Open Access Article
Open Access ArticleSynthesis of novel benzothiazole derivatives and investigation of their enzyme inhibitory effects against Alzheimer's disease†
Şevval Karacaab,
Derya Osmaniye bc,
Begum Nurpelin Sağlık
bc,
Begum Nurpelin Sağlık bc,
Serkan Leventbc,
Sinem Ilgın
bc,
Serkan Leventbc,
Sinem Ilgın d,
Yusuf Özkay
d,
Yusuf Özkay *bc,
Ahmet Çağrı Karaburunb,
Zafer Asım Kaplancıklı
*bc,
Ahmet Çağrı Karaburunb,
Zafer Asım Kaplancıklı b and
Nalan Gundogdu-Karaburunb
b and
Nalan Gundogdu-Karaburunb
aDepartment of Biochemistry, Faculty of Pharmacy, Anadolu University, 26470 Eskişehir, Turkey
bDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Anadolu University, 26470 Eskişehir, Turkey. E-mail: yozkay@anadolu.edu.tr
cCentral Analysis Laboratory, Faculty of Pharmacy, Anadolu University, 26470 Eskişehir, Turkey
dDepartment of Pharmaceutical Toxicology, Faculty of Pharmacy, Anadolu University, 26470 Eskişehir, Turkey
First published on 19th August 2022
Abstract
The use of dual acetylcholinesterase (AChE)–monoamine oxidase B (MAO-B) inhibitors is a new approach in the treatment of Alzheimer disease (AD). In this work, 14 new benzothiazoles (4a–4n) were designed and synthesized. In biological activity studies, the AChE, butyrylcholinesterase (BChE), MAO-A and MAO-B inhibitory potentials of all compounds were evaluated using the in vitro fluorometric method. Additionally, amyloid beta (Aβ)-aggregation inhibitory effects of active compounds were evaluated by means of an in vitro kit-based method. The biological evaluation showed that compounds 4a, 4d, 4f, 4h, 4k and 4m displayed significant activity against AChE and MAO-B enzymes. Compound 4f displayed inhibitory activity against AChE and MAO-B enzyme with IC50 values of 23.4 ± 1.1 nM and 40.3 ± 1.7 nM, respectively. It has been revealed that compound 4f may have the potential to inhibit AChE and MAO-B enzymes, as well as the ability to prevent the formation of beta amyloid plaques accumulated in the brains of patients suffering from AD. In silico studies also support the obtained biological activity findings. Compound 4f provided strong interactions with the active site of both enzymes. In particular, the interaction of compound 4f with flavin adenine dinucleotide (FAD) in the MAO-B enzyme active site is a promising and exciting finding.
1. Introduction
Alzheimer's disease (AD), a devastating neurodegenerative disease, causes deficits in memory, thought, and behavior.1 Many hypotheses have been proposed for the treatment of AD, including inhibition of cholinesterase, inhibition of amyloid protein (Aβ) formation, and inhibition of tau protein accumulation.2 Cholinergic signaling is known to play a critical role in cognitive performance and cholinergic signaling has been observed to gradually disappear in AD. For this purpose, a treatment line has been established to treat symptoms using cholinesterase inhibitors.3 Several FDA-approved anti-AD drugs currently available clinically (such as cholinesterase inhibitors: donepezil, rivastigmine, galantamine) provide only partial relief of symptoms and cannot stop or reverse disease progression.4 Donepezil works in Alzheimer's disease specifically as an acetylcholinesterase (AChE) inhibitor at mild to moderate levels. However, donepezil has side effects such as nausea, diarrhea, weakness, dizziness, urinary incontinence, and insomnia. Donepezil may also cause gastrointestinal and neurological adverse reactions such as speech difficulties, involuntary tremors, and vomiting.5–9 Therefore, there is a need to develop new molecules in this field.The enzymatic cavity of AChE is characterized by a nearly 20 Å deep narrow gorge, which is composed of two binding sites: the catalytic active site (CAS) at the bottom and the peripheral anionic site (PAS) near the entrance of the gorge.10
Plaques, or more likely their soluble oligomers, of Aβ peptides, whose aggregation and accumulation in the brain play a key role in the initiation and progression of AD, are neurotoxic. The discovery of the ability of PAS of AChE to induce β-amyloid aggregation has triggered an intense search for compounds that exhibit dual activity as inhibitors of AChE activity and Aβ aggregation.11
It has been proven that the level of monoamine oxidase-B (MAO-B) is increased in Alzheimer's plaques and blood platelets of AD and Parkinson (PD) patients. This increase in MAO-B is most likely due to the following; it is due to the transcriptional upregulation of MAO-B in the protein and its predominance in plaque-associated astrocytes in pathologically confirmed AD brains.12
Compounds that co-inhibit the three formations mentioned above (AChE, Aβ plaque, MAO-B) will contribute significantly to this field. Recently, multifactorial agents have come to the fore. Because considering the age of the patient group, the use of a single agent that inhibits all of them instead of separate agents will also facilitate. It has been reported that benzothiazole ring provides all three of these effects.13–15 Thus, it was chosen as a pharmacophore in the main structure of our compounds.
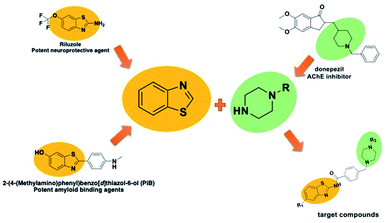
In addition to the above-mentioned properties of the benzothiazole ring, it also has use as a neuroprotective and amyloid-inhibiting agent.16,17 In addition, it is known that the secondary amine derivatives (piperazine or piperidine in the structure of donepezil) is necessary for interaction with the CAS region of the enzyme.3 Also, the presence of these groups in the chemical structure provide the flexibility and thus contribute to binding properly to the gorge of the enzyme active site. Within the scope of this study, the compounds were designed to be similar in length to donepezil and to localize to the enzyme active site. In the light of this information, both benzothiazole and piperazine (as a secondary amine derivatives) were used in the structure of the target compounds (Fig. 1).
2. Result and discussion
2.1. Chemistry
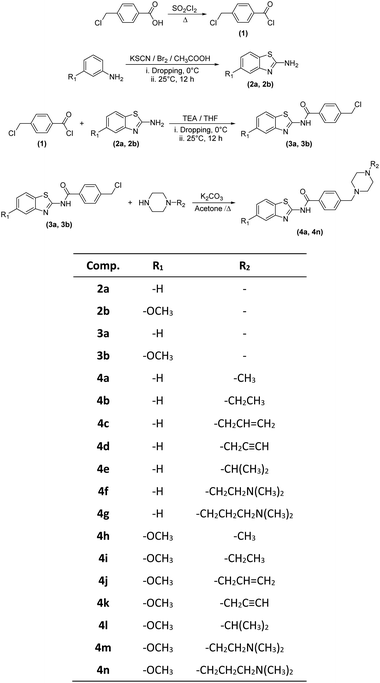
In the scope of the present study, it was aimed to synthesize new compounds containing benzothiazole and piperazine groups in the same chemical structure. For this purpose, 14 new compounds were synthesized. Target compounds were obtained using a four-step reaction method. The synthesis pathway of the compounds is presented in Scheme 1. Initially, 4-(chloromethyl) benzoyl chloride (1) was synthesized using thionyl chloride. Secondly, 2-aminobenzothiazoles (2a–2b) derivatives were synthesized by means of ring closure reaction using brome solution. Thirdly, 4 (chloromethyl)-N-(5,6-substituted benzo[d]thiazol-2-yl) benzamides (3a–3b) were obtained by means of reaction between 4-(chloromethyl) benzoyl chloride (1) and 2-aminobenzothiazoles (2a–2b). In the last step, compounds 3a, 3b and appropriate piperazine derivatives were reacted to synthesize target compounds (4a–4n). The structures of the obtained compounds were proved by 1H-NMR, 13C-NMR and HRMS spectroscopic methods (ESI†).When the chemical structures of the synthesized compounds were examined, the specific stretching band of the carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) was recorded between 1660–1681 cm−1. The C
O) was recorded between 1660–1681 cm−1. The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching band on this group was obtained in the range of 1442–1473 cm−1. All the synthesis products (4a–4n) contain a 1,4-disubstituted benzene ring in their structure. The specific out-of-plane deformation band of this ring was observed in the range of 812–871 cm−1.
