 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In vivo synthesized 34S enriched amino acid standards for species specific isotope dilution of proteins
Gerrit
Hermann
a,
Laura Hyrup
Møller
c,
Bente
Gammelgaard
c,
Jonas
Hohlweg
d,
Diethard
Mattanovich
d,
Stephan
Hann
b and
Gunda
Koellensperger
*a
aFaculty of Chemistry, Department of Analytical Chemistry, University of Vienna, Währingerstraße 38, 1090 Vienna, Austria. E-mail: gunda.koellensperger@univie.ac.at
bDepartment of Chemistry, University of Natural Resources and Life Sciences, Muthgasse 18, 1190 Vienna, Austria
cDepartment of Pharmacy, Analytical Biosciences, University of Copenhagen, Universitetsparken 2, DK-2100 Copenhagen, Denmark
dDepartment of Biotechnology, University of Natural Resources and Life Sciences, Muthgasse 18, 1190 Vienna, Austria
First published on 24th February 2016
Abstract
A generic quantification approach was introduced addressing the characterization of protein standards while fulfilling the principles of metrology. Traceable absolute quantification was achieved combining a proven biochemical method, i.e. protein hydrolysis followed by amino acid quantification with the concept of species specific isotope dilution analysis (IDA). The method relies on the determination of the two sulfur containing amino acids, cysteine and methionine by sulfur speciation analysis and is hence applicable to any protein containing sulfur. In vivo synthesis using 34S as sulfur source in yeast fermentations provided species specific isotopically enriched standards for IDA quantification of cysteine and methionine in the oxidized forms, methionine sulfone and cysteic acid. Reverse isotope dilution mass spectrometry (IDMS) characterization by inductively coupled plasma mass spectrometry (ICP-MS) combined to anion exchange showed that very high concentrated spike material could be produced with μmol amounts of proteinogenic sulfur containing amino acids per g cell dry weight. An enrichment of 34S to 96.3 ± 0.4% (n = 3) and 98.5 ± 0.4% (n = 3) for cysteic acid and methionine sulfone, respectively, was assessed. The established IDA method was validated for the absolute quantification of commercially available lysozyme and ceruloplasmin standards including the calculation of a total combined uncertainty budget.
Introduction
The development of reference methods and standards of high analytical order is a topical research theme in biomedical analysis of proteins representing a significant fraction of clinically addressed biomarkers.1,2 In the case of clinical analysis it is especially important to accurately quantify absolute concentrations of proteins. Indeed several directives, such as e.g. the directive 98/79/EC (ref. 3) on in vitro diagnostic medical devices clearly states that “the traceability of values assigned to calibrators and/or control materials must be assured through available reference measurement procedures and/or available reference materials of a higher order”. Furthermore, the standard EN ISO 17511:2003 (ref. 4) demands reference measurement systems including reference measurement procedures for the determination of analytes in samples of human origin. It is particularly important for medical laboratory measurements concerned with both patient care and health screening to give adequately comparable, reproducible and accurate results (ISO 15193:2009).In light of these directives, it would be highly desirable to establish methods ensuring measurement traceability, since it is required to standardize results irrespective of the measurement procedure and laboratory. As a matter of fact, up to date the concentration of most commercially available proteins and even protein standards is not assessed by methods providing traceability of values assigned to calibrants, following the principles of metrology. E.g. in most cases the purity of protein solutions is reported as SDS page- or as HPLC-purity. The given concentration is typically obtained by Bradford assays5 which are calibrated against a species unspecific calibrant, i.e. in most cases albumin. As a consequence, the traceability of many proteins is maintained by reference to the first standard produced, which may no longer exist, with values assigned by consensus. This reliance on the original standard preparations causes severe problems.
