Polarity-based fluorescence probes: properties and applications
Xiaojun
Qin
ab,
Xingye
Yang
b,
Lupei
Du
 b and
Minyong
Li
b and
Minyong
Li
 *bc
*bc
aSchool of Pharmacy, Guangxi Medical University, Nanning, Guangxi 530021, China
bDepartment of Medicinal Chemistry, Key Laboratory of Chemical Biology (MOE), School of Pharmacy, Cheeloo College of Medicine, Shandong University, Jinan, Shandong 250012, China. E-mail: mli@sdu.edu.cn
cState Key Laboratory of Microbial Technology, Shandong University, Jinan, Shandong 250100, China
First published on 9th August 2021
Abstract
Local polarity can affect the physical or chemical behaviors of surrounding molecules, especially in organisms. Cell polarity is the ultimate feedback of cellular status and regulation mechanisms. Hence, the abnormal alteration of polarity in organisms is closely linked with functional disorders and many diseases. It is incredibly significant to monitor and detect local polarity to explain the biological processes and diagnoses of some diseases. Because of their in vivo safe and real-time monitoring, several polarity-sensitive fluorophores and fluorescent probes have gradually emerged and been used in modern research. This review summarizes the fluorescence properties and applications of several representative polarity-sensitive fluorescent probes.
Introduction
Currently, fluorescent probes are widely used in academic research, medicine, and industry, such as physiological imaging, environmental monitoring, biomedical investigations, clinical diagnostics, drug discovery, and so on.1–3 Therefore, the synthesis and characterization of fluorescent probes have increasingly attracted research interest.It is well known that local environmental parameters (e.g., polarity, viscosity, pH, and temperature) in biosystems play a pivotal role in regulating transportation, diffusion, and intermolecular interactions.4 In most cases, these environmental parameters can control the physical or chemical behaviors of surrounding molecules. It has been found that abnormal changes in these parameters correlate closely with some physiological function disorders or diseases.5–7 It is essential to monitor these parameters, leading to the explanation of biological processes and even diagnosing of some diseases.8 With the wide application of fluorescence technology, many fluorescent probes have been designed and synthesized in response to the variation of these parameters in vitro and in vivo.
Cell polarity is the ultimate feedback of cellular status and regulation mechanisms. These complex mechanisms establish and maintain the functionality of particular domains in the cytoplasm and plasma membrane.9 The membrane and protein composition as well as function of these domains facilitate many cellular processes, such as localized membrane growth, cell directional migration and differentiation, immune response activation, and vectorial transport of molecules into a particular site.10 Studies revealed that these different physiological and pathological activities correlated closely with another cell status; when the cell status changed, polarity variation was also observed.11 Meanwhile, abnormal polarity changes are closely associated with disorders and diseases.8,12,13 Hence, monitoring cellular polarity is essential to understand physiological and pathological processes. Interestingly, several fluorescent probes in response to polarity have gradually emerged.
Environmentally sensitive fluorescent probes are mainly composed of an aromatic ring system with electron-donor and electron-acceptor groups; when these groups were as far apart as feasible, maximal effects were expected.14,15 Environmentally sensitive fluorophores can change their fluorescence properties in response to the polarity of local surroundings, including maximum excitation and emission wavelength (λex and λem), fluorescence quantum yield (Φf), and fluorescence lifetime.16
Solvatochromic dyes can vary their color in response to polarity, so a large class of environmentally sensitive probes were constructed based on these dyes. Typically, they belong to push–pull dyes that perform intramolecular charge transfer. After light absorption, the push–pull structure ensures that the charge is transferred to the acceptor group from the donor group, which creates a highly dipolar excited state. In more polar solvents, its emission shifts to longer wavelengths through the interaction of the highly dipolar excited state with the dipoles of the solvent.17 The second design concept of solvatochromic dyes is excited-state intramolecular proton transfer (ESIPT) which changes the relative intensity of two emissive tautomeric forms in response to the microenvironment. The ratio of intensities (normal/tautomeric state) increases with the solvent polarity and H-bond donor ability increase because the ESIPT reaction is inhibited in polar solvents, especially protic solvents.18 On the other hand, intramolecular rotation (conformation change) is a design concept of solvatochromic dyes. In polar media, molecular rotors undergo TICT, resulting in a poorly fluorescent state.19 Furthermore, some emerging concepts are reported in response to the microenvironment, such as aggregation-caused quenching, aggregation-induced emission and ground-state isomerization. Herein, several representative polarity-sensitive fluorescent probes and their applications are summarized. NBD and SBD (4,7-disubstituted benzofuran), derivatives with a benzofuran, have some advantages, including high fluorescence quantum yield (Φf), high reactivity, longer excitation and emission wavelengths, and low background interference.20 Many fluorescent reagents based on benzofuran have gradually emerged to achieve sensitive and selective determination of surroundings.