C stretching band on this group was obtained in the range of 1442–1473 cm−1. All the synthesis products (4a–4n) contain a 1,4-disubstituted benzene ring in their structure. The specific out-of-plane deformation band of this ring was observed in the range of 812–871 cm−1.
Aliphatic and aromatic protons in the synthesized compounds gave peaks in the 1H NMR spectra as expected. In the spectra of the obtained compounds, the total hydrogen number was determined, and the expected number of peaks was observed. In the synthesized compounds, there are two main substructures as non-substituted benzothiazole and 5-methoxy benzothiazole. When the 1H NMR results of the derivatives (4a–4g) containing the non-substituted benzothiazole structure are examined, the striking details are as follows.
Protons belonging to the benzothiazole substructure in compounds 4a–4g are a hydrogen triplet (δH 7.31–7.33 ppm), a hydrogen multiple (δH 7.42–7.46 ppm), and two hydrogen doublets (δH 7.76–7.78; 7.99–8.01 ppm). Protons belonging to the 1,4-disubstituted benzene ring were observed as an AB system [δH 7.40–7.48 (2H); 7.96–8.11 ppm (2H)].
Protons belonging to the 5-methoxybenzothiazole substructure in compounds 4h–4n are a hydrogen double doublet (δH 6.76–7.07 ppm), a hydrogen doublet (δH 6.99–7.62 ppm) and a hydrogen doublet (δH 7.29–7.82 ppm). Protons belonging to the 1,4-disubstitutedbenzene ring were observed as an AB system [δH 7.36–7.47 (2H); 7.95–8.10 ppm (2H)].
In the synthesized compounds, the common structural particles gave peaks in the 13C-NMR spectra as expected in general. In the spectra of the compounds, carbons belonging to the carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) gave a peak in range of 165.98–166.59 ppm and agreed with the literature data.18 The other aliphatic carbons were observed in the range of 11.93–79.86 ppm, while aromatic carbons were observed in the range of 102.78–166.59 ppm.
O) gave a peak in range of 165.98–166.59 ppm and agreed with the literature data.18 The other aliphatic carbons were observed in the range of 11.93–79.86 ppm, while aromatic carbons were observed in the range of 102.78–166.59 ppm.
2.2. Cholinesterase enzymes inhibition assay
The inhibitory activities of all obtained derivatives (4a–4n) against cholinesterase enzymes were evaluated using the previously described in vitro modified Ellman's spectrophotometric method.19–27 Activity tests were carried out in two steps. Enzyme inhibition tests were performed by preparing all compounds (4a–4n) and reference agents (donepezil and tacrine) at 10−3 and 10−4 M concentrations. The IC50 values of the selected compounds and standard agents were determined by non-linear regression analysis over the calculated % inhibition values at the concentrations between 10−3–10−9 M (by serial dilutions) with the help of GraphPad Prism Version 6 software. Enzyme inhibition results, IC50 values of test compounds and reference drugs are presented in Table 1 (the IC50 curves are presented in ESI, Fig. S53†).| Comp. | AChE % inhibition | AChE IC50 (nM) | BChE % inhibition | BChE IC50 (nM) | ||
|---|---|---|---|---|---|---|
| 10−3 M | 10−4 M | 10−3 M | 10−4 M | |||
| 4a | 93.3 ± 1.4 | 89.1 ± 1.1 | 56.3 ± 2.5 | 38.6 ± 0.7 | 26.5 ± 0.8 | >106 |
| 4b | 85.6 ± 1.3 | 47.6 ± 0.8 | >105 | 35.7 ± 0.9 | 24.4 ± 0.6 | >106 |
| 4c | 82.8 ± 1.5 | 48.2 ± 0.9 | >105 | 30.9 ± 0.7 | 20.2 ± 0.7 | >106 |
| 4d | 91.6 ± 1.3 | 86.5 ± 1.1 | 89.6 ± 3.2 | 37.1 ± 0.8 | 28.4 ± 0.9 | >106 |
| 4e | 83.9 ± 1.7 | 46.9 ± 0.8 | >105 | 39.1 ± 0.8 | 26.8 ± 0.7 | >106 |
| 4f | 96.4 ± 1.4 | 93.0 ± 1.3 | 23.4 ± 1.1 | 45.3 ± 0.9 | 32.8 ± 0.6 | >106 |
| 4g | 94.3 ± 1.0 | 90.6 ± 1.3 | 36.7 ± 1.4 | 41.5 ± 0.8 | 30.9 ± 0.8 | >106 |
| 4h | 92.3 ± 1.1 | 87.4 ± 1.3 | 64.9 ± 2.9 | 33.5 ± 0.8 | 27.0 ± 0.7 | >106 |
| 4i | 86.9 ± 1.2 | 48.0 ± 0.9 | >105 | 39.8 ± 0.9 | 25.2 ± 0.9 | >106 |
| 4j | 82.5 ± 1.5 | 47.6 ± 0.8 | >105 | 32.9 ± 0.8 | 22.6 ± 0.8 | >106 |
| 4k | 90.3 ± 1.5 | 84.7 ± 1.3 | 102.5 ± 4.8 | 30.4 ± 0.7 | 26.4 ± 0.7 | >106 |
| 4l | 81.7 ± 1.3 | 48.1 ± 0.9 | >105 | 38.1 ± 0.9 | 24.3 ± 0.7 | >106 |
| 4m | 95.3 ± 1.2 | 91.8 ± 1.3 | 27.8 ± 1.0 | 42.4 ± 0.8 | 31.9 ± 0.8 | >106 |
| 4n | 95.3 ± 1.7 | 90.0 ± 1.4 | 42.1 ± 1.8 | 40.9 ± 0.9 | 30.4 ± 0.9 | >106 |
| Donepezil | 99.1 ± 1.3 | 97.4 ± 1.3 | 20.1 ± 1.4 | — | — | — |
| Tacrin | — | — | — | 99.8 ± 1.4 | 98.7 ± 1.4 | 6.4 ± 0.2 |
In the enzyme inhibition test, it was observed that all compounds were more effective against AChE at 10−3 M concentrations. Also, compounds carrying dimethylaminoethyl and dimethylaminopropyl side chains showed inhibitory activity against AChE comparable to donepezil. Among the compounds, 4a, 4d, 4f, 4g, 4h, 4k, 4m and 4n showed more than 50% inhibition at 10−4 M concentration. The IC50 values of compounds 4a, 4d, 4f, 4g, 4h, 4k, 4m and 4n were calculated as 56.3 ± 2.5 nM, 89.6 ± 3.2 nM, 23.4 ± 1.1 nM, 36.7 ± 1.4 nM, 64.9 ± 2.9 nM, 102.5 ± 4.8 nM, 27.8 ± 1.0 nM, and 42.1 ± 1.8 nM, respectively. These compounds came to the fore because they have very close IC50 values to that of donepezil. The IC50 value of donepezil was found to be 20.1 ± 1.4 nM. Among these derivatives, the 4f was found to be the most active derivative in the series with an IC50 value of 23.4 ± 1.1 nM.
When the BChE inhibition test results were examined, it was observed that the reference compound tacrin had an inhibition activity of 99.8 ± 1.4% and 98.7 ± 1.4% at 10−3 and 10−4 M concentrations, respectively. However, none of the compounds showed significant inhibitory activity against BChE (Table 1).