In this work, we propose a traceable quantification method based on elemental speciation analysis, which could be generalized to a wide range of protein standards. The validity of elemental analysis serving as a reference method has been recognized by metrological institutions and was shown by several recent publications.2 For example transferrin was accurately quantified in a serum.6 Our work focused on sulfur speciation analysis as a generic absolute protein quantification method. The potential of elemental analysis addressing hetero-elements in proteins has been comprehensively reviewed.7 The most evident direct targeting approach would involve quantification of intact proteins by sulfur using size exclusion chromatography and ICP-MS detection.8,9 As a prerequisite, the amino acid sequence and hence the number of sulfur containing amino acids has to be known. Size exclusion chromatography is perfectly suited to separate inorganic and low molecular weight organic impurities from proteins. However, as a major drawback the size exclusion chromatography shows column recoveries depending on the protein, the column life time and the separation conditions. Accordingly, species unspecific quantification has to be corrected for column recovery and this column recovery – since it might be changing along the experiments has to be monitored.
Due to these limitations we turned to a proven biochemical method as an alternative strategy, which is protein quantification by analyzing the amino acid content of protein samples after complete decomposition of the proteins in its constituents, the amino acids. Amino acid analysis techniques formed an integral part in almost all biochemical structure determinations in earlier days. At that time, dedicated sample preparation methods addressing the recovery and extraction efficiency of individual amino acids were developed. For the two highly reactive sulfur compounds, methionine and cysteine, a sample preparation including oxidative protection prior to hydrolysis proved to be suitable among other methods. In a recent publication10 we implemented this approach, validating the procedure by external calibration using cysteic acid and methionine sulfone standards and determination by LC-ICP-MS. Applying a weak anion exchange chromatographic separation excellent column recoveries of 95 ± 2% for cysteic acid and 105 ± 10% for methionine sulfone standards were found. The limits of detection obtained by reaction cell technology ICP-MS were at 1 μM level for the two investigated sulfur containing compounds. The analytical technique was robust, since LC-ICP-MS showed a long term repeatability of 1.9% for methionine sulfone and 3.5% for cysteic acid.7
In this work we combine the proven biochemical method of amino acid based quantification for the first time with the concept of isotope dilution using sulfur speciation analysis. The objective was to introduce a traceable accurate quantification strategy for any protein standard containing sulfur amino acids. Moreover, we apply for the first time an in vivo synthesized 34S enriched yeast hydrolysate as internal standard for isotope dilution based quantification of proteins.
Experimental
Reagents and materials
The standard for lysozyme (from chicken egg white) with a purity >90% and the amino acid calibrants, namely methionine sulfone and cysteic acid, the reagents phenol purified by redistillation, ≥99%, and ammonium formate ≥ 99.995% trace metals basis were all purchased from Sigma Aldrich (Steinheim, Germany). Ceruloplasmin (Human Plasma) was purchased from Athens Research and Technology (Athens, USA). Formic acid 98–100% (Suprapur®), fuming hydrochloric acid 37% for analysis (EMSURE®), sulfur ICP standard traceable to SRM from NIST H2SO4 in H2O 1000 mg L−1 S CertiPUR® and hydrogen peroxide solution 31% Ultrapur were purchased from Merck (Darmstadt, Germany). Acetonitrile (LC-MS grade) was obtained from Fluka (Buchs, Switzerland). The ammonium acetate buffers applied as eluent for chromatography were derived from ammonium hydroxide solution ≥ 25% in H2O (TraceSELECT®, for trace analysis), (Sigma Aldrich, Steinheim, Germany) and ammonium acetate (Fractopur®), Merck, Darmstadt, Germany.All other reagents used throughout the study were of ultra-pure grade. Ultrapure HNO3 was prepared by double sub-boiling distillation of 65% nitric acid of p.a. grade (Merck) using a duoPUR quartz sub-boiling unit (MLS Lab Systems GmbH, Leutkirch, Germany). Ultra-pure water was used for the preparation of standards, aqueous buffers samples and model solutions by sub-boiling distillation of purified water (18.2 MU cm) obtained from an ultra clear system (SG water GmbH, Barsbüttel, Germany). The used hydrochloric acid was also subject to sub-boiling distillation in order to prevent sulfur contaminations. For calibration of amino acid analysis, the NIST standard reference material, SRM 2389a, amino acids in 0.1 mol L−1 hydrochloric acid, was used.
