate reader at 460 nm excitation and 528 nm emission wavelengths. Antioxidant activity was expressed as the fold increase in DCF fluorescence relative to untreated/unstimulated cells.
Cellular uptake of NR-labeled NPs
RAW 264.7 macrophages were treated with NR-labeled NPs to enable the visualization and quantification of the NPs’ cellular uptake. For imaging by confocal laser scanning microscopy (CLSM), cells were seeded in 4-well chamber slides (Nunc Lab-Tek II, Thermofisher, USA) at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and allowed to attach overnight. Cells were then treated with free NR or NR-labeled NPs diluted in complete culture medium at a concentration equivalent to 1 μg mL−1 NR for 1 h. Another group of cells was treated with the complete culture medium alone. After 1 h, the media was removed from the wells and the cells were washed with PBS three times. After the third wash, cells were fixed in 4% paraformaldehyde for 10 min at room temperature protected from light, followed by washing with PBS. After removing the remaining PBS, the chamber gasket was detached and the slide was mounted with ProLong™ Diamond Antifade Mountant with DAPI (Invitrogen, Thermofisher, USA) and covered with a glass coverslip. Imaging was performed using a Zeiss confocal LSM 780 (Zeiss, Germany). DAPI was detected at 405 nm excitation and 410–585 nm emission wavelengths, whereas NR was detected at 561 nm excitation and 566–691 emission wavelengths. Images were acquired using a Plan-Apochromat 63×/1.4 objective at a box size of 1912 × 1912 pixels and a pixel time of 3.38 μs.
000 cells per well and allowed to attach overnight. Cells were then treated with free NR or NR-labeled NPs diluted in complete culture medium at a concentration equivalent to 1 μg mL−1 NR for 1 h. Another group of cells was treated with the complete culture medium alone. After 1 h, the media was removed from the wells and the cells were washed with PBS three times. After the third wash, cells were fixed in 4% paraformaldehyde for 10 min at room temperature protected from light, followed by washing with PBS. After removing the remaining PBS, the chamber gasket was detached and the slide was mounted with ProLong™ Diamond Antifade Mountant with DAPI (Invitrogen, Thermofisher, USA) and covered with a glass coverslip. Imaging was performed using a Zeiss confocal LSM 780 (Zeiss, Germany). DAPI was detected at 405 nm excitation and 410–585 nm emission wavelengths, whereas NR was detected at 561 nm excitation and 566–691 emission wavelengths. Images were acquired using a Plan-Apochromat 63×/1.4 objective at a box size of 1912 × 1912 pixels and a pixel time of 3.38 μs.
For quantification of cellular uptake by fluorimetry, cells were seeded in 24-well plates at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in triplicate and allowed to attach overnight. The next day, cells were treated with free NR or NR-labeled NPs as described above for CLSM imaging. After the treatment, the cells were washed with PBS three times, 500 μL PBS was added to each well, and NR fluorescence was measured using a Synergy HTX multi-mode microplate reader at 540 nm excitation and 620 nm emission wavelengths. Results were expressed as the average fold increase in fluorescence relative to untreated cells.
000 cells per well in triplicate and allowed to attach overnight. The next day, cells were treated with free NR or NR-labeled NPs as described above for CLSM imaging. After the treatment, the cells were washed with PBS three times, 500 μL PBS was added to each well, and NR fluorescence was measured using a Synergy HTX multi-mode microplate reader at 540 nm excitation and 620 nm emission wavelengths. Results were expressed as the average fold increase in fluorescence relative to untreated cells.
In vivo anti-inflammatory activity of ROF NPs
| Group | Pretreatment | Inflammation induction | |
|---|---|---|---|
| I | Control | IP normal saline | — |
| II | Untreated | IP normal saline | 0.5 mL kg−1 SC formalin |
| III | DIC | 10 mg kg−1 IP DIC | 0.5 mL kg−1 SC formalin |
| IV | ROF | 10 mg kg−1 IP free ROF | 0.5 mL kg−1 SC formalin |
| V | ROF NPs | 10 mg kg−1 IP ROF NPs | 0.5 mL kg−1 SC formalin |
| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| tnf-α | Forward | AAATGGGCTCCCTCTCATCAGTTC |
| Reverse | TCTGCTTGGTGGTTTGCTACGAC | |
| il-1β | Forward | CACCTCTCAAGCAGAGCACAG |
| Reverse | GGGTTCCATGGTGAAGTCAAC | |
| β-actin | Forward | AAGTCCCTCACCCTCCCAAAAG |
| Reverse | AAGCAATGCTGTCACCTTCCC |
Statistical analysis
Experimental results were expressed as mean ± SD from at least three independent trials. All statistical analyses were performed in GraphPad Prism 7 software. Unless otherwise indicated, means were compared using one-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons test, or two-way ANOVA followed by Sidak's multiple comparisons test to determine significant differences between groups. Values of p < 0.05 were considered statistically significant.Results
Isolation of ROF from T. polium and spectral characterization
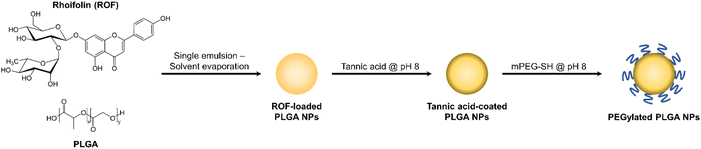
The spectroscopic data for ROF is presented in the ESI.† The IR spectrum of the compound (Fig. S1†) showed absorption bands at 1602 and 1586 cm−1 for the aromatic rings of the flavone, and at 3275 and 1654 cm−1 which are characteristic of –OH and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, respectively. High-resolution mass spectrometry (HRMS) of the compound (Fig. S2†) showed a peak at m/z 601.15278, which belongs to the [M + Na]+ ion. The UV/Vis spectra of the compound in methanol showed two maxima, at 333 nm and 226 nm for band I and band II, respectively. Addition of NaOMe led to a bathochromic shift of 58 nm without the appearance of a new band, indicating the presence of free –OH at C-4′ and the absence of –OH at C-7. The presence of free –OH on C-5 was indicated by a bathochromic shift of 54 nm in AlCl3. The shift was not affected by the addition of HCl, justifying the absence of ortho dihydroxy substitution of ring B. The 1H and 13C NMR spectra showed signals corresponding to apigenin and two sugar moieties. The 1H NMR (DMSO-d6) spectrum (Fig. S3†) showed two doublet peaks each integrating to one proton at 6.41 and 6.74 ppm (J = 1.9 Hz) due to the meta coupled protons on ring A (H-8 and H-6, respectively). H-3 appeared as a singlet at 6.83 ppm. The presence of the para substituted B ring was indicated by the two doublet peaks each integrating to 2 protons at 6.92 and 7.91 ppm (J = 8.7 Hz), referring to H-3′,5′ and H-2′,6′, respectively. The presence of rhamnose in the sugar moiety was indicated by the three protons doublet at 1.03 ppm for the methyl group. The β-linkage of the diglucoside was confirmed by the high value of coupling constant of the two anomeric protons, which appeared at 3.80 ppm (J = 10.8 Hz) and 5.03 ppm (J = 7.0 Hz), which correspond to H′′′ and H′′, respectively. The 13C NMR (DMSO-d6) spectrum (Fig. S4†) and the DEPT experiment of the compound (Fig. S5†) showed the presence of 25 carbon signals corresponding to 27 carbons, classified as one –CH3, one –CH2, 17 –CH, and 8 quaternary carbons. Thirteen of the C-signals are due to the apigenin moiety, and the remaining 12 C-signals are due to the sugar moiety. The spectral data of ROF were in full agreement with literature values.45