2.3. MAO enzymes inhibition assay
The inhibition effect of synthesized compounds on MAO isoenzymes was evaluated by using the in vitro fluorometric method described previously by our research group.28–34 The IC50 values of the selected compounds and standard agents were determined by non-linear regression analysis over the calculated % inhibition values at the concentrations between 10−3–10−9 M (by serial dilutions) with the help of GraphPad Prism Version 6 software. The results are presented in Table 2 and ESI, Fig. S54.† In MAO-A inhibition test, the reference agent moclobemide showed an inhibition of 94.1 ± 2.8% at 10−3 M concentration. However, none of the synthesized compounds (4a–4n) showed greater than 50% inhibition in this concentration.| Comp | MAO-A % inhibition | MAO-A IC50 (nM) | MAO-B % inhibition | MAO-B IC50 (nM) | ||
|---|---|---|---|---|---|---|
| 10−3 M | 10−4 M | 10−3 M | 10−4 M | |||
| 4a | 55.3 ± 0.9 | 38.6 ± 0.7 | >105 | 91.0 ± 1.1 | 88.7 ± 1.3 | 67.4 ± 3.1 |
| 4b | 46.5 ± 0.8 | 36.4 ± 0.8 | >106 | 73.3±1.2 | 42.3 ± 0.8 | > 105 |
| 4c | 48.3 ± 0.9 | 34.6 ± 0.8 | >106 | 69.6 ± 1.2 | 40.3 ± 0.9 | > 105 |
| 4d | 57.6 ± 0.9 | 39.7 ± 0.9 | >105 | 89.5 ± 1.2 | 84.0 ± 1.1 | 109.7 ± 4.3 |
| 4e | 44.7 ± 0.9 | 33.8 ± 0.7 | >106 | 72.7 ± 1.1 | 44.6 ± 0.8 | >105 |
| 4f | 67.9 ± 1.1 | 41.3 ± 0.9 | >105 | 95.3 ± 1.3 | 91.3 ± 1.1 | 40.3 ± 1.7 |
| 4g | 64.9 ± 1.1 | 40.1 ± 0.9 | >105 | 74.2 ± 1.0 | 42.5 ± 0.9 | >105 |
| 4h | 60.5 ± 0.9 | 38.5 ± 0.6 | >105 | 90.6 ± 1.4 | 85.7 ± 1.4 | 85.1 ± 3.8 |
| 4i | 43.8 ± 0.9 | 30.6 ± 0.7 | >106 | 75.6 ± 1.2 | 41.7 ± 0.9 | >105 |
| 4j | 45.2 ± 0.9 | 32.9 ± 0.7 | >106 | 74.5 ± 1.1 | 43.5 ± 0.8 | >105 |
| 4k | 58.6 ± 0.9 | 39.9 ± 0.8 | >105 | 88.4 ± 1.5 | 85.9 ± 1.1 | 124.3 ± 5.8 |
| 4l | 40.6 ± 0.9 | 34.0 ± 0.9 | >106 | 77.9 ± 1.4 | 40.7 ± 0.9 | >105 |
| 4m | 61.3 ± 0.9 | 37.9 ± 0.8 | >105 | 93.3 ± 1.3 | 90.9 ± 1.5 | 56.7 ± 2.2 |
| 4n | 60.1 ± 0.8 | 38.8 ± 0.7 | >105 | 70.3 ± 1.3 | 41.0 ± 0.9 | >105 |
| Moc. | 94.1 ± 2.8 | 82.1 ± 2.7 | 6061.3 ± 262.5 | — | — | — |
| Selegilin | — | — | — | 98.6 ± 2.1 | 94.9 ± 1.1 | 37.4 ± 1.6 |
In the MAO-B inhibition test, compounds 4a, 4d, 4f, 4h, 4k and 4m showed more than 50% inhibition at 10−3 and 10−4 M concentrations. Compounds were observed to be more effective on MAO-B compared to MAO-A–A enzyme. The IC50 values of 4a, 4d, 4f, 4h, 4k and 4m were found as 67.4 ± 3.1 nM, 109.7 ± 4.3 nM, 40.3 ± 1.7 nM, 85.1 ± 3.8 nM, 124.3 ± 5.8 nM and 56.7 ± 2.2 nM, respectively. The IC50 value of the reference drug selegiline was calculated as 37.4 ± 1.6 nM. Accordingly, among the synthesized compounds, the 4f is the most active derivative with the similar IC50 value to that of selegiline. Therefore, the compound 4f, exhibiting inhibitory activity profile on both AChE and MAO-B enzymes, can be suggested as a dual enzyme inhibitor against AD.
2.4. Inhibition of beta amyloid 1–42 (Aβ42) aggregation
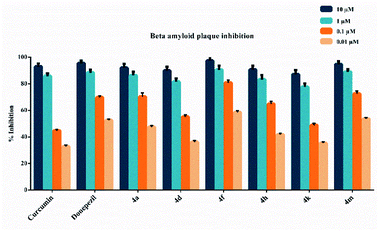
The beta amyloid aggregation inhibitory activities of compounds 4a, 4d, 4f, 4h, 4k and 4m, which show high inhibitory activity against AChE and MAO-B enzymes, were investigated by using a fluorometric beta amyloid 1–42 (Aβ42) ligand screening assay (BioVision, Milpitas, CA, USA). If an Aβ42 ligand is present by the procedure, this reaction is inhibited/destroyed, thereby reducing or completely eliminating fluorescence. Percent inhibition of beta amyloid 1–42 (Aβ42) peptide aggregation is given in Fig. 2. The compounds were tested at concentrations of 10 μM, 1 μM, 0.1 μM, and 0.01 μM, and the Aβ42 inhibitor included in the kit was used as a standard. All test compound concentrations were applied in quadruplicate in plates.According to Fig. 2, compounds 4a, 4d, 4f, 4h, 4k and 4m, exhibited greater than 50% inhibition (91.9 ± 3.1%, 89.8 ± 3.0%, 97.3 ± 3.2%, 90.7 ± 2.9%, 87.1 ± 3.2% and 94.7 ± 2.1%, respectively) at 10 μM concentration. They also exhibited more than 50% inhibition at a concentration of 1 μM. They showed inhibition as 86.4 ± 2.7%, 81.5 ± 2.5%, 90.6 ± 3.1%, 83.2 ± 3.3%, 77.7 ± 2.7% and 88.9 ± 2.0%, respectively. Compounds 4a, 4d, 4f, 4h, 4k and 4m showed inhibition as 70.4 ± 2.8%, 55.2 ± 1.4%, 80.8 ± 2.1%, 64.8 ± 1.9%, 48.9 ± 1.3% and 72.5 ± 2.2%, respectively at a concentration of 0.1 μM. Their inhibition potency was 47.4 ± 1.1%, 36.2 ± 1.0%, 58.6 ± 1.1%, 41.8 ± 1.0%, 35.4 ± 1.0% and 53.6 ± 1.0%, respectively at a concentration of 0.01 μM. The IC50 values of curcumin and donepezil were calculated as 105.5 ± 4.1 nM and 236.5 ± 10.5 nM, respectively. Besides, compounds 4a, 4d, 4f, 4h, 4k and 4m displayed IC50 values of 256.1 ± 11.0 nM, 241.8 ± 9.7 nM, 167.5 ± 8.0 nM, 295.5 ± 13.0 nM, 324.7 ± 15.1 nM and 198.8 ± 8.8 nM, respectively. Compounds 4f and 4m exhibited a higher inhibition potential compared to compounds 4a, 4d, 4h, and 4k. As a result, it has been revealed that compounds 4f and 4m may have a potential to inhibit AChE and MAO-B enzymes, as well as the ability to prevent the formation of beta amyloid plaques accumulated in the brains of patients suffering from AD.