Cooled centrifugation (4 °C) was performed with a Mikro 200 R (Hettich Zentrifugen; 2424B rotor). For evaporation of the samples a Genevac EZ-2 Series Personal Evaporator was used. For incubation of the samples at 100 °C for 24 h a Heraeus T5042K oven was employed.
Oxidation of amino acids and subsequent hydrolysis of proteins
Oxidative treatment and protein hydrolysis is described elsewhere.10 In short, protein samples were oxidized by performic acid prior to hydrolysis. The performic acid derivatisation reagent was prepared by mixing 36 mL of formic acid, 4 mL of H2O2 and 200 μL of phenol and incubation for 30 min at room temperature until the typical yellow colour of performic acid was reached. Then, 500 μL of performic acid for protein standards and yeast samples were added either to 10 mg of protein or 50 mg of yeast sample. The samples were mixed until they were fully covered with acid, then transferred to a water bath and incubated at 65 °C for 15 min. During this time, a homogenization step by mixing after 7 min was applied. Afterwards, the samples were cooled on ice for 5 min and evaporated to dryness. Finally, 1 mL of 6 M HCl was added and the samples were hydrolysed at 100 °C for 24 h.Separation and detection of oxidized amino acids by SAX-ICP-MS
The measurement of the sulfur isotopes by elemental speciation analysis was carried out on two different ICP-MS systems, namely, the Perkin Elmer Elan DRC II combined with a Perkin Elmer Series 200 HPLC system and the ICP-QQQMS in MS/MS mode (Agilent Technologies, 8800) combined with an Agilent 1200 HPLC system. For both systems oxygen was applied as reaction gas. The chromatographic separation was based on a PRP X100 strong anion exchange (SAX) column (250 mm × 2.1 mm, 5 μm, Hamilton) applying conditions described elsewhere.10 This separation is employs ammonium acetate, pH 8 gradient elution with a flow rate of 0.4 mL min−1 at a temperature of 20 °C. The injection volume was set to 8 μL.Synthetic medium for yeast cultivation
The synthetic cultivation medium for preculture and cultivation in shake flasks contained per liter: 22 g glucose, 22 g citric acid, 3.93 g (NH4)2HPO4, 0.16 g MgCl2·7H2O, 0.68 g KCl, 0.027 g CaCl2·2H2O, 2 mL biotin stock solution (0.2 g L−1), 0.12 g Na2SO4 and 4.6 mL PTM2 trace salt stock solution. The pH was set to 5.0 with KOH. The trace salt stock solution PTM2 contained (per liter): 63.3 g FeCl2·6H2O, 20.0 g ZnCl2, 5.77 g CuCl2 2H2O, 3.94 g MnCl2·4H2O, 0.82 g CoCl2·6H2O, 0.2 g Na2MoO4·2H2O, 0.08 g NaI, 0.02 g H3BO3 and 5 mL HCl (25%).For the labeling approach, Na2SO4 in cultivation medium was replaced by enriched 34S sodium sulfate (isotopic distribution: <0.1% 32S, 1.1% 33S, 98.8% 34S and <0.05% 36S) from Isoflex USA.