O groups, respectively. High-resolution mass spectrometry (HRMS) of the compound (Fig. S2†) showed a peak at m/z 601.15278, which belongs to the [M + Na]+ ion. The UV/Vis spectra of the compound in methanol showed two maxima, at 333 nm and 226 nm for band I and band II, respectively. Addition of NaOMe led to a bathochromic shift of 58 nm without the appearance of a new band, indicating the presence of free –OH at C-4′ and the absence of –OH at C-7. The presence of free –OH on C-5 was indicated by a bathochromic shift of 54 nm in AlCl3. The shift was not affected by the addition of HCl, justifying the absence of ortho dihydroxy substitution of ring B. The 1H and 13C NMR spectra showed signals corresponding to apigenin and two sugar moieties. The 1H NMR (DMSO-d6) spectrum (Fig. S3†) showed two doublet peaks each integrating to one proton at 6.41 and 6.74 ppm (J = 1.9 Hz) due to the meta coupled protons on ring A (H-8 and H-6, respectively). H-3 appeared as a singlet at 6.83 ppm. The presence of the para substituted B ring was indicated by the two doublet peaks each integrating to 2 protons at 6.92 and 7.91 ppm (J = 8.7 Hz), referring to H-3′,5′ and H-2′,6′, respectively. The presence of rhamnose in the sugar moiety was indicated by the three protons doublet at 1.03 ppm for the methyl group. The β-linkage of the diglucoside was confirmed by the high value of coupling constant of the two anomeric protons, which appeared at 3.80 ppm (J = 10.8 Hz) and 5.03 ppm (J = 7.0 Hz), which correspond to H′′′ and H′′, respectively. The 13C NMR (DMSO-d6) spectrum (Fig. S4†) and the DEPT experiment of the compound (Fig. S5†) showed the presence of 25 carbon signals corresponding to 27 carbons, classified as one –CH3, one –CH2, 17 –CH, and 8 quaternary carbons. Thirteen of the C-signals are due to the apigenin moiety, and the remaining 12 C-signals are due to the sugar moiety. The spectral data of ROF were in full agreement with literature values.45
Preparation and characterization of ROF-loaded PLGA NPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PLGA ratio (Table 3). All NPs exhibited a particle size between 150–260 nm with relatively low polydispersities. Entrapment efficiency (EE%) and drug loading (DL%) were used to determine the percentage amount of drug successfully entrapped into the NPs. ROF-loaded NPs prepared at different ROF
PLGA ratio (Table 3). All NPs exhibited a particle size between 150–260 nm with relatively low polydispersities. Entrapment efficiency (EE%) and drug loading (DL%) were used to determine the percentage amount of drug successfully entrapped into the NPs. ROF-loaded NPs prepared at different ROF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PLGA ratio showed an EE% ranging between 26.3% and 45.0% and a DL% ranging between 1.4% and 9.0%. It was observed that the EE increased with the increase in both PLGA and ROF amounts. The highest EE of 45.0% was obtained for formulation NP3 containing the highest amounts of ROF and PLGA (10 mg and 50 mg, respectively). It was also observed that DL increased proportionally with the increase in the amount of ROF added. For example, when a constant amount of 50 mg PLGA was maintained, DL increased from 2.0% to 9.0% upon increasing the amount of ROF from 2.5 mg to 10 mg. Also, when a constant amount of 20 mg PLGA was maintained, DL was increased from 1.8% to 5.4% as the amount of ROF was increased from 1 mg to 4 mg. Similarly, when a constant amount of 10 mg PLGA was maintained, DL was increased from 1.4% to 5.3% by increasing the amount of drug from 0.5 mg to 2 mg. These findings may be explained based on the change in viscosity of the emulsion produced during preparation of the NPs. For example, increasing the amount of PLGA increased the viscosity of the organic phase, resulting in high resistance to diffusion of ROF from the organic phase to the aqueous phase, thereby encapsulating more ROF in the PLGA NPs. Furthermore, increased ROF amount in the organic phase led to an increase in EE% and DL% which may be due to the more available ROF molecules that could interact with the PLGA polymer chains.47 The particle size of all NP formulations as measured by DLS ranged between 153 and 263 nm, with PDI values ranging between 0.14 and 0.25 (Table 3), denoting narrow particle size distributions. Based on the EE and DL results, NP3 was chosen for further investigation.
PLGA ratio showed an EE% ranging between 26.3% and 45.0% and a DL% ranging between 1.4% and 9.0%. It was observed that the EE increased with the increase in both PLGA and ROF amounts. The highest EE of 45.0% was obtained for formulation NP3 containing the highest amounts of ROF and PLGA (10 mg and 50 mg, respectively). It was also observed that DL increased proportionally with the increase in the amount of ROF added. For example, when a constant amount of 50 mg PLGA was maintained, DL increased from 2.0% to 9.0% upon increasing the amount of ROF from 2.5 mg to 10 mg. Also, when a constant amount of 20 mg PLGA was maintained, DL was increased from 1.8% to 5.4% as the amount of ROF was increased from 1 mg to 4 mg. Similarly, when a constant amount of 10 mg PLGA was maintained, DL was increased from 1.4% to 5.3% by increasing the amount of drug from 0.5 mg to 2 mg. These findings may be explained based on the change in viscosity of the emulsion produced during preparation of the NPs. For example, increasing the amount of PLGA increased the viscosity of the organic phase, resulting in high resistance to diffusion of ROF from the organic phase to the aqueous phase, thereby encapsulating more ROF in the PLGA NPs. Furthermore, increased ROF amount in the organic phase led to an increase in EE% and DL% which may be due to the more available ROF molecules that could interact with the PLGA polymer chains.47 The particle size of all NP formulations as measured by DLS ranged between 153 and 263 nm, with PDI values ranging between 0.14 and 0.25 (Table 3), denoting narrow particle size distributions. Based on the EE and DL results, NP3 was chosen for further investigation.