2.5. Cytotoxicity assay
Compound 4f, exhibiting potent AchE–MAO-B inhibition profile, were further tested for cytotoxicity against NIH/3T3 cell line by MTT assay and its IC50 value was calculated. Compound 4f showed an IC50 value of 72.9 ± 2.7 μM against NIH/3T3 cells. This result suggests that it has not cytotoxic potential at its effective concentrations against AChE and MAO-B enzymes.2.6. Molecular docking studies on AChE and MAO-B enzymes
To determine the possible interactions of compound 4f, which has the highest inhibition on AChE and MAO-B enzymes, with the relevant enzyme active sites. Docking studies were carried out on crystal structures of AChE (PDB code: 4EY7)35 and MAO-B (PDB f: 2V5Z)36 enzymes. In the studies, Glide 7.1 (ref. 37) program was used and the most probable poses were produced with GlideScore SP. For validations of docking methods, studies were performed firstly under the same conditions with suitable ligands. At this step, the obtained compatible interaction results of the reference ligands were compared with the literature. Thus, the docking protocols were confirmed, and further studies were carried out for selected active compound 4f (the docking poses of reference ligands are presented in ESI, Fig. S52†). Docking poses related to AChE and MAO-B enzyme are given in Fig. 3–5 and Fig. 6–9, respectively.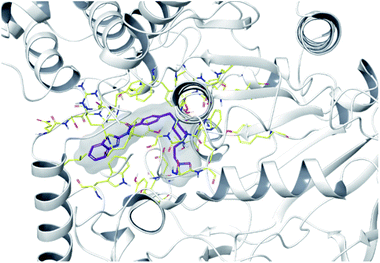 | ||
| Fig. 3 The localization of compound 4f to the AChE enzyme active site (PDBID: 4EY7). | ||
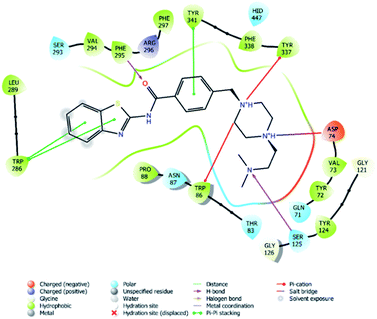 | ||
| Fig. 4 2D interaction of compound 4f at binding region of AChE (PBDID: 4EY7). | ||
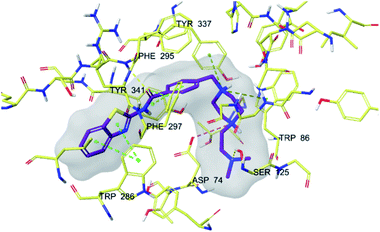 | ||
| Fig. 5 3D interaction of compound 4f at binding region of AChE (PBDID: 4EY7). Maroon carbons: compound 4f; yellow carbons: binding site residues; yellow dashes: H-bond; blue dashes: aromatic H-bond; green dashes: π–π interaction; dark green dashes: π–cation; pink dashes: salt bridge. | ||
 | ||
| Fig. 6 The localization of compound 4f to the MAO-B enzyme active site (PDBID: 2V5Z). | ||
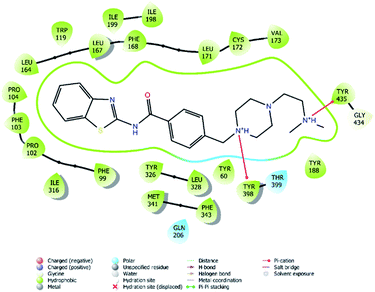 | ||
| Fig. 7 2D interaction of compound 4f at binding region of MAO-B (PBDID: 2V5Z). | ||
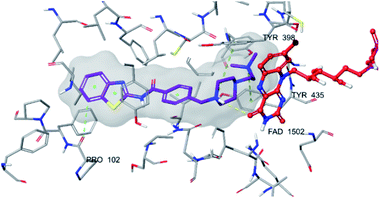 | ||
| Fig. 8 3D interaction of compound 4f at binding region of MAO-B (PBDID: 2V5Z). Maroon carbons: compound 4f; red carbons: FAD protein, grey carbons: binding site residues; yellow dashes: H-bond; blue dashes: aromatic H-bond; green dashes: π–π interaction; dark green dashes: π–cation. | ||
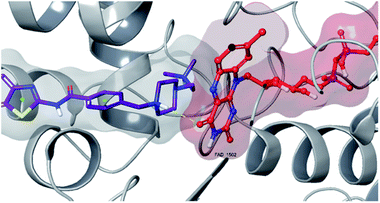 | ||
| Fig. 9 3D interaction of compound 4f with FAD at binding region of MAO-B (PBDID: 2V5Z). | ||
In many molecular modeling studies, it has been determined that donepezil interacts with both regions of the AChE enzyme and settles very well in the passage with its dual binding site (DBS) feature.38–40 Cheung et al. (2012) examined the binding sites of donepezil to enzyme active sites and explained the interactions of donepezil with active sites. In the CAS region, the benzyl ring shows π–π interaction with the indole ring of Trp86. The carbonyl group has an important interaction. Hydrogen bonding is established between the carbonyl oxygen of indanone and the amino group of Phe395. In the PAS region, the indanone structure binds with the indole in Trp286 by π–π interaction. In addition, the piperidine ring is in van der Waals interaction with both the CAS and PAS domains.35 The docking study of compound 4f with the AChE indicates that it bounds very strongly to the enzyme active site by forming various interactions. Like donepezil, it is seen that the compound 4f binds to both the CAS and PAS regions of the enzyme. It settles in the gate and thus can interact fully, taking advantage of the features of the DBS. As can be seen in Fig. 4 and 5, both the phenyl and thiazole parts of the benzothiazole ring in compound 4f form two π–π interactions with the indole ring of amino acid Trp286. It is seen that carbonyl group is also very important in terms of polar interactions. It forms a hydrogen bond with the amino group of Phe295 and an aromatic-hydrogen bond with the phenyl of Phe297. These interactions of benzothiazole and carbonyl show that compound 4f strongly settle-downs in the PAS region, as donepezil does. It has been determined that the benzene ring in the middle of the structure forms a π–π interaction with the phenyl of the Tyr341. It is seen that nitrogen atom in the 1st position of piperazine forms two cation–π interactions with the indole of Trp86 and the phenyl of Try337. It has been determined that nitrogen atom in the 4th position of piperazine forms a salt bridge with the carbonyl of Asp74. Finally, it has been determined that the terminal nitrogen atom in the dimethylaminoethyl side chain forms a hydrogen bond with the hydroxyl of Ser125. These interactions with Asp74, Trp86, Ser125, Tyr337 and Tyr341 supply very strong and conformable binding to the CAS region of the AChE enzyme active site. All above data explain the strong inhibitory activity of compound 4f on AChE enzyme in vitro.
Structural difference between compounds 4a–4g and 4h–4m is the presence of the methoxy group at 5th position of benzothiazole ring in compounds 4h–4m. Hence, molecular docking studies were carried out with compound 4m, carrying methoxy substituent, to explain how the inclusion of this group in the structure influenced binding to the AChE enzyme active site. The docking pose of compound 4m on AChE enzyme was given in ESI, Fig. S51.† According to this docking pose compound 4m showed the same interactions with Trp286, Phe295 and Tyr341 as explained in interactions for compound 4f. The presence of methoxy group monopolized the binary π–π interaction with Trp286. At the same time, the Trp86 interaction, which is an important interaction in the CAS region of the enzyme has been lost due to growth in the molecule. The other interactions observed in the docking pose of 4f could not be identified for compound 4m. These findings reveal that the methoxy substituent at the 5th position of the benzothiazole ring reduced the binding potency to the active region and also biological activity. It can be suggested that the methoxy group effects the conformations of the compounds and thus the derivatives with this moiety show different interactions and less binding profile to enzyme active site.
Fig. 6–8 show the docking poses of compound 4f with the MAO-B enzyme. As seen in the poses, compound 4f is strongly and favorably located in the active site of the MAO-B enzyme, as in the AChE enzyme. The benzothiazole ring forms an aromatic-hydrogen bond with the carbonyl of Pro102. It is seen that the terminal nitrogen atom of dimethylaminoethyl side chain establishes a cation–π interaction with the phenyl of Try435. The nitrogen in the 1st position of the piperazine forms the same interaction with the phenyl of Tyr398. In addition, it has been determined that the piperazine ring forms a hydrogen bond with the carbonyl adjacent to the N5 of FAD molecule located in the enzyme active site (see Fig. 6 and 9). This interaction reveals that compound 4f binds strongly and appropriately to the substrate binding site of MAO-B enzyme active site by interacting with FAD. All available data clearly indicate the strong MAO-B enzyme inhibition profile of compound 4f.
As a result, compound 4f forms a hydrogen bond with the cofactor FAD at the MAO-B active site, which is a significant reason for inhibitory potential of this compound (Fig. 9).
3. Experimental
3.1. Chemistry
All reagents were purchased from commercial suppliers and were used without further purification. Melting points (M. p.) were determined on the Mettler Toledo-MP90 Melting Point System and were uncorrected. 1H-NMR (nuclear magnetic resonance) Bruker DPX 300 FT-NMR spectrometer; 13C-NMR, Bruker DPX 75 MHz spectrometer (Bruker Bioscience, Billerica, MA, USA). In the 1H NMR spectra, the splitting patterns were designated as follows: s: singlet; brs: broad singlet; d: doublet; dd: doublet of doublets; t: triplet; m: multiplet; q: quartet. The coupling constants (J) were expressed in Hertz (Hz). Mass spectra were recorded on a LCMS-IT-TOF (Shimadzu, Kyoto, Japan) using ESI.3.1.2.1. Benzo[d]thiazol-2-amine (2a). Yield: 70%, cream powder. 1H-NMR (300 MHz, DMSO-d6): δ = 6.99 (1H, t, J = 7.5 Hz, benzothiazole), 7.20 (1H, t, J = 7.4, benzothiazole), 7.34 (1H, d, J = 7.9 Hz, benzothiazole), 7.50 (2H, s, –NH2), 7.64 (1H, d, J = 7.7 Hz, benzothiazole). HRMS (m/z): [M + H]+ calcd for C7H6N2S: 151.0324; found 151.0330.