Shake flask cultivations
Precultures (100 mL) with “S-natural abundance” medium were inoculated with Pichia pastoris CBS7435 wildtype strain and cultivated over night (25 °C, 180 rpm). Main cultures on “S-natural abundance” medium and medium for 34S labelling (100 mL) were inoculated with washed precultures for a starting optical density (OD) (600 nm) of 25 and then cultivated 24 h (25 °C, 180 rpm). Finally, samples were taken for yeast dry mass (YDM) determination and cells were harvested, washed twice with reverse osmosis (RO) water (2500 relative centrifugal force (RCF); 20 °C) and stored at −20 °C until analysis.Sample preparation and spiking strategy
All sample preparation steps were performed gravimetrically. The spike material, i.e. the hydrolyzed 34S yeast was prepared out of 1 g dry cell material. The material was split in 20 aliquots and subsequently subject to the oxidation and hydrolysis procedure and finally pooled. For the IDMS quantification (including IDMS and reverse IDMS), the cysteic acid and methionine sulfone standards as well as the protein samples were spiked with the hydrolysates of the 34S enriched yeast prior to the oxidation and subsequent hydrolysis procedure. The spike levels were based on preliminary screening experiments.Separation of amino acids by HILIC and detection via ESI MS/MS
The separation procedure was carried out on a LC system consisting of a Thermo Scientific CTC PAL autosampler and a Thermo Scientific Accela 1250 pump. For HILIC separation of the amino acids, a Nucleodur® silica-based column (100 mm × 2.0 mm, 1.8 μm particle size) equipped with a guard column (20 mm × 2.0 mm, 1.8 μm particle size), both purchased from Macherey-Nagel (Düren, Germany), were used applying a gradient of 10 mM ammonium formate, pH 3, 25, (0–100%). The column temperature was set to 40 °C. The injection volume was 5 μl. Tandem mass spectrometric analysis was carried out on a Thermo Scientific TSQ Vantage MS/MS. The capillary temperature was 370 °C and the vaporizer temperature was 400 °C. The collision gas pressure was 1.5 mTorr and the spray voltage was set to 3300 V. The elution times and SRM transitions is published elsewhere.11 IDMS quantification of methionine sulfone and cysteic acid was based on the principle by Fasset and Paulsen.12Calculation of uncertainty
The determination of combined uncertainties was based on the EURACHEM/CITAC guide “Quantifying Uncertainty in Analytical Measurement”.13Overview on experimental workflow (Fig. 1)
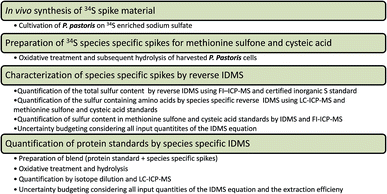 | ||
| Fig. 1 Experimental workflow of the introduced species specific IDMS method addressing protein quantification by the sulfur containing amino acids. | ||
Results and discussion
Sulfur speciation analysis
Protein quantification addressing the amino acids was considered a mature technique already decades ago. The combination with isotope dilution mass spectrometry however, was only rarely considered. Purpose of this study is the integration of an elemental speciation concept to the field of traceable absolute protein quantification, namely quantification by species specific isotope dilution via the sulfur containing amino acids. As described elsewhere,10 oxidative treatment with performic acid secured a uniform oxidation of all the sulfur containing amino acids to methionine sulfone and cysteic acid, respectively during protein hydrolysis. A proof of principle study showed that methionine could be recovered from a lysozyme standard with 89.4 ± 10%, while cysteine showed a recovery of 99.3 ± 3%.10 These recovery figures were assessed by external calibration using methionine sulfone and cysteic acid standards. Strong anion exchange chromatography enabled separation of the amino acids, methionine, cysteine, the corresponding oxidized forms methionine sulfone, cysteic acid and sulfate. Separation of underivatized and derivatized amino acids was essential to optimize the oxidative protection during protein hydrolysis. In the following chromatography of hydrolyzed samples, no traces of native methionine or cysteine could be detected.In the present study, sulfur speciation analysis was performed by ICP-MS detection using oxygen as reaction gas (hence measuring S as SO). Reaction cell technology was applied in single quadrupole as well as triple quadrupole ICP-MS configuration. It has to be mentioned that the major goal was not ultimate sensitivity; accordingly no effort was put into finding the ideal LC flow regime of the LC-ICP-MS hyphenation in this regard. Both instruments revealed absolute limits of detection in the low pmol to sub-pmol range for sulfur when combined with the implemented ion exchange chromatography. The QQQ-MS approach was superior regarding detection limit by a factor of 3–4 when 32S16O was considered, and superior by at least a factor of 30 for 34S16O measurement. As a consequence, superior isotope ratio measurement precision was achieved measuring the 34S/32S ratio in methionine sulfone and cysteic acid, respectively by LC-ICP-QQQ-MS. As can be readily observed in Table 1, the ratios assessed by LC-ICP-QQQ-MS analysis of 60 μM methionine sulfone and cysteic acid standards revealed an excellent standard uncertainty of 0.6% (n = 4). Consequently, only this approach was further considered.