| NP | PLGA (mg) | ROF (mg) | Particle size (nm) | PDI | EE (%) | DL (%) |
|---|---|---|---|---|---|---|
| NP1 | 50 | 2.5 | 168 ± 16 | 0.19 ± 0.02 | 39.8 ± 16.5 | 2.0 ± 0.8 |
| NP2 | 50 | 5 | 181 ± 24 | 0.19 ± 0.01 | 36.1 ± 21.7 | 3.6 ± 2.2 |
| NP3 | 50 | 10 | 182 ± 8 | 0.15 ± 0.01 | 45.0 ± 4.3 | 9.0 ± 0.9 |
| NP4 | 20 | 1 | 153 ± 17 | 0.25 ± 0.09 | 35.8 ± 4.9 | 1.8 ± 0.2 |
| NP5 | 20 | 2 | 178 ± 47 | 0.21 ± 0.05 | 31.8 ± 12.6 | 3.2 ± 1.3 |
| NP6 | 20 | 4 | 223 ± 17 | 0.23 ± 0.11 | 27.2 ± 13.6 | 5.4 ± 2.7 |
| NP7 | 10 | 0.5 | 213 ± 66 | 0.19 ± 0.06 | 28.9 ± 12.5 | 1.4 ± 0.6 |
| NP8 | 10 | 1 | 263 ± 27 | 0.22 ± 0.13 | 37.0 ± 22.1 | 3.7 ± 2.2 |
| NP9 | 10 | 2 | 255 ± 38 | 0.18 ± 0.01 | 26.3 ± 8.7 | 5.3 ± 1.7 |
| NP | Particle size (nm) | PDI | Zeta potential (mV) |
|---|---|---|---|
| PLGA NPs (NP3) | 182 ± 8 | 0.15 ± 0.01 | −27 ± 8 |
| PLGA@TA NPs | 197 ± 4 | 0.16 ± 0.01 | −30 ± 4 |
| PLGA@PEG NPs (ROF NPs) | 204 ± 2 | 0.14 ± 0.02 | −28 ± 3 |
XPS was employed to verify successful PEGylation of ROF NPs by comparing the surface chemistry of uncoated PLGA, PLGA@TA, and PLGA@PEG NPs. XPS survey and high-resolution scans of the NPs before and after PEGylation are provided in the ESI,† and the relative composition of the C 1s peak components is depicted in Table 5. Uncoated PLGA NPs produced a C/O ratio of 1.05, which is lower than the theoretical value for PLGA of 1.25. This was most likely attributed to the presence of extra oxygens arising from incomplete removal of PVA during NP preparation and/or surface exposed ROF molecules. On the other hand, PLGA@TA NPs exhibited an increase in the C/O ratio to 1.70, which is close to the theoretical value for TA of 1.65. The increase in the C/O ratio was accompanied by an increase in the % of the C–C/C–H components from 54.2 to 66.0%, and a decrease in the % of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O component from 21.8 to 10.3%, confirming successful TA coating. PEGylation resulted in further changes to the NPs’ surface chemistry. Notably, PLGA@PEG NPs showed a decrease in the C/O ratio from 1.70 to 1.04, reflecting a rise in the % of oxygen species on the NP surface. Additionally, there was a marked decrease in the % of the C–C/C–H components from 66.0 to 34.6% and an increase in the % of the C–O/C–OH components from 23.7 to 42.6%. These changes strongly support the successful immobilization of PEG on the NPs’ surface mediated by the TA adhesive layer, as a facile surface modification approach to produce sterically stabilized ROF NPs.
O component from 21.8 to 10.3%, confirming successful TA coating. PEGylation resulted in further changes to the NPs’ surface chemistry. Notably, PLGA@PEG NPs showed a decrease in the C/O ratio from 1.70 to 1.04, reflecting a rise in the % of oxygen species on the NP surface. Additionally, there was a marked decrease in the % of the C–C/C–H components from 66.0 to 34.6% and an increase in the % of the C–O/C–OH components from 23.7 to 42.6%. These changes strongly support the successful immobilization of PEG on the NPs’ surface mediated by the TA adhesive layer, as a facile surface modification approach to produce sterically stabilized ROF NPs.
| NP | Atomic composition (%) | C/O | Composition of C 1s components (%) | |||
|---|---|---|---|---|---|---|
| C 1s | O 1s | C–C/C–H | C–O/C–OH | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
||
| PLGA | 51.2 | 48.8 | 1.05 | 54.2 | 24.0 | 21.8 |
| PLGA@TA | 63.0 | 37.0 | 1.70 | 66.0 | 23.7 | 10.3 |
| PLGA@PEG | 50.9 | 49.1 | 1.04 | 34.6 | 42.6 | 22.8 |
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching band observed in ROF NPs between 1650–1750 cm−1 was assigned to the ester groups of PLGA, and the C–O stretching bands appearing in both ROF and ROF NPs were attributed to the multiple C–OH groups in ROF and the ethylene oxide units of PEG, respectively. The NPs were further examined using TEM, which confirmed their spherical morphology with an approximate diameter of 110 nm (Fig. 2B). Note that particle diameters measured from electron microscopy images are typically smaller than DLS readings due to the collapsed state of the NPs under vacuum during TEM imaging.38 The colloidal stability of ROF NPs upon storage at 4 °C was evaluated by measuring the change in particle size over time. As shown in Fig. 2C, there was no significant increase in size up to 4 months compared to freshly prepared NPs. Serum stability of NPs is also crucial, since small particles may react with different blood components and aggregate during systemic administration. To examine the serum stability of ROF NPs, the fresh NP dispersion was incubated in DMEM supplemented with 10% FBS at 37 °C, withdrawing samples every 24 h up to 96 h. As shown in Fig. 2C, there was no statistical difference in terms of fold increase in size compared to freshly prepared NPs.
O stretching band observed in ROF NPs between 1650–1750 cm−1 was assigned to the ester groups of PLGA, and the C–O stretching bands appearing in both ROF and ROF NPs were attributed to the multiple C–OH groups in ROF and the ethylene oxide units of PEG, respectively. The NPs were further examined using TEM, which confirmed their spherical morphology with an approximate diameter of 110 nm (Fig. 2B). Note that particle diameters measured from electron microscopy images are typically smaller than DLS readings due to the collapsed state of the NPs under vacuum during TEM imaging.38 The colloidal stability of ROF NPs upon storage at 4 °C was evaluated by measuring the change in particle size over time. As shown in Fig. 2C, there was no significant increase in size up to 4 months compared to freshly prepared NPs. Serum stability of NPs is also crucial, since small particles may react with different blood components and aggregate during systemic administration. To examine the serum stability of ROF NPs, the fresh NP dispersion was incubated in DMEM supplemented with 10% FBS at 37 °C, withdrawing samples every 24 h up to 96 h. As shown in Fig. 2C, there was no statistical difference in terms of fold increase in size compared to freshly prepared NPs.
Sustained release from the drug delivery system is a desirable property to maintain a constant therapeutic concentration of the drug for a prolonged period of time. Therefore, in vitro release of ROF from the optimized ROF NPs was investigated in PBS pH 7.4 at 37 °C to mimic physiologic conditions. Release was also conducted in acetate buffer pH 5.0 to examine the effect of pH on modulating drug release. As shown in Fig. 2D, under both pH conditions, the release behavior of ROF from the NPs followed a biphasic pattern, where an initial fast release was observed during the first 6 h, followed by a slower sustained release pattern over 96 h. In PBS pH 7.4, approximately 54.3% of ROF was released from the NPs within the first 6 h, 80.2% was released after 24 h, 90.8% was released after 48 h, 97.6% was released after 72 h, and 99.0% was released after 96 h. Release in acetate buffer pH 5.0 followed similar kinetics, except that 84.3% of ROF was released at 24 h, which was significantly higher than the amount released in PBS pH 7.4 (p < 0.05). No differences were observed at all the other time points, reflecting the slow degradation rate of PLGA and indicating that drug release was mainly governed by diffusion.50
Antioxidant activity of ROF NPs
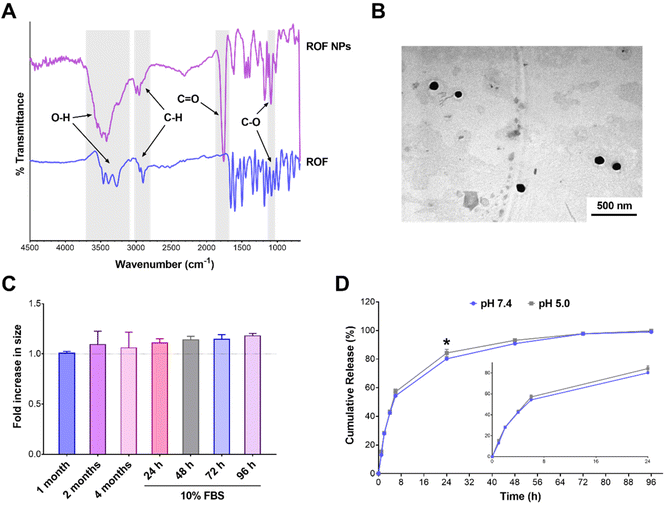
The antioxidant and anti-inflammatory properties of polyphenols have been largely attributed to their chemical structures. The highly conjugated aromatic system with multiple hydroxyl groups makes these compounds good hydrogen donors or electron donors, neutralizing free radicals and other reactive species.51 The antioxidant activity of the optimized ROF NPs was compared to free ROF using the ABTS assay. Different amounts of ROF and ROF NPs were incubated with the ABTS radical solution, and the decrease in absorbance of the ABTS radical was used as an indication of the radical scavenging activity. As shown in Fig. 3A, ROF NPs exhibited a dose-dependent increase in scavenging activity which was comparable to free ROF, indicating that the antioxidant activity of free ROF was not compromised during NP preparation.The antioxidant and free radical scavenging activity of ROF NPs was further evaluated in a cell-based assay using RAW 264.7 macrophages. Cells were first treated with various concentrations of free ROF and ROF NPs for 24 h and cell viability was determined (Fig. S8†). Then, cells were treated with nontoxic concentrations of free ROF or ROF NPs for 24 h, followed by inducing oxidative stress using a hydrogen peroxide derivative, tBHP, and monitoring the extent of cellular ROS generated as indicated by the fluorescence signal of the DCFDA probe. Interestingly, even though ROF in its free and NP form exhibited similar antioxidant activities in the cell-free assay (Fig. 3A), the two forms showed different behaviors in the cell-based assay. As depicted in Fig. 3B, tBHP caused a significant increase in cellular ROS in untreated/stimulated cells. Pretreating the cells with free ROF before tBHP stimulation did not have a significant effect on the elevated ROS levels even at the highest ROF concentration (40 μM). Conversely, cells pretreated with 10 and 20 μM ROF NPs maintained a cellular ROS level that was similar to that of untreated/unstimulated control cells. Moreover, at 40 μM, ROF NPs were associated with a significant reduction (p < 0.0001) in the cellular ROS level compared to the untreated/unstimulated controls, denoting their ability to reduce intrinsic oxidative stress. Additionally, the ROS levels in cells pretreated with ROF NPs were significantly lower than the levels obtained with equivalent concentrations of free ROF across the three tested concentrations.
Cellular uptake of NR NPs
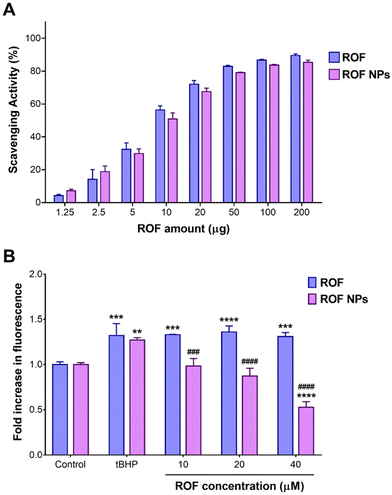
Cellular uptake experiments were conducted using NR-labeled NPs to verify whether the observed difference in cellular ROS scavenging activity was attributed to enhanced cellular uptake of the NP formulation compared to the free drug. CLSM images of RAW 264.7 cells after 1 h of incubation with free NR and NR NPs are depicted in Fig. 4A, and the corresponding cell-associated fluorescence values are shown in Fig. 4B. Cells treated with NR NPs displayed significantly stronger intracellular red fluorescence signals compared to free NR, indicating faster uptake of the NPs compared to the free dye, which correlates with the superior intracellular antioxidant activity of the NPs compared to the free drug.In vivo anti-inflammatory activity of ROF NPs
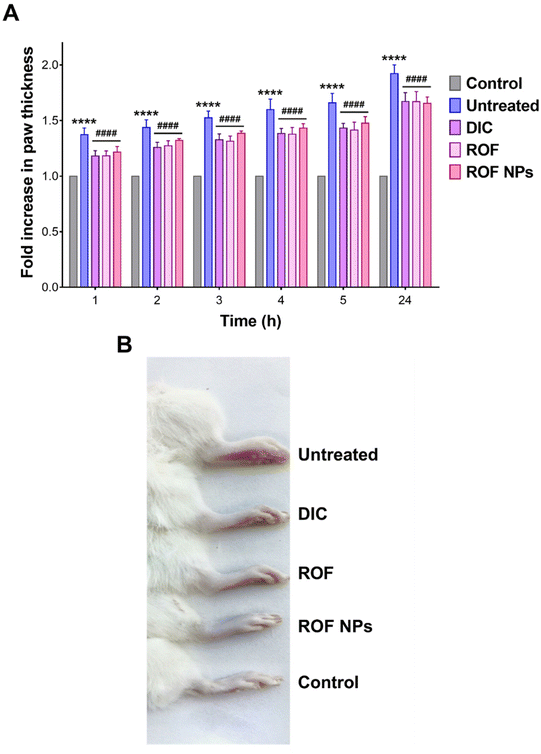
The anti-inflammatory activity of ROF NPs was evaluated in a formalin-induced rat paw edema model and compared to free ROF and a reference anti-inflammatory drug (DIC). Treatments were administered 1 h prior to paw edema induction and edema progression was monitored up to 24 h by measuring the fold increase in paw thickness relative to the 0 h time point. Note that the different agents were administered as a pretreatment in accordance with previous studies on ROF in animal models of inflammation.17,18 As shown in Fig. 5A, the untreated, DIC, ROF, and ROF NPs animal groups exhibited a significant time-dependent increase in paw thickness after formalin injection compared to the control group (p < 0.0001), consistent with paw edema formation. The greatest fold increase in paw thickness was observed in untreated rats, which showed a steady rise from 1.4-fold (1 h post formalin injection) to 1.9-fold (24 h post formalin injection). This increase in paw thickness indicated a successful induction and persistence of paw edema over the period of the study.Although all groups that received pretreatment with either DIC, free ROF, or ROF NPs were still associated with an increase in paw thickness compared to the control, this increase was significantly lower than the increase associated with the untreated group at all time points (p < 0.0001). This indicates that the pretreatment with DIC, ROF, and ROF NPs significantly inhibited edema formation and progression compared to the untreated group, confirming the anti-inflammatory activity of these drugs. Note that no significant difference was observed between DIC, free ROF, or ROF NPs across all time points, which strongly supports the potent anti-inflammatory activity of ROF alone and in the form of NPs. Morphological changes, including swelling and erythema of the rat paw in the untreated, DIC, ROF, and ROF NPs groups compared to the control group after 24 h of edema induction are presented in Fig. 5B.
The anti-inflammatory activity of ROF NPs compared to the different experimental groups was further evaluated by examining the histopathological changes in the paw tissues after 24 h of paw edema induction. The main changes are summarized in Table 6 and representative images of H & E-stained dermal layers of the paw tissues are shown in Fig. 6. Untreated animals showed significant infiltration of lymphocytes, neutrophils, and mast cells, in addition to severe vasculitis. The DIC group was associated with moderate lymphocyte infiltration, mild neutrophil infiltration, occasional appearance of mast cells, and minimal vasculitis. ROF was associated with mild lymphocyte infiltration and occasional appearance of mast cells. However, moderate neutrophil infiltration and moderate vasculitis were also observed. On the other hand, ROF NPs displayed mild lymphocyte and neutrophil infiltration with occasional appearance of mast cells and minimal vasculitis, denoting an enhancement in the anti-inflammatory activity of ROF when administered as ROF NPs.
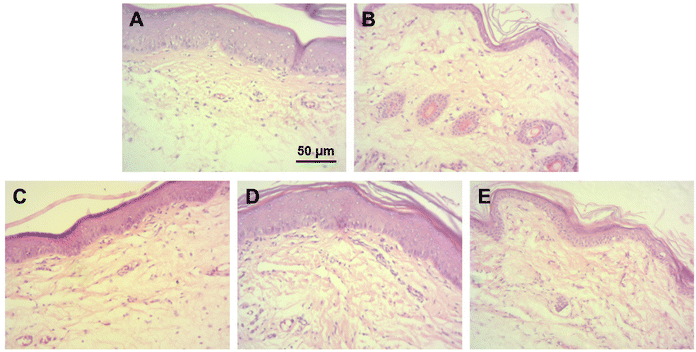 | ||
| Fig. 6 Histological examination of paw tissues after 24 h of paw edema induction. Representative images from (A) control, (B) untreated, (C) DIC, (D) ROF, and (E) ROF NPs groups. | ||
| Group | Lymphocytes | Neutrophils | Mast cells | Vasculitis |
|---|---|---|---|---|
| Control | − | − | − | − |
| Untreated | +++ | +++ | +++ | +++ |
| DIC | ++ | + | − | − |
| ROF | + | ++ | − | ++ |
| ROF NPs | + | + | − | − |
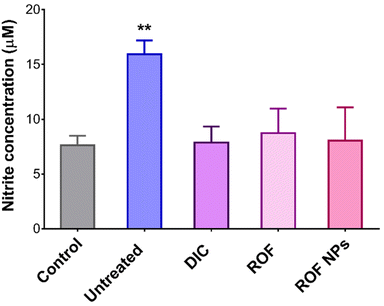
The in vivo antioxidant activity of ROF NPs was also evaluated by measuring the NO levels in the inflamed paw tissues using the Griess assay for nitrite determination. As shown in Fig. 7, the untreated group displayed 2.1-fold increase in nitrite levels compared to the control group, while the groups treated with DIC, free ROF, and ROF NPs maintained normal nitrite levels, which correlates with their potent anti-inflammatory activity.
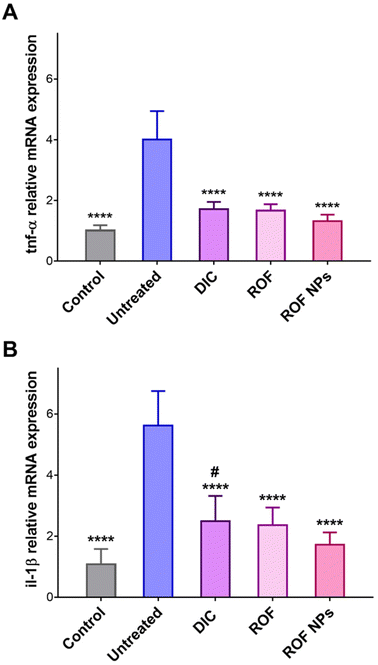
The effect of ROF NPs on slowing the progression of paw edema and the underlying inflammatory response was further supported by analyzing the gene expression of two pro-inflammatory cytokines, tnf-α and il-1β, in paw tissues extracted after 24 h of paw edema induction. TNF-α and IL-1β are important pro-inflammatory mediators as they are involved in the initiation and amplification of the inflammatory response.52,53 RT-PCR was performed to determine the relative mRNA expression of the pro-inflammatory mediators’ genes in the inflamed paw tissues. As shown in Fig. 8, the untreated group showed a significant increase (p < 0.0001) in both tnf-α and il-1β relative mRNA expression compared to the control, DIC, ROF, and ROF NPs groups. Moreover, groups pretreated with ROF and ROF NPs showed no significant difference in tnf-α and il-1β mRNA expression compared to the control group. Interestingly, the DIC pretreated group exhibited significantly higher (p < 0.05) expression of il-1β mRNA and no significant difference in tnf-α mRNA expression compared to the control group. However, the gene expression of the two cytokines was still significantly lower compared to the untreated group. The marked reduction in gene expression of the pro-inflammatory mediators confirms the anti-inflammatory activity of ROF NPs, which was comparable to free ROF and DIC.
Discussion
Poor aqueous solubility is a major challenge encountered in the development of drug candidates. Various studies have reported on polymeric NPs as drug delivery systems for different classes of drugs to enhance their solubility, efficacy and bioavailability.54–58 In this study, we developed an NP formulation for ROF, a promising bioactive flavonoid, based on PLGA. PLGA-based polymeric NPs have been previously employed to enhance the bioavailability, biological activity, and water solubility of various natural products. For example, Derman et al. prepared quercetin-encapsulated PLGA NPs by the single emulsion/solvent evaporation method and the NPs showed enhanced water solubility, biocompatibility, and sustained antioxidant activity of encapsulated quercetin compared to the free drug.59 The same technique was reported for the preparation of curcumin-loaded PLGA NPs to improve encapsulation efficiency, cytocompatibility with intestinal cells, and oral bioavailability of curcumin.60 In another study, Feng et al. reported that apigenin, the aglycone of ROF, loaded into PEG–PLGA NPs had a stronger anti-inflammatory activity and longer in vivo circulation time than unencapsulated apigenin.61 In another report, apigenin, was loaded in PEG–PLGA polymeric NPs to improve its solubility and bioavailability. Results demonstrated that apigenin NPs exerted an anti-inflammatory effect by inhibiting the expression and production of inflammatory cytokines and proved to be more effective in inhibiting lung fibrosis compared to free apigenin.62PLGA NPs experience high rates of uptake and opsonization by the reticuloendothelial system (RES) due to their hydrophobic nature. PEGylation has been shown to be a suitable technique to enhance the colloidal stability and water solubility of polymeric NPs, prolong their systemic circulation, reduce their immunogenicity, and decrease their accumulation in the RES organs.48,63 In this work, we used a simple and rapid technique to PEGylate ROF NPs by relying on the adhesive properties of oxidized polyphenols such as TA, avoiding the use of coupling reagents and complex purification steps. Physicochemical characterization of the optimized ROF NPs confirmed their composition and morphology. The NPs maintained excellent colloidal stability during storage and in the presence of serum, which may be attributed to the PEGylated NP surface. Similar results were obtained by our group for PEGylated polymeric nanocapsules entrapping cirsiliol, another bioactive flavonoid isolated from T. polium.33 As for drug release, the relatively fast release observed during the initial hours could be explained by drug desorption from the surface of the NPs, and the sustained release pattern in the later hours may be due to slow drug diffusion and erosion of the polymeric matrix.64 It is possible that the presence of the PEG coating increased the wettability of the hydrophobic PLGA surface, improved water permeation and drug diffusion through the polymer matrix, and consequently facilitated drug release.65 Moreover, the ABTS antioxidant assay confirmed that loading ROF in polymeric NPs may be an appropriate approach to maintain its antioxidant activity while controlling its release. The results were consistent with those of the previous study involving cirsiliol-loaded polymeric nanocapsules.33
Having shown promising attributes as a delivery system for ROF, the anti-inflammatory activity of ROF NPs was evaluated in RAW 264.7 macrophages, by monitoring the levels of cellular ROS following stimulation with a hydrogen peroxide derivative, tBHP. ROF NPs significantly enhanced the ROS scavenging activity of free ROF and even reduced the intrinsic oxidative stress at higher concentrations. These observations may be attributed to enhanced cellular uptake of the NP formulation, similar to previous studies involving PLGA NPs as carriers for anti-inflammatory drugs.66,67 The results were further corroborated by the cellular uptake experiments, which demonstrated faster uptake of NR-labeled NPs compared to the free dye.
Several animal models have been developed for the assessment of anti-inflammatory activities of novel drugs. Among the various animal models for inflammation, the formalin-induced paw edema model in rats has close similarity with inflammation in humans. Inflammation induced by formalin is biphasic. Bradykinin, and substance-P are involved in the early phase, whereas histamine, 5-hydroxytryptamine (5-HT), prostaglandins, and bradykinin are involved in the later phase.68 IL-1β and TNF-α are the primary cytokines in mediating acute inflammatory reactions at the site of their production. IL-1β can stimulate T-cell proliferation, induce fever by enhancing prostaglandin E2 (PGE2) synthesis, and trigger vasodilation and increase of vascular permeability by enhancing the release of histamine from mast cells. TNF-α can also induce the acute phase response and fever, either by stimulating PGE2 synthesis or inducing the production and release of IL-1β. TNF-a and IL-1β further exert secondary inflammatory effects by stimulating IL-6 synthesis and activating the innate immune response.52
Recent studies have shown that the anti-inflammatory activity of ROF is mediated by affecting the production and secretion of inflammatory mediators such as TNF-α, IL-1β, and IL-6. Moreover, ROF can suppress the expression of nuclear factor kappa-B (NF-κB) signaling pathway18 and reduce PGE2 levels17 and the oxidative stress associated with RA.19 Therefore, the anti-inflammatory activity of ROF NPs in the formalin-induced rat paw edema model was evaluated by monitoring the increase in paw thickness and the associated histopathological changes, and by measuring the NO and gene expression levels of tnf-α and il-1β in the inflamed paw tissues. Our results confirmed the potent anti-inflammatory activity of free ROF, which was comparable to the reference NSAID, DIC. ROF NPs exhibited an equal inhibitory effect on the progression of paw edema, tissue oxidative stress, and the gene expression levels of the pro-inflammatory mediators. However, ROF NPs performed better than the free drug in terms of mitigating the histopathological changes associated with formalin-induced paw edema, signifying that the NPs could successfully deliver ROF to the site of inflammation.
Conclusion
In this work, the poorly soluble bioactive flavonoid ROF was isolated from T. polium and successfully entrapped in PLGA NPs by the emulsification/solvent evaporation method, followed by TA-mediated NP PEGylation. The effect of drug/polymer ratio and polymer concentration on NP size, polydispersity, DL, and EE was investigated to obtain the optimal NP formulation. The optimal NPs showed an average size of 204 nm with a narrow size distribution and an EE of 45%. The NPs also showed long term stability for up to 4 months and excellent stability in the presence of serum for up to 96 h. The drug release profile exhibited a biphasic pattern involving a fast initial release followed by a slower sustained release pattern for up to 96 h regardless of the pH. Antioxidant assays using ABTS demonstrated that ROF NPs maintained similar free radical scavenging activity compared to free ROF. Cellular ROS assays conducted in RAW 264.7 macrophages revealed the superiority of ROF NPs in reducing oxidative stress compared to free ROF, which correlated with the faster cellular uptake of the NPs. The in vivo anti-inflammatory activity of ROF NPs was investigated in a formalin-induced rat paw edema model, which confirmed the role of ROF NPs in mitigating paw edema and the histopathological signs of inflammation, most likely by ROF-mediated inhibition of oxidative stress and gene expression of pro-inflammatory cytokines such as TNF-α and IL-1β. Even though the in vivo anti-inflammatory activity of ROF NPs was similar to free ROF in most anti-inflammatory assays, the NP formulation offers advantages such as solubility enhancement and controlled release, which will likely prove valuable for repeated dosing by improving the pharmacokinetics and biodistribution of the free compound. Collectively, the results demonstrate the promising potential of the developed ROF NP formulation as a nanomedicine for the treatment of inflammation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work received funding from Al-Zaytoonah University of Jordan (grants no. 15/28/2017-2018 and 29/11/2020-2021), and the Arab-German Young Academy of Sciences and Humanities (AGYA). AGYA draws on financial support of the German Federal Ministry of Education and Research (BMBF) grant no. 01DL20003. Isolation and spectral characterization of ROF was performed at the University of Jordan, School of Science, Department of Chemistry. The XPS data collection was performed at the Engineering and Physical Sciences Research Council (EPSRC) National Facility for XPS (“HarwellXPS”), operated by Cardiff University and University College London, under contract no. PR16195. The authors thank Dr Shaoliang Guan (School of Chemistry, Cardiff University) for assistance with XPS analysis, Dana Al-Qudah (Cell Therapy Center, University of Jordan) for assistance with CLSM imaging, and Dr Imad Hamdan (Alpha Medical Labs Pathology Consultations Center, Amman, Jordan) for assistance with histological examination.References
- F. R. Greten and S. I. Grivennikov, Immunity, 2019, 51, 27–41 CrossRef CAS PubMed.
- M. A. Alblihed, Mol. Biol. Rep., 2020, 47, 6611–6620 CrossRef CAS PubMed.
- D. Laveti, R. Hemalatha, R. Sistla, V. Naidu, V. Talla, V. Verma, N. Kaur and R. Nagpal, Inflammation Allergy: Drug Targets, 2013, 12(5), 349–361 CrossRef CAS PubMed.
- D. Furman, J. Campisi, E. Verdin, P. Carrera-Bastos, S. Targ, C. Franceschi, L. Ferrucci, D. W. Gilroy, A. Fasano, G. W. Miller, A. H. Miller, A. Mantovani, C. M. Weyand, N. Barzilai, J. J. Goronzy, T. A. Rando, R. B. Effros, A. Lucia, N. Kleinstreuer and G. M. Slavich, Nat. Med., 2019, 25, 1822–1832 CrossRef CAS PubMed.
- R. M. Haley and H. A. von Recum, Exp. Biol. Med., 2019, 244, 433–444 CrossRef CAS PubMed.
- S. Bindu, S. Mazumder and U. Bandyopadhyay, Biochem. Pharmacol., 2020, 180, 114147 CrossRef CAS PubMed.
- K. H. Maniar, I. A. Jones, R. Gopalakrishna and C. T. Vangsness Jr., Expert Opin. Pharmacother., 2018, 19, 93–102 CrossRef CAS PubMed.
- M. Salice, F. Rizzello, C. Calabrese, L. Calandrini and P. Gionchetti, Expert Rev. Gastroenterol. Hepatol., 2019, 13, 557–561 CrossRef CAS PubMed.
- T. Volmer, T. Effenberger, C. Trautner and R. Buhl, Eur. Respir. J., 2018, 52(4), 1800703 CrossRef PubMed.
- N. Yahfoufi, N. Alsadi, M. Jambi and C. Matar, Nutrients, 2018, 10, 1618 CrossRef PubMed.
- A. Attiq, J. Jalil, K. Husain and W. Ahmad, Front. Pharmacol., 2018, 976 CrossRef PubMed.
- S. M. Zaidi, T. M. Al-Qirim and N. Banu, Drugs R&D, 2005, 6, 157–165 CrossRef CAS PubMed.
- R. A. Khalaf, Curr. Top. Med. Chem., 2016, 16, 2549–2561 CrossRef PubMed.
- M. Y. Ansari, N. Ahmad and T. M. Haqqi, Biomed. Pharmacother., 2020, 129, 110452 CrossRef CAS PubMed.
- I. Jantan, M. A. Haque, L. Arshad, H. Harikrishnan, A. W. Septama and Z.-A. Mohamed-Hussein, J. Nutr. Biochem., 2021, 93, 108634 CrossRef CAS PubMed.
- J. Refaat, S. Yehia, M. Ramadan and M. Kamel, Int. J. Pharmacogn., 2015, 2, 102–109 CAS.
- O. Eldahshan and S. Azab, J. Appl. Pharm. Sci., 2012, 2, 74–79 Search PubMed.
- J. Fang, Z. Cao, X. Song, X. Zhang, B. Mai, T. Wen, J. Lin, J. Chen, Y. Chi, T. Su and F. Xiao, Inflammation, 2020, 43, 2191–2201 CrossRef CAS PubMed.
- S. Peng, C. Hu, X. Liu, L. Lei, G. He, C. Xiong and W. Wu, Braz. J. Med. Biol. Res., 2020, 53(6), e9489 CrossRef CAS PubMed.
- O. Eldahshan, Br. J. Pharm. Res., 2013, 3, 46–53 CrossRef.
- A. Faridi Esfanjani and S. M. Jafari, Colloids Surf., B, 2016, 146, 532–543 CrossRef CAS PubMed.
- R. Conte, V. Marturano, G. Peluso, A. Calarco and P. Cerruti, Int. J. Mol. Sci., 2017, 18, 709 CrossRef PubMed.
- M. Howard, B. J. Zern, A. C. Anselmo, V. V. Shuvaev, S. Mitragotri and V. Muzykantov, ACS Nano, 2014, 8, 4100–4132 CrossRef CAS PubMed.
- B. Begines, T. Ortiz, M. Pérez-Aranda, G. Martínez, M. Merinero, F. Argüelles-Arias and A. Alcudia, Nanomaterials, 2020, 10, 1403 CrossRef CAS PubMed.
- E. Calzoni, A. Cesaretti, A. Polchi, A. Di Michele, B. Tancini and C. Emiliani, J. Funct. Biomater., 2019, 10, 4 CrossRef CAS PubMed.
- M. Khalid and H. S. El-Sawy, Int. J. Pharm., 2017, 528, 675–691 CrossRef PubMed.
- K. Park, S. Skidmore, J. Hadar, J. Garner, H. Park, A. Otte, B. K. Soh, G. Yoon, D. Yu, Y. Yun, B. K. Lee, X. Jiang and Y. Wang, J. Controlled Release, 2019, 304, 125–134 CrossRef CAS PubMed.
- M. Mir, N. Ahmed and A. u. Rehman, Colloids Surf., B, 2017, 159, 217–231 CrossRef CAS PubMed.
- G. Lewin, A. Maciuk, A. Moncomble and J.-P. Cornard, J. Nat. Prod., 2013, 76, 8–12 CrossRef CAS PubMed.
- K. Qabaha, T. Hijawi, A. Mahamid, H. Mansour, A. Naeem, J. Abbadi and F. Al-Rimawi, Asian J. Chem., 2021, 33, 881–884 CAS.
- M. Khazaei, S. N. Nematollahi-Mahani, T. Mokhtari and F. Sheikhbahaei, J. Contemp. Med. Sci., 2018, 4, 1–6 CAS.
- S. Bahramikia and R. Yazdanparast, Phytother. Res., 2012, 26, 1581–1593 CrossRef CAS PubMed.
- E. Al-Shalabi, M. Alkhaldi and S. Sunoqrot, J. Drug Delivery Sci. Technol., 2020, 56, 101544 CrossRef CAS.
- K. Kızılbey, ACS Omega, 2019, 4, 555–562 CrossRef.
- S. A. Abouelmagd, F. Meng, B.-K. Kim, H. Hyun and Y. Yeo, ACS Biomater. Sci. Eng., 2016, 2, 2294–2303 CrossRef CAS PubMed.
- L. Pang, Y. Pei, G. Uzunalli, H. Hyun, L. T. Lyle and Y. Yeo, Pharm. Res., 2019, 36, 65 CrossRef PubMed.
- J. Shen and D. J. Burgess, Drug Delivery Transl. Res., 2013, 3, 409–415 CrossRef CAS PubMed.
- S. Sunoqrot, E. Al-Shalabi, A. G. Al-Bakri, H. Zalloum, B. Abu-Irmaileh, L. H. Ibrahim and H. Zeno, ACS Omega, 2021, 6, 2767–2776 CrossRef CAS PubMed.
- L. Wang, M. L. Xu, J. H. Hu, S. K. Rasmussen and M.-H. Wang, Food Chem. Toxicol., 2011, 49, 149–154 CrossRef CAS PubMed.
- G. Li, J. Zhou, M. Sun, J. Cen and J. Xu, J. Funct. Foods, 2020, 74, 104196 CrossRef CAS.
- A. Soyocak, H. Kurt, D. T. Cosan, F. Saydam, I. Calis, U. Kolac, Z. O. Koroglu, I. Degirmenci, F. S. Mutlu and H. Gunes, Hum. Exp. Toxicol., 2019, 38, 1296–1301 CrossRef CAS PubMed.
- C.-W. Chang, W.-T. Chang, J.-C. Liao, Y.-J. Chiu, M.-T. Hsieh, W.-H. Peng and Y.-C. Lin, J. Evidence-Based Complementary Altern. Med., 2012, 135379 Search PubMed.
- A. Peinnequin, C. Mouret, O. Birot, A. Alonso, J. Mathieu, D. Clarençon, D. Agay, Y. Chancerelle and E. Multon, BMC Immunol., 2004, 5, 3 CrossRef PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- A. K. Yadav, D. Yadav, K. Shanker, R. K. Verma, A. K. Saxena and M. M. Gupta, Chromatographia, 2009, 69, 653–658 CrossRef CAS.
- F. Masood, Mater. Sci. Eng., C, 2016, 60, 569–578 CrossRef CAS PubMed.
- N. Sharma, P. Madan and S. Lin, Asian J. Pharm. Sci., 2016, 11, 404–416 CrossRef.
- N. Bertrand, P. Grenier, M. Mahmoudi, E. M. Lima, E. A. Appel, F. Dormont, J.-M. Lim, R. Karnik, R. Langer and O. C. Farokhzad, Nat. Commun., 2017, 8, 777 CrossRef PubMed.
- T. S. Sileika, D. G. Barrett, R. Zhang, K. H. A. Lau and P. B. Messersmith, Angew. Chem., Int. Ed., 2013, 52, 10766–10770 CrossRef CAS PubMed.
- J. Yoo and Y.-Y. Won, ACS Biomater. Sci. Eng., 2020, 6, 6053–6062 CrossRef CAS PubMed.
- H. Zhang and R. Tsao, Curr. Opin. Food Sci., 2016, 8, 33–42 CrossRef.
- C. A. Feghali and T. M. Wright, Front. Biosci., 1997, 2, d12–d26 CrossRef CAS PubMed.
- A. A. M. Abd-Allah, N. El-Deen, W. A. M. Mohamed and F. M. Naguib, Iran. J. Basic Med. Sci., 2018, 21, 97–107 Search PubMed.
- D. Beconcini, F. Felice, Y. Zambito, A. Fabiano, A. M. Piras, M. H. Macedo, B. Sarmento and R. Di Stefano, Pharmaceutics, 2019, 11, 500 CrossRef CAS PubMed.
- G. M. Pontes-Quero, L. Benito-Garzón, J. Pérez Cano, M. R. Aguilar and B. Vázquez-Lasa, Pharmaceutics, 2021, 13, 290 CrossRef CAS PubMed.
- S. Sunoqrot and L. Abujamous, J. Drug Delivery Sci. Technol., 2019, 52, 670–676 CrossRef CAS.
- S. Sunoqrot, M. Alfaraj, A. a. M. Hammad, V. Kasabri, D. Shalabi, A. A. Deeb, L. Hasan Ibrahim, K. Shnewer and I. Yousef, Pharmaceutics, 2020, 12, 811 CrossRef CAS PubMed.
- B. Yang, Y. Dong, F. Wang and Y. Zhang, Molecules, 2020, 25, 4613 CrossRef CAS PubMed.
- S. Derman, D. Uzunoglu, T. Acar, T. Ozbek Arasoglu, S. Ucak, V. Ozalp and B. Mansuroglu, Iran. J. Pharm. Res., 2020, 19, 424–435 CAS.
- A. Umerska, C. Gaucher, F. Oyarzun-Ampuero, I. Fries-Raeth, F. Colin, M. G. Villamizar-Sarmiento, P. Maincent and A. Sapin-Minet, Antioxidants, 2018, 7, 46 CrossRef PubMed.
- W.-m. Feng, H.-h. Guo, T. Xue, X. Wang, C.-w. Tang, B. Ying, H. Gong and G. Cui, RSC Adv., 2015, 5, 83628–83635 RSC.
- J. Zhang, L. Chao, X. Liu, Y. Shi, C. Zhang, L. Kong and R. Li, Exp. Lung Res., 2017, 43, 359–369 CrossRef CAS PubMed.
- Z. Zhao, A. Ukidve, V. Krishnan and S. Mitragotri, Adv. Drug Delivery Rev., 2019, 143, 3–21 CrossRef CAS PubMed.
- N. Kamaly, B. Yameen, J. Wu and O. C. Farokhzad, Chem. Rev., 2016, 116, 2602–2663 CrossRef CAS PubMed.
- N. M. Khalil, T. C. F. d. Nascimento, D. M. Casa, L. F. Dalmolin, A. C. d. Mattos, I. Hoss, M. A. Romano and R. M. Mainardes, Colloids Surf., B, 2013, 101, 353–360 CrossRef CAS PubMed.
- F. Y. Siu, S. Ye, H. Lin and S. Li, Int. J. Nanomed., 2018, 13, 4133–4144 CrossRef CAS PubMed.
- J. Pinto, M. Ahmad and B. R. Guru, J. Biomater. Sci., Polym. Ed., 2019, 30, 1188–1211 CrossRef CAS PubMed.
- K. R. Patil, U. B. Mahajan, B. S. Unger, S. N. Goyal, S. Belemkar, S. J. Surana, S. Ojha and C. R. Patil, Int. J. Mol. Sci., 2019, 20, 4367 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Spectral characterization of rhoifolin, survey and high-resolution XPS spectra, cell viability assays. See DOI: https://doi.org/10.1039/d2bm00309k |
| This journal is © The Royal Society of Chemistry 2022 |