3.1.2.2. 5-Methoxybenzo[d]thiazol-2-amine (2b). Yield: 71%, beige powder. 1H-NMR (300 MHz, DMSO-d6): δ = 3.72 (3H, s, –OCH3), 6.79–6.81 (1H, m, benzothiazole), 7.22–7.29 (4H, m, benzothiazole + NH2). HRMS (m/z): [M + H]+ calcd for C8H8N2SO: 181.0430; found 181.0436.
3.1.3.1. N-(Benzo[d]thiazol-2-yl)-4-(chloromethyl)benzamide (3a)41. Yield: 81%, beige powder. 1H-NMR (300 MHz, DMSO-d6): δ = 4.85 (2H, s, –CH2–), 7.33 (1H, t, J = 7.3 Hz, benzothiazole), 7.47 (1H, t, J = 7.8 Hz, benzothiazole), 7.62 (2H, d, J = 7.1 Hz, 1,4-disubstitutedbenzene), 7.79 (1H, d, J = 7.9 Hz, benztohiazole), 8.02 (1H, d, J = 7.9 Hz, benzothiazole), 8.14 (2H, d, J = 7.0 Hz, 1,4-disubstitutedbenzene), 12.97 (1H, s, –NH). HRMS (m/z): [M + H]+ calcd for C15H11N2SOCl: 303.0353; found 303.0348.
3.1.3.2. 4-(Chloromethyl)-N-(5-methoxybenzo[d]thiazol-2-yl)benzamide (3b). Yield: 79%, beige powder. 1H-NMR (300 MHz, DMSO-d6): δ = 3.82 (3H, s, –OCH3), 4.86 (2H, s, –CH2–), 7.06 (1H, dd, J1 = 2.3 Hz, J2 = 8.6 Hz, benzothiazole), 7.59–7.63 (3H, m, benzothiazole + 1,4-disubstitutedbenzene), 7.68 (1H, d, J = 8.8 Hz, benzothiazole), 8.12 (2H, d, J = 8.1 Hz, 1,4-disubstitutedbenzene). HRMS (m/z): [M + H]+ calcd for C16H13N2SO2Cl: 333.0459; found 333.0464.
3.1.4.1. N-(Benzo[d]thiazol-2-yl)-4-((4-methylpiperazine-1-yl)methyl)benzamide (4a). Yield: 87%, cream powder. M. P. = 145.3–147.0 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.14 (3H, s, –CH3), 2.36 (8H, br.s, piperazine), 3.52 (2H, s, –CH2–), 7.32 (1H, t, J = 4.03 Hz, benzothiazole), 7.43–7.44 (1H, m, benzothiazole), 7.46 (2H, d, J = 6.82 Hz, 1,4-disubstitutedbenzene), 7.77 (1H, d, J = 7.92 Hz, benzothiazole), 8.00 (1H, d, J = 7.56 Hz, benzothiazole), 8.11 (2H, d, J = 8.26 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 46.14, 53.04, 55.14, 62.01, 120.68, 122.16, 124.00, 126.56, 128.78, 129.23, 131.25, 132.00, 144.16, 148.81, 159.76, 166.42. HRMS (m/z): [M + H]+ calcd for C22H22N2O2: 367.1587; found 367.1595.
3.1.4.2. N-(Benzo[d]thiazol-2-yl)-4-((4-ethylpiperazine-1-yl)methyl)benzamide (4b). Yield: 85%, beige powder. M. P. = 149.9–151.9 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 0.97 (3H, t, J = 7.14 Hz, –CH3), 2.27–2.38 (10H, m, piperazine + –CH2–), 3.54 (2H, s, –CH2–), 7.33 (1H, t, J = 7.55 Hz, benzothiazole), 7.43–7.46 (1H, m, benzothiazole), 7.47 (2H, d, J = 7.95 Hz, 1,4-disubstitutedbenzene), 7.78 (1H, d, J = 7.99 Hz, benzothiazole), 8.01 (1H, d, J = 7.95 Hz, benzothiazole), 8.11 (2H, d, J = 8.18 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 12.45, 52.06, 52.82, 53.18, 62.03, 120.71, 122.19, 124.06, 126.60, 128.78, 129.24, 131.14, 131.97, 144.23, 148.77, 159.62, 166.37. HRMS (m/z): [M + H]+ calcd for C21H24N4OS: 381.1744; found 381.1754.
3.1.4.3. 4-((4-Allylpiperazine-1-yl)methyl)-N-(benzo[d]thiazol-2-yl)benzamide (4c). Yield: 81%, yellow powder. M. P. = 150.1–152.3 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.38 (8H, br.s, piperazine), 2.92 (2H, d, J = 6.36 Hz, –CH2–), 3.54 (2H, s, –CH2–), 5.08–5.18 (2H, m, –CH2), 5.72–5.84 (1H, m, CH), 7.31 (1H, t, J = 4.03 Hz, benzothiazole), 7.42–7.45 (1H, m, benzothiazole), 7.46 (2H, d, J = 8.24 Hz, 1,4-disubstitutedbenzene), 7.76 (1H, d, J = 7.86 Hz, benzothiazole), 7.99 (1H, d, J = 7.95 Hz, benzothiazole), 8.11 (2H, d, J = 8.25 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 53.04, 53.15, 61.34, 62.04, 117.85, 120.61, 122.13, 123.88, 126.50, 128.78, 129.19, 131.52, 132.05, 136.08, 144.03, 148.89, 160.08, 166.59. HRMS (m/z): [M + H]+ calcd for C22H24N4OS: 393.1744; found 393.1754.
3.1.4.4. N-(Benzo[d]thiazol-2-yl)-4-((4-(prop-2-yn-1-yl)piperazine-1-yl)methyl)benzamide (4d). Yield: 79%, cream powder. M. P. = 134.7–136.2 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.41 (4H, br.s., piperazine), 2.47 (4H, br.s., piperazine), 3.14 (1H, s, –CH), 3.25 (2H, s, –CH2), 3.54 (2H, s, –CH2), 7.31 (1H, t, J = 4.03 Hz, benzothiazole), 7.43 (1H, br.s., benzothiazole), 7.47 (2H, d, J = 7.63 Hz, 1,4-disubstitutedbenzene), 7.76 (1H, d, J = 7.89 Hz, benzothiazole), 7.99 (1H, d, J = 7.96 Hz, benzothiazole), 8.11 (2H, d, J = 8.24 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 46.43, 51.60, 53.00, 61.97, 76.12, 79.86, 120.67, 122.14, 123.99, 126.56, 128.80, 129.21, 131.31, 132.00, 144.12, 148.79, 159.78, 166.47. HRMS (m/z): [M + H]+ calcd for C22H22N4OS: 391.1587; found 391.1599.
3.1.4.5. N-(Benzo[d]thiazol-2-yl)-4-((4-isopropylpiperazine-1-yl)methyl)benzamide (4e). Yield: 82%, cream powder. M. P. = 140.3–142.0 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 1.07 (6H, d, J = 6.51 Hz, –CH3 + –CH3), 2.48 (4H, br.s., piperazine), 2.55 (1H, br.s., piperazine), 2.61–2.74 (1H, m, –CH), 3.54 (2H, s, –CH2), 7.26–7.32 (3H, m, benzothiazole), 7.40 (2H, d, J = 8.16 Hz, 1,4-disubstitutedbenzene), 7.84–7.87 (1H, m, benzothiazole), 7.96 (2H, d, J = 8.29 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 18.65, 48.60, 53.42, 54.45, 62.50, 120.64, 121.42, 123.92, 126.06, 127.93, 129.56, 130.68, 131.97, 144.26, 147.84, 159.68, 165.79. HRMS (m/z): [M + H]+ calcd for C22H26N4OS: 395.1900; found 395.1912.