| LC-ICP-DRCMS | LC-ICP-MS/MS | |||
|---|---|---|---|---|
| LOD (3 s), μmol L−1 | 32S/34S | LOD (3 s), μmol L−1 | 32S/34S | |
| Standard uncertainty/% (n = 4)a | Standard uncertainty/% (n = 4)a | |||
| a Individually prepared standards. | ||||
| Met. Sulf. 32S | 0.45 | 0.14 | ||
| Cys. Ac. 32S | 0.40 | 0.25 | ||
| Met. Sulf. 34S | 16 | 0.56 | ||
| Cys. Ac. 34S | 55 | 1.0 | ||
| Met. Sulf. | 2.8 | 0.6 | ||
| Cys. Ac. | 2.5 | 0.6 | ||
Characterization of in vivo synthesized spike material
In recent years, in vivo synthesized isotopically enriched standards has become an indispensable tool for metabolite profiling.14 These so called targeted metabolomics strategies involve the production of fully 13C labelled biomass utilizing different organisms. In our group a routine was established producing 13C fully labelled P. pastoris metabolome as internal standard.15 The use of such standards demonstrated to be essential in sample preparation development and for accurate absolute quantification of metabolites.16 In this work the concept of in vivo synthesis was transferred to 34S enrichment focusing on the use of the labelled proteome as spike material. As described in the experimental section 34S enriched P. pastoris was produced in cultivations with 34S enriched sulfate as sole sulfur source. The obtained biomass was prepared according to the previously established procedure10 yielding 34S enriched methionine sulfone and cysteic acid.In a first step the spike material, i.e. the hydrolyzed 34S yeast extract was characterized by reverse IDMS using methionine sulfone and cysteic acid standards. For this purpose the 34S yeast preparation was spiked with the oxidized forms of the two sulfur amino acids prior to analysis. LC-ICP-QQQ-MS measurements revealed isotopic abundances of 34S enriched up to 96.3 ± 0.4% (n = 3) and 98.5 ± 0.4% (n = 3) for cysteic acid and methionine sulfone, respectively. Table 2 gives the obtained concentration levels of the spike material. A total expanded uncertainty was assessed (1) traced to the concentration of methionine sulfone and cysteic acid standards as given by the certificate of analysis (based on titration versus NaOH and HClO4) and (2) considering their quantity as determined by their sulfur content (based on the certificate of an inorganic sulfur standard). The total sulfur determinations of the standards were carried out by reverse IDMS and flow injection analysis. Considering total sulfur, concentrations of 103 ± 3% and 105 ± 3% (weight%) for cysteic acid and methionine sulfone standards, respectively were assessed. The concentration obtained by titration was 99.0–101.0% for both calibrants. Accordingly, the calculated uncertainty of the spike quantification was higher in the case of tracing the standard concentration to the sulfur content. As can be readily seen in Table 2, the experimental repeatability (% RSD) of spike preparation was below the calculated total expanded uncertainties proving that the quantification procedure was under control.
| Concentration in yeast hydrolysate, nmol g−1 | Standard uncertainty, % (n = 3)a | Total expanded uncertainty, % (k = 2), traced to certificate of analysis | Total expanded uncertainty, % (k = 2) traced to sulfur content | ||
|---|---|---|---|---|---|
| a Individually prepared standards. | |||||
| Methionine sulfone | 1670 | 5 | 6 | 12 | |
| Cysteic acid | 1820 | 2 | 5 | 8 | |
The determination of the total sulfur content in the 34S yeast hydrolysate, again by reversed IDMS using single element sulfur standards and flow injection analysis revealed a sulfur concentration of 5.7 μmol g−1 (3% RSD; n = 3), which implied that the amount of sulfur recovered as proteinogenic amino acids was 62%, the rest corresponding to untransformed inorganic sulfate. The total sulfur concentration in the dry yeast material given as μmol sulfur per g cell dry weight was 81 μmol g−1.