3.1.4.6. N-(Benzo[d]thiazol-2-yl)-4-((4-(2-(dimethylamino)ethyl)piperazine-1-yl)methyl)benzamide (4f). Yield: 78%, yellow powder. M. P. = 175.7–177.2 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.25 (6H, s, –CH3 + –CH3), 2.46 (4H, br.s., piperazine), 2.48 (4H, br.s., piperazine), 2.89 (2H, s, –CH2), 2.96 (2H, s, –CH2), 3.54 (2H, s, –CH2), 7.27–7.30 (2H, m, benzothiazole), 7.37–7.40 (1H, m, benzothiazole), 7.41 (2H, d, J = 8.30 Hz, 1,4-disubstitutedbenzene), 7.84–7.87 (1H, m, benzothiazole), 7.96 (2H, d, J = 8.23 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 45.90, 45.94, 53.03, 53.59, 56.65, 56.89, 62.45, 120.69, 121.42, 123.91, 126.04, 127.91, 129.50, 130.71, 132.02, 144.94, 147.94, 159.49, 165.68. HRMS (m/z): [M + H]+ calcd for C23H29N5OS: 424.2166; found 424.2167.
3.1.4.7. N-(Benzo[d]thiazol-2-yl)-4-((4-(3-(dimethylamino)propyl)piperazine-1-yl)methyl)benzamide (4g). Yield: 85%, light brown powder. M. P. = 72.9–74.4 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 1.48–1.56 (2H, m, –CH2–), 2.13 (6H, s, –CH3 + –CH3), 2.20–2.27 (4H, m, piperazine), 2.35 (8H, br.s., –CH2– + –CH2– + piperazine), 3.52 (2H, s, –CH2–), 7.31 (1H, t, J = 7.57 Hz, benzothiazole), 7.42–7.44 (1H, m, benzothiazole), 7.45 (2H, d, J = 8.11 Hz, 1,4-disubstitutedbenzene), 7.76 (1H, d, J = 7.95 Hz, benzothiazole), 7.99 (1H, d, J = 7.64 Hz, benzothiazole), 8.11 (2H, d, J = 8.20 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 24.67, 45.40, 53.17, 53.26, 56.35, 57.60, 62.04, 120.64, 122.15, 123.94, 126.54, 128.77, 129.23, 131.37, 132.01, 144.06, 148.84, 159.94, 166.51. HRMS (m/z): [M + H]+ calcd for C24H31N5OS: 438.2322; found 438.2333.
3.1.4.8. N-(5-Methoxybenzo[d]thiazol-2-yl)-4-((4-methylpiperazine-1-yl)methyl)benzamide (4h). Yield: 85%, light brown powder. M. P. = 137.1–138.9 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.31 (3H, s, –CH3), 2.47 (8H, br.s., piperazine), 3.54 (2H, s, –CH2–), 3.87 (3H, s, –CH3–), 6.86 (1H, dd, J = 3.82 Hz, benzothiazole), 7.20 (1H, d, J = 8.89 Hz, benzothiazole), 7.32 (1H, d, J = 2.51 Hz, benzothiazole), 7.41 (2H, d, J = 8.27 Hz, 1,4-disubstitutedbenzene), 7.95 (2H, d, J = 8.29 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 46.02, 52.91, 55.06, 55.99, 61.93, 105.09, 105.95, 113.32, 115.51, 118.52, 121.45, 128.71, 129.28, 144.13, 154.70, 156.69, 165.16. HRMS (m/z): [M + H]+ calcd for C21H24N4O2S: 397.1693; found 397.1707.
3.1.4.9. 4-((4-Ethylpiperazine-1-yl)methyl)-N-(5-methoxybenzo[d]thiazol-2-yl)benzamide (4i). Yield: 87%, dark brown powder. M. P. = 129.8–130.9 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 1.11 (3H, t, J = 7.17 Hz, –CH3), 2.41–2.48 (10H, m, –CH2– + piperazine), 3.55 (2H, s, –CH2–), 3.87 (3H, s, –CH3–), 6.83 (1H, dd, J1 = 3.63 Hz, J2 = 9.0 Hz, benzothiazole), 7.13 (1H, d, J = 8.83 Hz, benzothiazole), 7.31 (1H, d, J = 2.13 Hz, benzothiazole), 7.40 (2H, d, J = 7.96 Hz, 1,4-disubstitutedbenzene), 7.95 (2H, d, J = 8.00 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 11.93, 52.31, 52.71, 53.05, 55.79, 62.46, 103.88, 115.24, 121.29, 127.94, 129.51, 130.78, 133.20, 142.00, 144.15, 156.75, 157.67, 165.63. HRMS (m/z): [M + H]+ calcd for C22H26N4O2S: 411.1849; found 411.1854.
3.1.4.10. 4-((4-Allylpiperazine-1-yl)methyl)-N-(5-methoxybenzo[d]thiazol-2-yl)benzamide (4j). Yield: 90%, brown powder. M. P. = 173.9–175.5 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.38 (8H, piperazine), 2.92 (2H, d, J = 6.34 Hz, –CH2–), 3.54 (2H, s, –CH2–), 3.83 (3H, s, –CH3–), 5.09–5.19 (2H, m, –CH2), 5.75–5.84 (1H, m, –CH–), 7.07 (1H, dd, J1 = 3.82 Hz, J2 = 6.0 Hz, benzothiazole), 7.47 (2H, d, J = 8.41 Hz, 1,4-disubstitutedbenzene), 7.62 (1H, d, J = 2.57 Hz, benzothiazole), 7.68 (1H, d, J = 8.82 Hz, benzothiazole), 8.10 (2H, d, J = 8.26 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 53.07, 54.85, 55.57, 57.03, 61.99, 104.02, 106.13, 116.50, 120.35, 122.50, 127.63, 130.32, 131.06, 135.03, 137.04, 142.93, 144.13, 157.33, 165.94. HRMS (m/z): [M + H]+ calcd for C23H26N4O2S: 423.1849; found 423.1860.
3.1.4.11. N-(5-Methoxybenzo[d]thiazol-2-yl)-4-((4-(prop-2-yn-1-yl)piperazine-1-yl)methyl)benzamide (4k). Yield: 86%, brown powder. M. P. = 99.9–101.8 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.49 (4H, br.s., piperazine), 2.59 (4H, br.s., piperazine), 2.93 (1H, d, J = 21.96 Hz, –CH), 3.32 (2H, s, –CH2–), 36.53 (2H, s, –CH2–), 3.86 (3H, s, –CH3), 6.79 (1H, dd, J = 3.79 Hz, J2 = 9.0 Hz, benzothiazole), 7.06 (1H, d, J = 8.89 Hz, benzothiazole), 7.30 (1H, d, J = 2.49 Hz, benzothiazole), 7.38 (2H, d, J = 8.22 Hz, 1,4-disubstitutedbenzene), 7.95 (2H, d, J = 8.24 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 46.80, 51.83, 52.91, 52.96, 54.81, 62.35, 71.62, 102.78, 103.83, 115.21, 121.22, 122.30, 128.05, 129.42, 141.90, 144.04, 156.73, 157.96, 165.87. HRMS (m/z): [M + H]+ calcd for C23H24N4O2S: 421.1693; found 421.1693.
3.1.4.12. 4-((4-Isopropylpiperazine-1-yl)methyl)-N-(5-methoxybenzo[d]thiazol-2-yl)benzamide (4l). Yield: 89%, brown powder. M. P. = 126.3–128.1 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 1.05 (5H, d, J = 6.52 Hz, –CH3 + –CH3), 2.46 (4H, br.s., piperazine), 2.53 (4H, br.s., piperazine), 2.61–2.69 (1H m, –CH), 3.51 (2H, s, –CH2–), 3.85 (3H, s, –CH3), 6.76 (1H, dd, J1 = 3.82 Hz, J2 = 9.0 Hz, benzothiazole), 6.99 (1H, d, J = 8.90 Hz, benzothiazole), 7.29 (1H, d, J = 2.47 Hz, benzothiazole), 7.36 (2H, d, J = 8.26 Hz, 1,4-disubstitutedbenzene), 7.95 (2H, d, J = 8.29 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 18.66, 48.59, 53.46, 54.43, 55.71, 55.76, 62.53, 103.76, 115.22, 121.20, 128.20, 129.53, 130.85, 133.10, 141.84, 144.07, 156.69, 158.10, 165.98. HRMS (m/z): [M + H]+ calcd for C23H28N4O2S: 425.2006; found 425.2013.
3.1.4.13. 4-((4-(2-(Dimethylamino)ethyl)piperazine-1-yl)methyl)-N-(5-methoxybenzo[d]thiazol-2-yl)benzamide (4m). Yield: 81%, orange powder. M. P. = 60.1–62.0 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 2.15 (6H, s, –CH3 + –CH3), 2.36–2.52 (10H, m, piperazine + –CH2–), 3.45 (2H, t, J = 3.45 Hz, –CH2–), 3.53 (2H, s, –CH2–), 3.83 (3H, s, –CH3), 7.06 (1H, dd, J = 3.81 Hz, J2 = 9.0 Hz, benzothiazole), 7.47 (2H, d, J = 8.28 Hz, 1,4-disubstitutedbenzene), 7.61 (1H, d, J = 2.56 Hz, benzothiazole), 7.68 (2H, d, J = 8.83 Hz, benzothiazole), 8.10 (2H, d, J = 8.29 Hz, 1,4-disubstitutedbenzene). 13C-NMR (75 MHz, DMSO-d6): δ = 45.83, 45.87, 53.15, 53.53, 56.15, 56.84, 62.02, 120.32, 121.39, 127.61, 128.69, 129.24, 131.22, 133.33, 142.99, 144.03, 156.62, 157.54, 166.06. HRMS (m/z): [M + H]+ calcd for C24H31N5O2S: 454.2271; found 454.2280.
3.1.4.14. 4-((4-(3-(Dimethylamino)propyl)piperazine-1-yl)methyl)-N-(5-methoxybenzo[d]thiazol-2-yl)benzamide (4n). Yield: 83%, brown powder. M. P. = 137.3–139.1 °C. 1H-NMR (300 MHz, DMSO-d6): δ = 1.47–1.57 (2H, m, –CH2-), 2.12 (6H, s, –CH3 + –CH3), 2.19 (2H, t, J = 6.60 Hz, –CH2–), 2.24 (2H, t, J = 3.66 Hz, –CH2–), 2.37 (8H, br.s., piperazine), 3.53 (2H, s, –CH2–), 3.83 (3H, s, –CH3), 7.06 (1H, dd, J1 = 3.81 Hz, J2 = 9.0 Hz, benzothiazole), 7.46 (2H, d, J = 8.27 Hz, 1,4-disubstitutedbenzene), 7.61 (1H, d, J = 2.55 Hz, benzothiazole), 7.67 (1H, d, J = 8.83 Hz, benzothiazole), 8.10 (2H, d, J = 8.26 Hz, 1,4-disubstitutedbenzene).13C-NMR (75 MHz, DMSO-d6): δ = 24.80, 45.49, 53.18, 53.27, 56.04, 56.41, 57.67, 62.04, 105.05, 115.40, 121.36, 128.69, 129.22, 131.30, 133.35, 143.02, 143.99, 156.60, 157.66, 166.09. HRMS (m/z): [M + H]+ calcd for C25H33N5O2S: 468.2428; found 468.2436.
3.2. Cholinesterase enzymes inhibition assay
The in vitro AChE and BChE inhibition potencies of the synthesized compounds (4a–4n) were evaluated according to the modified Ellman's spectrophotometric method.20 The reagents and materials used in the enzyme inhibition assay were supplied commercially by Sigma-Aldrich (St Louis, MO, USA) and Fluka (Steinheim, Germany). The cholinesterase enzyme inhibition procedure was applied as reported in our previous research papers.19,21–27 Human acetylcholinesterase (CAS no.: 9000-81-1) and human butyrylcholinesterase (CAS no.: 9001-08-5) enzymes were used as enzymes.3.3. MAO enzymes inhibition assay
The in vitro MAO inhibition test was performed using the available fluorometric method and the percentages and IC50 values of obtained compounds were calculated as previously described by our research group.28–343.4. Inhibition of beta amyloid 1–42 (Aβ42) aggregation
The test procedure was created based on the protocol of the beta amyloid 1–42 (Aβ42) ligand screening assay (BioVision, Milpitas, CA, USA), based on the fluorometric method.3.5. Cytotoxicity assay
The NIH/3T3 mouse embryonic fibroblast cell line (ATCC® CRL-1658™, London, UK) was used for cytotoxicity assays. The incubation period of NIH/3T3 cells was based on the supplier's recommendation. NIH/3T3 cells were seeded at 1 × 104 cells into each well of 96-well plates. MTT assay was carried out in accordance with the standards previously described.19,213.6. Molecular docking
Molecular docking studies were performed using an in silico procedure to define the binding modes of active compound in the active regions of AChE and MAO-B. X-ray crystal structures of the AChE (PDB ID: 4EY7)35 and MAO-B (PDB ID: 2V5Z)43 were retrieved from the Protein Data Bank server (https://www.pdb.org, accessed 01 May 2021). The structures of the enzymes were built using the Schrödinger Maestro44 interface and were then submitted to the Protein Preparation Wizard protocol of the Schrödinger Suite 2020. The ligand was prepared using the LigPrep module45 to correctly assign the protonation states as well as the atom types. Bond orders were assigned, and hydrogen atoms were added to the structures. The grid generation was performed using the Glide module,37 and docking runs were conducted in standard precision (SP) docking mode.4. Conclusion
In this study, compounds 4a–4n were synthesized and screened for potential inhibitory activities against AChE, BChE, MAO-A, and MAO-B enzymes. As a result of the studies, compound 4f showed significant inhibition on AChE and MAO-B enzymes. The IC50 value of donepezil was found to be 20.1 ± 1.4 nM against AChE, whereas IC50 value of the most active compound 4f was determined as 23.4 ± 1.1 nM. In other words, it showed activity comparable to donepezil. Additionally, the IC50 value of the reference drug selegiline on MAO-B enzyme was calculated as 37.4 ± 1.6 nM. Accordingly, among the synthesized compounds, the most active compound 4f displayed similar IC50 value (40.3 ± 1.7 nM) to that of selegiline. Moreover, β-amyloid 1–42 (Aβ42) inhibitor screening studies were performed for active compounds. As a result of these studies, it was observed that especially compound 4f and 4m are important agents for dual-acting drug design by inhibiting β-amyloid plaque formation in addition to AChE and MAO-B enzyme inhibitions. Data of molecular docking studies expressed the significant interactions with AChE and MAO-B enzymes, and thus supported the in vitro dual inhibitory property of compound 4f. The findings show that compound 4f has a promising potential in the treatment of AD and PD with its dual inhibition profile.Author contributions
Conceptualization, Ş. K., D. O., and Z. A. K.; methodology, Ş. K., D. O., B. N. S. and S. L.; software, D. O. and B. N. S.; validation, D. O. and B. N. S.; formal analysis, D. O. and S. L.; investigation, Ş. K., B. N. S., D. O., S. L., S. I. and Z. A. K.; resources, Y. Ö., A. Ç. K., Z. A. K. and N. G. K.; data curation, Ş. K., D. O. and B. N. S.; writing-original draft preparation, Ş. K., D. O., B. N. S., S. L., S. I., Y. Ö., A. Ç. K., Z. A. K. and N. G. K.; writing-review and editing, B. N. S., D. O. and Y. Ö.; visualization, D. O. and Z. A. K.; supervision, Y. Ö., A. Ç. K., Z. A. K. and N. G. K.; project administration, Ş. K.; D. O., B. N. S. Y. Ö.; funding acquisition, B. N. S. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by Anadolu University Scientific Research Projects Commission under the grant no: 2101S001. As the authors of this study, we thank Anadolu University Faculty of Pharmacy Central Analysis Laboratory for their support and contributions.References
- Ü. D. Özkay, O. D. Can, Y. Özkay and Y. Özturk, Pharmacol. Rep., 2012, 64, 834–847 CrossRef.
- T. Liu, S. Chen, J. Du, S. Xing, R. Li and Z. Li, Eur. J. Med. Chem., 2021, 113973 Search PubMed.
- G. F. Makhaeva, S. V. Lushchekina, N. V. Kovaleva, T. Y. Astakhova, N. P. Boltneva, E. V. Rudakova, O. G. Serebryakova, A. N. Proshin, I. V. Serkov and T. P. Trofimova, Bioorg. Chem., 2021, 112, 104974 CrossRef CAS PubMed.
- B. Ren, C. Guo, R.-Z. Liu, Z.-Y. Bian, R.-C. Liu, L.-F. Huang and J.-J. Tang, Eur. J. Med. Chem., 2022, 228, 114031 CrossRef CAS PubMed.
- P. H. Syaifie, A. W. Hemasita, D. W. Nugroho, E. Mardliyati and I. Anshori, 2021.
- N. Dunn, G. Pearce and S. Shakir, J. Psychopharmacol., 2000, 14, 406–408 CrossRef CAS PubMed.
- M. Hashimoto, T. Imamura, S. Tanimukai, H. Kazui and E. Mori, Lancet, 2000, 356, 568 CrossRef CAS.
- M. M. Carrasco, L. Agüera, P. Gil, A. Moríñigo and T. Leon, Alzheimer Dis. Assoc. Disord., 2011, 25, 333–340 CrossRef CAS PubMed.
- H.-C. Li, K.-X. Luo, J.-S. Wang and Q.-X. Wang, Medicine, 2020, 99, 19443 CrossRef PubMed.
- Z. Zhang, J. Guo, M. Cheng, W. Zhou, Y. Wan, R. Wang, Y. Fang, Y. Jin, J. Liu and S.-S. Xie, Eur. J. Med. Chem., 2021, 213, 113154 CrossRef CAS PubMed.
- G. F. Makhaeva, N. V. Kovaleva, N. P. Boltneva, S. V. Lushchekina, E. V. Rudakova, T. S. Stupina, A. A. Terentiev, I. V. Serkov, A. N. Proshin and E. V. Radchenko, Bioorg. Chem., 2020, 94, 103387 CrossRef CAS PubMed.
- R. K. P. Tripathi and S. R. Ayyannan, Med. Res. Rev., 2019, 39, 1603–1706 CrossRef CAS PubMed.
- A. Kumar and A. K. Mishra, Mini-Rev. Med. Chem., 2021, 21, 314–335 CrossRef PubMed.
- E. A. Abd El-Meguid, A. M. Naglah, G. O. Moustafa, H. M. Awad and A. M. El Kerdawy, Bioorg. Med. Chem. Lett., 2022, 128529 CrossRef CAS PubMed.
- A. Catalano, A. Rosato, L. Salvagno, D. Iacopetta, J. Ceramella, G. Fracchiolla, M. S. Sinicropi and C. Franchini, Antibiotics, 2021, 10, 803 CrossRef CAS PubMed.
- A. Doble, Neurology, 1996, 47, 233S–241S CrossRef PubMed.
- A. D. Cohen and W. E. Klunk, Neurobiol. Dis., 2014, 72, 117–122 CrossRef CAS PubMed.
- A. U. Baran, Turk. J. Chem., 2013, 37, 927–935 CrossRef CAS.
- B. N. Sağlık, S. Ilgın and Y. Özkay, Eur. J. Med. Chem., 2016, 124, 1026–1040 CrossRef PubMed.
- G. L. Ellman, K. D. Courtney, V. Andres Jr and R. M. Featherstone, Biochem. Pharmacol., 1961, 7, 88–95 CrossRef CAS PubMed.
- Ü. D. Özkay, Ö. D. Can, B. N. Sağlık, U. A. Çevik, S. Levent, Y. Özkay, S. Ilgın and Ö. Atlı, Bioorg. Med. Chem. Lett., 2016, 26, 5387–5394 CrossRef PubMed.
- U. Acar Cevik, S. Levent, B. Nurpelin Saglık, Y. Ozkay and Z. Asım Kaplancıklı, Lett. Drug Des. Discovery, 2017, 14, 528–539 CrossRef.
- S. Levent, U. Acar Çevik, B. N. Sağlık, Y. Özkay, Ö. D. Can, Ü. D. Özkay and Ü. Uçucu, Phosphorus, Sulfur Silicon Relat. Elem., 2017, 192, 469–474 CrossRef CAS.
- W. Hussein, B. N. Sağlık, S. Levent, B. Korkut, S. Ilgın, Y. Özkay and Z. A. Kaplancıklı, Molecules, 2018, 23, 2033 CrossRef PubMed.
- U. Acar Cevik, B. N. Saglik, S. Levent, D. Osmaniye, B. Kaya Cavuşoglu, Y. Ozkay and Z. A. Kaplancikli, Molecules, 2019, 24, 861 CrossRef PubMed.
- D. Osmaniye, B. N. Sağlık, U. Acar Çevik, S. Levent, B. Kaya Çavuşoğlu, Y. Özkay, Z. A. Kaplancıklı and G. Turan, Molecules, 2019, 24, 2392 CrossRef CAS.
- F. Tok, B. Koçyiğit-Kaymakçıoğlu, B. N. Sağlık, S. Levent, Y. Özkay and Z. A. Kaplancıklı, Bioorg. Chem., 2019, 84, 41–50 CrossRef CAS PubMed.
- B. N. Sağlık, B. K. Çavuşoğlu, D. Osmaniye, S. Levent, U. A. Çevik, S. Ilgın, Y. Özkay, Z. A. Kaplancıklı and Y. Öztürk, Bioorg. Chem., 2019, 85, 97–108 CrossRef PubMed.
- Ö. D. Can, D. Osmaniye, Ü. D. Özkay, B. N. Sağlık, S. Levent, S. Ilgın, M. Baysal, Y. Özkay and Z. A. Kaplancıklı, Eur. J. Med. Chem., 2017, 131, 92–106 CrossRef PubMed.
- N. Ö. Can, D. Osmaniye, S. Levent, B. N. Sağlık, B. Korkut, Ö. Atlı, Y. Özkay and Z. A. Kaplancıklı, Eur. J. Med. Chem., 2018, 144, 68–81 CrossRef CAS PubMed.
- S. Ilgın, D. Osmaniye, S. Levent, B. N. Sağlık, U. Acar Çevik, B. K. Çavuşoğlu, Y. Özkay and Z. A. Kaplancıklı, Molecules, 2017, 22, 2187 CrossRef PubMed.
- N. Ö. Can, D. Osmaniye, S. Levent, B. N. Sağlık, B. Inci, S. Ilgın, Y. Özkay and Z. A. Kaplancıklı, Molecules, 2017, 22, 1381 CrossRef PubMed.
- F. Tok, Z. Uğraş, B. N. Sağlık, Y. Özkay, Z. A. Kaplancıklı and B. Koçyiğit-Kaymakçıoğlu, Bioorg. Chem., 2021, 112, 104917 CrossRef CAS PubMed.
- F. Tok, B. N. Sağlık, Y. Özkay, S. Ilgın, Z. A. Kaplancıklı and B. Koçyiğit-Kaymakçıoğlu, Bioorg. Chem., 2021, 105038 CrossRef CAS PubMed.
- J. Cheung, M. J. Rudolph, F. Burshteyn, M. S. Cassidy, E. N. Gary, J. Love, M. C. Franklin and J. J. Height, J. Med. Chem., 2012, 55, 10282–10286 CrossRef CAS PubMed.
- C. Binda, J. Wang, L. Pisani, C. Caccia, A. Carotti, P. Salvati, D. E. Edmondson and A. Mattevi, J. Med. Chem., 2007, 50, 5848–5852 CrossRef CAS PubMed.
- L. Schrödinger, Schrödinger, LLC, New York, NY, USA, 2020 Search PubMed.
- M. Alipour, M. Khoobi, A. Foroumadi, H. Nadri, A. Moradi, A. Sakhteman, M. Ghandi and A. Shafiee, Bioorg. Med. Chem., 2012, 20, 7214–7222 CrossRef CAS PubMed.
- Z. F. Al-Rashid and R. P. Hsung, Bioorg. Med. Chem. Lett., 2015, 25, 4848–4853 CrossRef CAS PubMed.
- D. Genest, C. Rochais, C. Lecoutey, J. Sopkova-de Oliveira Santos, C. Ballandonne, S. Butt-Gueulle, R. Legay, M. Since and P. Dallemagne, MedChemComm, 2013, 4, 925–931 RSC.
- Y. Zong, J. Wang, P. An, G. Yue, Y. Pan and X. Wang, Appl. Organomet. Chem., 2017, 31, e3762 CrossRef.
- L. Yurttaş, Z. A. Kaplancıklı and Y. Özkay, J. Enzyme Inhib. Med. Chem., 2012, 28, 1040–1047 CrossRef PubMed.
- S.-Y. Son, J. Ma, Y. Kondou, M. Yoshimura, E. Yamashita and T. Tsukihara, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 5739–5744 CrossRef CAS PubMed.
- Maestro, Schrödinger, LLC, New York, NY, USA, 2020 Search PubMed.
- LigPrep. Schrödinger, Schrödinger, LLC, New York, NY, USA, 2020 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra03803j |
| This journal is © The Royal Society of Chemistry 2022 |