Absolute quantification of model protein
Finally, a quantification exercise was carried out addressing accurate quantification of lysozyme preparations by species specific isotope dilution. For this purpose the 34S yeast extract was spiked to lysozyme prior to sample preparation. As can be observed in Table 3 the resulting protein concentrations (given as nmol protein per g protein sample intake (dry weight)) considering the two amino acids, respectively are in excellent agreement. The strategy resulted in experimental repeatability expressed as standard uncertainties for n = 3 individually prepared samples of ≤5%.| Quantified via | Concentration/nmol protein per g | Standard uncertainty/RSD% (n = 3) | Expanded uncertainty (k = 2)a/% | Concentration/weight% |
|---|---|---|---|---|
| a For the calculation of the total combined uncertainty a factor E was introduced to the isotope dilution equation accounting for the extraction efficiency which is not corrected for by the ID approach. This factor was assumed to be 1, associated with a standard uncertainty of 4%. | ||||
| Met. Sulf. | 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280 |
5 | 13 | 78 |
| Cys. Ac. | 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 820 820 |
2 | 14 | 77 |
The finally observed total combined uncertainty was 13 and 14% (coverage factor k = 2) considering the two amino acids. The budget was calculated assuming an uncertainty for the spike concentration of 5% and 2% for methionine sulfone and cysteic acid, respectively. Moreover, a factor E was introduced in the model equation otherwise given by the IDMS. The factor E, with the value of 1 accounted for the extraction efficiency, since this not corrected for by the ID approach. For the calculation, it was assumed that the extraction efficiency (and hence the factor E) was associated with an uncertainty of 4%. Both input quantities, the spike concentration and the extraction efficiency contributed to the highest degree to the calculated total expanded uncertainty. The uncertainty of the spike concentration determination was the highest contribution ranging at 40% and the sample preparation contributed with 30%.
For cross validation the protein standard was additionally investigated by hydrolysis followed by HILIC-MS/MS analysis addressing 6 amino acids, namely alanine, leucine, arginine, lysine, proline and serin. The HILIC-MS/MS approach is described in detail elsewhere.11 A lysozyme concentration of 77 ± 10% (weight%, n = 3 replicate samples) was found. Evidently there is no universal hydrolysis procedure applicable to all amino acids. Hence, implementing simply acid hydrolysis with only 6 M HCl as reagent resulted in a comparatively high experimental uncertainty of amino acid quantification. It should be noted that, internal standardization by 13C fully labelled yeast was carried out only prior to HILIC-MS/MS and not upon sample preparation. However, within the obtained uncertainties, the average concentrations of lysozyme obtained by the complementary approaches were in good agreement.
Finally, the enriched spiking material was used for IDMS quantification of ceruloplasmin. It appears from Table 4 that the biomarker was quantified with a relative standard uncertainty of 5%, disregarding which amino acid was chosen for quantification. The importance of traceable protein standards becomes clear considering the state of the art of the art of ceruloplasmin determination, a clinical relevant biomarker.2,17 In this specific case poor comparability of results resulting from different immunological measurement platforms have been reported.17 Accordingly, traceable protein standards for calibration would significantly improve this situation.
Conclusion
Amino acid quantification based on IDMS provided a valuable tool for protein or peptide quantification. However it failed as method in complex biological samples (due to limited selectivity and sensitivity). The method offers the introduction of traceable protein standards, which are of utmost importance in clinical analysis (and the implied requirements of reference methods of higher metrological order). As a matter of fact, the use of species specific isotope dilution poses a major advantage as there is no need to produce spike material for each investigated protein when amino acids are considered for quantification. In principle, any protein can be quantified provided it is present at sufficiently high concentration with a HPLC or SDS page purity >95% and contains at least one of the sulfur bearing amino acids. The value of such a generic approach for the production of metrological valid protein standards cannot be underestimated.Acknowledgements
This work was funded by the European Association of National Metrology Institutes (EURAMET, Project HLT05-REG4). Wirtschaftsagentur Wien and EQ BOKU VIBT GmbH is acknowledged for funding mass spectrometry instrumentation.Notes and references
- N. Greenberg, Clin. Chim. Acta, 2014, 432, 49–54 CrossRef CAS PubMed.
- C. Swart, Anal. Bioanal. Chem., 2013, 405, 5697–5723 CrossRef CAS PubMed.
- Directive 98/79/EC, EUR-Lex – 31998L0079-EN, In vitro diagnostic medical devices, 27 October 1998.
- ISO 17511:2003, In vitro diagnostic medical devices – Measurement of quantities in biological samples – Metrological traceability of values assigned to calibrators and control materials, ISO, Geneva, Switzerland, 2003 Search PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- C. Frank, O. Rienitz, R. Jährling, D. Schiel and S. Zakel, Metallomics, 2012, 4, 1239–1244 RSC.
- A. Sanz-Medel, M. Montes-Bayón, J. Bettmer, M. Luisa Fernández-Sanchez and J. Ruiz Encinar, TrAC, Trends Anal. Chem., 2012, 40, 52–63 CrossRef CAS.
- J. Giner Martínez-Sierra, F. Moreno Sanz, P. Herrero Espílez, R. Santamaria-Fernandez, J. M. Marchante Gayón and J. I. García Alonso, J. Anal. At. Spectrom., 2010, 25, 989–997 RSC.
- J. Giner Martínez-Sierra, O. Galilea San Blas, J. M. Marchante Gayón and J. I. García Alonso, Spectrochim. Acta, Part B, 2015, 108, 35–52 CrossRef.
- E. Rampler, T. Dalik, G. Stingeder, S. Hann and G. Koellensperger, J. Anal. At. Spectrom., 2012, 27, 1018–1023 RSC.
- R. Guerrasio, C. Haberhauer-Troyer, D. Mattanovich, G. Koellensperger and S. Hann, Anal. Bioanal. Chem., 2014, 406, 915–922 CrossRef CAS PubMed.
- J. D. Fassett and P. J. Paulsen, Anal. Chem., 1989, 61, 643A–649A CrossRef CAS.
- Eurachem/CITAC guide: Quantifying Uncertainty in Analytical Measurement, ed. S. L. R. Ellison and A. Williams, third edition, 2012 ISBN 978-0-948926-30-3, available from http://www.eurachem.org Search PubMed.
- S. Aljoscha Wahl, R. M. Seifar, A. Ten Pierick, C. Ras, J. C. van Dam, J. J. Heijnen and W. M. van Gulik, Quantitative metabolomics using ID-MS, 2014, vol. 1191 Search PubMed.
- S. Neubauer, C. Haberhauer-Troyer, K. Klavins, H. Russmayer, M. G. Steiger, B. Gasser, M. Sauer, D. Mattanovich, S. Hann and G. Koellensperger, J. Sep. Sci., 2012, 35, 3091–3105 CrossRef CAS PubMed.
- K. Klavins, S. Neubauer, A. Al Chalabi, D. Sonntag, C. Haberhauer-Troyer, H. Russmayer, M. Sauer, D. Mattanovich, S. Hann and G. Koellensperger, Anal. Bioanal. Chem., 2013, 405, 5159–5169 CrossRef CAS PubMed.
- I. Infusino, C. Valente, A. Dolci and M. Panteghini, Anal. Bioanal. Chem., 2010, 397, 521–525 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |