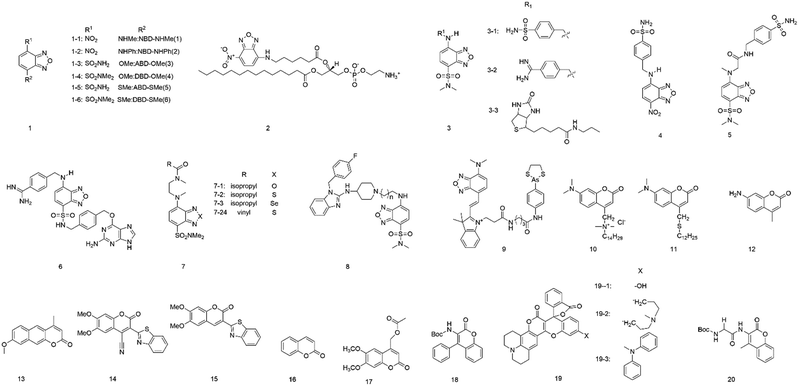
To further understand the benzofuran derivatives, Imai and co-workers21 studied the fluorescence characteristics (e.g., λem, λex and Φf) of six 4,7-disubstituted benzofuran compounds (1, Fig. 1). They demonstrated that the maximum excitation, absorption wavelength, and maximum emission wavelength were determined by the energy gaps between the HOMO and the LUMO at the geometry of the ground and excited states. The intersystem crossing frequency (S1 → T2) and internal conversion influenced the fluorescence quantum yield (Φf). This study revealed that the semi-empirical molecular orbital calculation could predict the fluorescence characteristics of these compounds.
In 2015, Marcus and colleagues22 presented the incorporation of lipid chains into polypropylene fibers using lipids labelled with the fluorophore 7-nitro-2-1,3-benzoxadiazol-4-yl (NBD). The NBD fluorophore could be attached to the headgroup (NBD-PE) or the acyl chain (acyl NBD-PE) (2) of a lipid. When fibers were modified with the acyl NBD-PE, the excitation wavelength changed from 470 nm to 510 nm, and the emission shifted 32 nm toward the red edge of the absorption band, indicating that the NBD molecule is motionally restricted. Fibers modified with NBD-PE or NBD-Cl show no emission change. Overall, these results explained that the acyl chain of the lipids incorporates into the polypropylene fiber structure at the free volume.
Tobita et al.16 synthesized a series of environment-sensitive fluorophores based on 2,1,3-benzoxadiazole (3). All the probes showed obvious solvatochromism in various solvents. The probe 3-1 showed high fluorescence in n-hexane (Φf, 0.91; λem, 520 nm) but was quenched in water (Φf, 0.027; λem, 616 nm) with excitation at 449 nm. They also found a similar variation in fluorescence properties for another two fluorophores, in which the oxygen atom of probe 3-1 has been replaced by sulfur (3-2) and selenium atom (3-3). The fluorophore 3-2 was highly fluorescent in n-hexane (Φf, 0.81; λem, 537 nm) but was almost non-fluorescent in water (Φf, 0.037; λem, 616 nm), similarly to 3-3 (Φf, 0.24, λem, 591 nm in n-hexane; Φf, 0.0046, λem, 672 nm in water). Studies of the variation in fluorescence properties for these fluorophores indicated that the nonradiative relaxation of the excited fluorophores was accelerated by the polarity of the environment and hydrogen bonding with solvent molecules.
To selectively detect proteins, Tan et al.23 developed environment-sensitive fluorescent probes (4–7) based on 4-sulfamonyl-7-aminobenzoxadiazole (SBD) and incorporated them with different ligands. Upon binding the ligand with the hydrophobic ligand-binding domain of the target protein, these fluorescent probes turned on and showed fluorescence enhancement, but not other environment-sensitive fluorophores (e.g., NBD and dansyl). This method based on SBD fluorophore is a powerful tool for the detection of enzymes and non-enzymatic proteins.
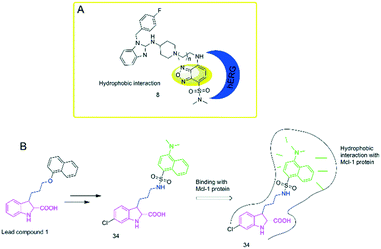
To study the hydrophobic interaction between the ligands and the human ether-a-go-go-related gene (hERG) potassium channel, Du's group24 developed an environment-sensitive turn-on probe (8) based on solvatochromic SBD fluorophores (Fig. 2A). They found that the probe had a high binding affinity with the hERG potassium channel (IC50 = 41.65 nM). The probe can be helpful for hERG channel imaging or detecting the cellular distribution of hERG channel blockers.
In 2017, Jiang and co-workers25 developed a powerful fluorescent probe based on a benzoxadiazole fluorophore (9). The probe can selectively target mitochondria and showed a sensitive response to the local environment. Upon binding to vicinal dithiol-containing proteins (VDPs) in mitochondria, the probe showed strong fluorescence but was almost non-fluorescent in an aqueous solution. It may provide a useful tool for mitochondrial VDP monitoring in situ.
Coumarin derivatives
Coumarin (10), derived from 1,2-benzopyrone, is a well-known laser dye that emits the blue-green region and shows hydrophobicity. Coumarins show fascinating and unique fluorescence properties, including high sensitivity to the local environment (e.g., polarity). Their fluorescence emission shows a large red-shift of 30 nm in a non-polar solvent from a polar solvent.26,27 Coumarins have several advantages, such as strongly polarity-dependent Stokes shifts, a significant change in dipole moment on excitation, and very high fluorescence quantum yields. However, these different derivatives based on coumarin can show other fluorescence behaviors when they are exposed to conditions that undergo the same changes.28In 1996, Epand et al.29 investigated two coumarin-based fluorescent probes (11 and 12) in model membrane systems. Both probes were found to be sensitive in response to the polarity of the solvent. TAMAC contains a quaternary ammonium function, which helps position it at a fixed location in the membrane interface. DTMAC does not have a quaternary ammonium function that fixes to the membrane interface, and it is more sensitive to membrane interfacial polarity.
Kumbhakar's group28 investigated the photophysical properties of coumarin-120 (13) in different solvents. The photophysical properties of coumarin-120 in non-polar solvents were different from those in medium- to high-polarity solvents. In moderate- to higher-polarity solvents, the Stokes shifts, fluorescence quantum yields, and fluorescence lifetimes showed a more or less linear correlation with the solvent polarity function. In contrast, in non-polar solvents, all these parameters have unusually deviated.
Bizzarri and co-workers15 developed a new solvatochromic probe (14), which is structured as a “push–pull” system. The probe comprises an electron-rich naphthyl ring with an electron-poor benzothiazine group, which is conjugated to a central coumarin core. These features confer interesting spectroscopic and solvatochromic properties to the fluorophore.
In 2010, Signore and co-workers30 developed coumarin derivatives (15 and 16) having a donor-(coumarin core)-acceptor structure, with alkyl ether or naphthyl groups as the donor and benzothiazine and cyano groups as the electron acceptor. These compounds were almost non-emissive in water but showed strong fluorescence intensity in less polar media. These derivatives were attractive fluorophores because of having high quantum yields (ca. 0.90) and significant Stokes shifts (up to 160 nm). Based on their fluorescence properties, they can be engineered in response to the local environment polarity.
Castanheira et al.31 studied the photophysical properties of two coumarin derivatives (17 and 18) and their related dipeptides in varying polarity solvents. These coumarin derivatives were incorporated in different liposome formulations (e.g., egg-PC, DPPC, DPPG, and mixed DPPC/DPPG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)) and were mainly located in lipid bilayer in response to the transition between the rigid gel phase and the fluid liquid-crystalline phase.
1)) and were mainly located in lipid bilayer in response to the transition between the rigid gel phase and the fluid liquid-crystalline phase.
Recently, Sekar and colleagues32,33 constructed three coumarin–rhodamine hybrid derivatives (19). These derivatives showed different fluorescence properties according to the rigidity of their substituents in solvents with varying viscosity and polarity. All these derivatives showed excellent fluorescence with increasing polarity and viscosity of the solvent. 19-2 showed low sensitivity to viscosity but higher than that of 19-1. It was also found that high polarity and viscosity together induced emission enhancement more efficiently than only a high viscosity.
With the perspective of its use in protein studies, the fluorescence properties of 6,7-dimethoxy-coumarin (20) were investigated in Aerosol OT (AOT) reverse micelles at various water media using the time-dependent fluorescence shift method by Sykora et al.33 It was found that the compound showed a sensitive response to the surrounding polarity and AOT reverse micelles using TDFS. The polarity and/or hydration could be reflected using this relatively small probe. It can be a powerful tool to study proteins because the probe can combine with the unnatural amino acid approach or serve as a substrate of the hydrolase enzyme family.
ANS (1-anilino-8-naphthalene-sulfonate) and derivatives
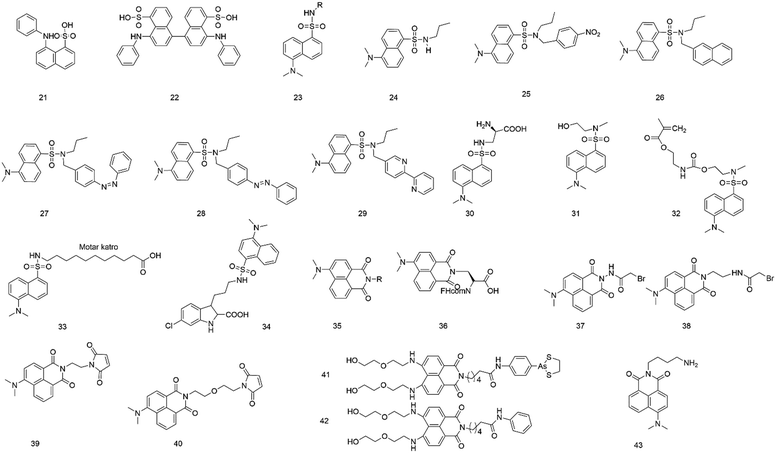
It is well-known that ANS (anilinonaphthalene-sulfonic acid) (21, Fig. 3) derivatives are solvatochromic fluorescent dyes that show maximal emission below 500 nm, but they have the ability to label proteins and image biological membranes.4 Upon binding to hydrophobic surfaces, probes with ANS show an increasing fluorescence.34–36 The fluorophore ANS also shows emission change in response to the medium polarity because photoinduced intramolecular charge transfer produces a polar excited state. It was found that ANS generally exhibits a low fluorescence quantum yield in an aqueous solution but becomes highly fluorescent in a non-polar solution.37In 1975, the fluorescence property of ANS (21) was studied after binding to bacterial luciferase, indicating that the fluorophore binding site in proteins was hydrophobic pockets.38 After binding to bacterial luciferase, the fluorophore changed both its maximum fluorescence excitation and the absorption from 353 nm to 370 nm. The fluorescence emission maximum changed from 540 to 480 nm, and the fluorescence quantum yield changed from 0.003 to 0.39.13
To investigate equilibrium folding intermediates (e.g., the molten globule state), ANS (21) and 1,1′-bis(4-anilino-5-naphthalenesulphonic acid) (bis-ANS, 22) were employed because the hydrophobic exposure increased after the formation of the latter.39 Due to the ability to bind to the intermediate state, these probes can be a powerful tool to investigate in a multiple-state folding transition of folding intermediates.
In 2003, Maitra and colleagues40 measured the dynamics of polarity-sensitive fluorophore ANS (21) partitioned between the bulk aqueous phase and hydrophobic pockets in a nonpolymeric hydrogel derived from tripodal cholamide. In the sol state, ANS showed faster rotational motion. However, restrained rotation was observed for ANS in the gel state because the sulfonate group of ANS bound with water molecules and hydrophilic parts of the gelator molecule. It was also demonstrated that the dynamics of ANS in the gel state would depend on the chemical nature of the dye.
Chitosan hydrogel beads have been widely used in biological and industrial fields, serving as interesting natural product-based materials that can accommodate a sizeable number of small molecules. Mandal et al.41 employed ANS (21) to study the details of the local environment when small guest molecules were sorbed by the hydrogel beads. Because of undergoing twisted intramolecular charge transfer, ANS was sensitive in response to the polarity of the medium.
To determine the critical aggregation concentration or critical micellar concentration of the bolalipid tetracosane-1,2,4-bis(phosphocholine), Blume's group42 selected 1,8-ANS and bis-ANS as probes, which can bind to the surface firmly and gave consistent results. Viles's research showed that 1,8-ANS and bis-ANS (21 and 22) could be used to report fiber formation kinetics. It showed that bis-ANS and ANS are appropriate dyes for the detection of oligomers and protofibers.
Dansyl amine
The fluorophore dansyl amine (23), an ANS derivative, is obtained by reacting dansyl chloride with amine groups. The fluorophore has several advantages: emission in the visible region, exceptionally high quantum yield of fluorescence, and significant Stokes shift avoiding autoabsorption effects.43 Also, its fluorescence intensity and emission maximum are found to vary with the polarity of the environment. Hence, it is widely used to label proteins and study microscopic interactions.44,45In 2000, Bright et al.46 synthesized and characterized three fluorescent dendrimers based on the dansyl amine group (23). They studied the photophysical properties of these dendrimers in an aqueous solution. They also investigated the host–guest interactions of the dendrimers, in which the dansyl residue was a guest, and β-cyclodextrin (β-CD) and polyclonal anti-dansyl antibodies were hosts. When the dendrimer generation increased, the dansyl residue was progressively shielded from the solvent, leading to marked changes in fluorescence behaviors.
To investigate the excited state behavior of fluorophores based on the dansyl amine group, Ceroni and colleagues47 synthesized several dyads (24–29) and studied their photochemical, photophysical, and electrochemical properties in acetonitrile solution. By changing the units linked to the dansyl amine group, the variation of features were observed, including sensitization of the dansyl fluorescence, quenching of the dansyl fluorescence by electron or energy transfer, and sensitized emission and reaction of the other components of the dyad. The results may provide a new design for supramolecular sensors, photoactive dendrimers and switching elements.
The ability to introduce fluorophores into proteins provides a powerful tool to study protein structure and function. In 2006, Schultz et al.48 reported the selective and efficient introduction of a low-molecular-weight fluorophore (dansyl-containing amino acid) (30) into proteins at defined sites of Saccharomyces cerevisiae by biosynthetic methods. The fluorescent amino acid was genetically encoded in Saccharomyces cerevisiae using an amber nonsense codon and an orthogonal tRNA-aminoacyl–tRNA synthetase pair. The fluorophore was introduced into human superoxide dismutase to monitor the unfolding of the protein upon the addition of guanidinium chloride. This strategy can facilitate studies of protein structure and function.
Buruiana's group49 discussed the fluorescence properties of two derivatives (31 and 32) based on dansyl amine. The results revealed that the fluorescence spectra of probe 31 showed a maximum at 440 nm and a small shoulder at 520 nm in DMF solution, but probe 32 showed a strong emission at 529 nm and had a shoulder located at 433 nm. In addition, the acid-sensing ability of these derivatives in DMF was tested by fluorescence titration with increasing HCl and toluene sulphonic acid (APTS) concentrations to acquire information about the mechanism of the fluorescence enhancement of “turn-on” chemosensors.
It is well known that the cytoplasm of cells is aqueous as a potentially significant barrier for many lipophilic drugs to reach their sites of action. The structures of poorly water-soluble fatty acids (FAs) in complex with fatty acid-binding proteins (FABPs) have been investigated and described. To describe the binding site of other lipophilic ligands (e.g., drugs), Scanlon et al.50 used an environmentally sensitive fluorophore (33), 11-dansylamino undecanoic acid (DAUDA), to investigate drug binding to FABP in human intestinal FABP (hIFABP). The choice of the fluorophore was based on the affinity of the fluorophore for FABP and the change in emission spectra after binding. Also, DAUDA could be replaced with most drugs that are bound to hIFABP. The studies showed that hIFABP could bind to different lipophilic compounds via various interaction modes at two separate sites. At the FA site, they bound with residues at the bottom of the hIFABP cavity via polar interactions. Also, they were characterized by favorable binding enthalpy, whereas at the portal site, the binding appeared to be largely driven by entropy via hydrophobic interactions.
To further explore the hydrophobic binding domain in the Mcl-1 protein surface, the Li lab developed a “turn-on” fluorescent probe (34) based on dansyl amine.51 The probe is composed of an inhibitory as a recognition group of Mcl-1 protein, a dansyl amine moiety as a fluorophore, and an alkyl chain with three carbon atoms as the linker moiety (Fig. 2B). It was found that the probe showed a selective binding affinity to Mcl-1 protein (Ki = 2.6 μM). In addition, the probe can provide a powerful tool to image Mcl-1 protein and monitor the cellular distribution of relating inhibitors in Mcl-1 protein. This can facilitate the studies of Mcl-1 protein.
Beck et al.52 designed and prepared an antibody–fluorophore conjugate (AFC) based on dansyl amine fluorophore (23).The method can be an analytical method at all stages of antibody–drug conjugate (ADC) discovery, preclinical and clinical development, and so on.
4-DMN (4-N,N-dimethylamino-1,8-naphthalimide)
4-DMN (35), is a yellow-green-emitting fluorophore, derived from 1,8-naphthalimides and then substituted in the C4 carbon atom with a dimethylamino moiety. It can emit strong fluorescence through a photoinduced electron transfer (PET) mechanism53 and has some advantages such as better chemical stability, longer excitation wavelength (408 nm), and facile synthesis.54 The fluorescence intensity is greatly enhanced in hydrophobic environments or non-polar solvents. The fluorescence quantum yield of this fluorophore is highly responsive to changes in the local solvent environment.55 Therefore, it is a well-known polarity-based fluorophore with unique spectral properties.To study protein–protein interactions, Imperiali and co-workers54 developed a fluorescent amino acid (36) based on polarity-based fluorophore 4-DMN. Its fluorescence quantum yield was highly sensitive to the local solvent environment. The fluorescent amino acid was incorporated into peptides through standard solid-phase peptide synthesis and was identified by calcium-activated calmodulin. Upon binding the probe with peptides, the enhancement of fluorescence emission was more than 900-fold. This provided a powerful tool for the study of protein–protein interactions.
In 2009, this lab55 reported a series of thiol-reactive agents (37–40) derived from 4-DMN. They evaluated and compared these 4-DMN derivatives with several commercially well-established solvatochromic fluorophores using the calcium-binding protein calmodulin system, leading to the monitoring of many critical factors, attributed to their successful application of investigating biomolecular interactions. Upon binding to calcium, a fluorescent calmodulin was produced, and then a greater than 100-fold enhancement of emission intensity could be observed.
In addition, Zhu et al.56 designed and synthesized 4-DMN-based fluorescent probes (41 and 42) as a tool for rapidly detecting and imaging vicinal dithiol-containing proteins (VDPs) both in vitro and in living cells. The 4-DMN fluorophores served as a directly environment-sensitive reporter to rapidly detect the location of VDPs. These results offer a non-invasive approach for identifying VDPs in situ and further studying their potential roles in cell function.
Peptoids, oligomers of N-substituted glycine, have been widely used as peptidomimetics and nanomaterials for study and diverse applications. Palla and colleagues57 reported an approach that exploited the exceptional environmentally sensitive fluorescence properties of 4-DMN to study peptide structure. The 4-DMN derivate (43) was a competent submonomer for peptoid synthesis, and it was incorporated sequence-specifically into the peptoids for this study. Upon modifying the peptides, their fluorescence emission intensity changed in response to their local polarity within a putative structure. This method for the study of peptide structure can be a powerful complement to existing analysis tools.
PRODAN and derivatives
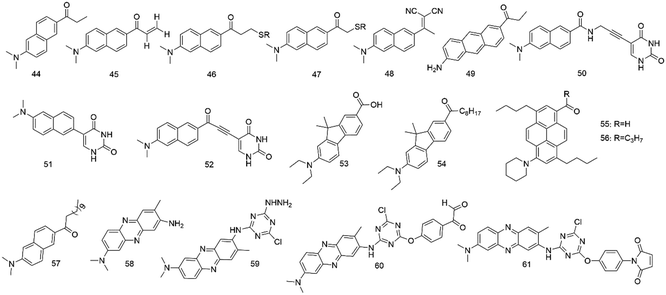
The physical and chemical properties of 6-propionyl-2-(dimethylamino) naphthalene (PRODAN) were first studied by Gregorio Weber in 1979.14 Studies revealed that the fluorescence of PRODAN (44, Fig. 4) was highly sensitive to solvent polarity. The maximum of emission varies with the solvent, extending from 401 nm (in cyclohexane solution) to 531 nm (in water solution). It was also found that these fluorophores showed the maximum emission in solvents of medium polarity (e.g., dimethylformamide), whereas a several-fold decrease was observed in cyclohexane. They also discussed the shift of these fluorophores in various solvents; the results further indicated that these fluorophores can be used as an environmental probe. Therefore, PRODAN fluorophores have increasingly attracted research interest as protein labeling agents.In 2013, Kaneshina et al.58 used PRODAN (44) and differential scanning calorimetry to examine the thermotropic phase behavior of diacylphosphatidylcholine (CnPC)-cholesterol binary bilayers (n = 14–16) by fluorescence spectroscopy. When PRODAN molecules were partitioned in a relatively hydrophilic region of the bilayer, the bilayer structure can vary with the changes in polarity around PRODAN, and the mentioned technique was helpful in visualization of the structural change. However, the conformational changes of the acyl chains were detected by scanning calorimetry.
Mukhopadhyay and co-workers59 used fluorescence spectroscopy to investigate the polarity-sensitive fluorescence probe PRODAN (44) binding with erythroid and nonerythroid spectrin. They found that both bound to PRODAN with high affinities (Kd, 0.50 and 0.17 μM). Studies of apparent dielectric constants revealed that the polarity of the binding site forms was highly hydrophobic.
Potter et al.60 synthesized 6-acryloyl-2-methylaminonaphthalene (ACRYLODAN) (45), which selectively labels thiol moieties in proteins with the acryloyl group. Upon reacting with thiols in protein, enhancement of fluorescence quantum yield for this agent was observed. The results showed that probe 45 was a powerful tool to study hydrophobic domains, dipolar relaxation processes, and conformation changes of the protein.
To obtain a consistent view of the nucleotide-induced conformational changes around Cys697 (SH2) and Cys707 (SH1) in skeletal myosin subfragment-1 (S-1), Hiratsuka and co-workers61 used two PRODAN fluorophores 46 and 47 to label the two thiols in S-1.
Barrio and colleagues62 reported another polarity-sensitive fluorophore (48). They found that it showed emission from 470 to 610 nm in hydrophobic and viscous surroundings due to its intramolecular rotational relaxation dependent on solvent polarity and viscosity. When using visible fluorescence microscopy, the auto-fluorescence from cells or tissues did not interfere with the fluorescence. In addition, in an aqueous environment, the fluorescence quantum yield was meager because of facile differentiation from the lipid or protein-bound state to the aqueous medium.
Twieg and colleagues63 synthesized Anthradan (49), a long-wavelength analogue of PRODAN, showing an additional spectral red shift that can avoid the autofluorescence of some biological systems. Anthradan displays polarity-sensitive emission and high quantum yields comparable to that of PRODAN. Furthermore, Anthradan has been successfully used for single-molecule level imaging.
To monitor various structures and dynamics of DNA and its surroundings, Saito and colleagues64 reported several fluorescent nucleosides with a uracil base and a fluorophore (50–52) tethered by rigid linkers. These probes had outstanding absorption and fluorescence emission spectra. The studies also showed that the fluorescence of nucleoside 48 with high CT character was very sensitive to solvent polarity. However, nucleoside 49 showed absorption and emission maxima with a longer wavelength because of the extension of the DNA conjugate system. These fluorescent nucleosides with unique absorption and fluorescence properties can be powerful probes to sensitively monitor the microenvironment changes around specific DNA sites.
Klymchenko et al.65 developed novel fluorescent dyes (53 and 54) by substituting PRODAN's naphthalene core with fluorene, showing excellent spectroscopic properties compared to PRODAN. Hocek et al.66 developed fluorene-linked nucleoside and DNA as a probe for sensing interactions. In 2013, Konishi and colleagues67 synthesized pyrene analogues of PRODAN. In apolar and polar solvents, they showed extremely high fluorescence quantum yields. In 2016, Klymchenko and colleagues68 reported a push–pull pyrene (PA, 55), changing its emission maximum as a lipid order function, but its spectroscopic properties outperformed those of Laurdan that was a PRODAN derivative. In 2020, they synthesized and characterized a push–pull dye PK (56) that was a ketone analogue of PA.69 The results showed that PA faded continuously and displayed emission color changes in long-term cell experiments, but PK exhibited a stable signal without emission color changes. In 2019, they also reported a push–pull dioxaborine probe, showing that its emission altered from the far red to the NIR region; it was used to map the polarity of biomembranes and lipid droplets under stress in cells.70
In phospholipid bilayers, Laurdan (57) displays solvent dipolar relaxation because there is a small number of water molecules at the membrane interface.71 Laurdan typically needs multiphoton excitation, so it is suitable for tissue imaging, such as whole and living zebrafish embryos. Gaus and colleagues72 employed Laurdan and di-4-aneppDHQ to study localized lipid environment imaging and membrane lipid order. The method directly examines cellular membrane organization in only around 4 h.
Neutral red
Neutral red (58), a lipophilic phenazine dye, is a photodynamic photosensitizer and has been widely used in various biological systems.73–75 The effects of polarity on the fluorescence properties of neutral red have been investigated in several neat and mixed solvents. The fluorescence behavior of neutral red strongly depends on the polarity of the solvents; its fluorescence maximum changes from 540 nm in pure dioxane to 637 nm in water. The fluorescence quantum yield increases from 0.17 in dioxane to 0.3 after the addition of 7% water, and then decreases, reaching 0.02 in pure water.73 Because of having two closely spaced electronic excited states, neutral red exhibited dual solvatochromic behavior. The fluorescence of neutral red was mainly emitted from the localized excited state in low-polarity solvents, whereas it was produced by a charge transfer state in high-polarity solvents. It was also demonstrated that the solvatochromic shifts affected the dipole moments of the localized and charge transfer states, the fluorescence quantum yields, and the lifetimes of neutral red.76The main application of neutral red is its use as a vital stain. In 1996, Santus and colleagues73 studied the lysosomal microenvironment with neutral red (58) living cells. Neutral red has been considered a typical lysosomotropic substance. Upon incubating human skin fibroblasts with neutral red, the maximum fluorescence immediately showed at the 575 nm region with excitation at the 35 nm region. It was found that this fluorescence originated from large fluorescent tracytoplasmic spots and was mainly distributed in the perinuclear region, implying staining of the endoplasmic reticulum–Golgi complex. As time goes by, this fluorescence shifted to 606 nm, demonstrating that the lysosomotropic substance gradually diffused to the more hydrated and acidic position inside lysosomes.
Because neutral red (58) has several advantages (e.g., rapidly penetrates root tissues, affinity to suberin and lignin, and accumulates in the vacuoles), it is also used to study various aspects of plant root biology using confocal laser scanning microscopy (CLSM).77 Upon short-term application of neutral red, the cellular actin cytoskeleton and the microtubule system and the dynamics of root growth were found to remain unchanged. Therefore, neutral red was useful for imaging proto- and metaxylem elements, casparian bands in the endodermis, and vacuoles in cells of living roots.
To detect the local polarity of the N-terminal domain in protein, Li and co-workers78 developed a polarity-sensitive fluorescent probe based on neutral red (59). The fluorescent probe included an s-triazine ring as a backbone, a neutral red moiety as a polarity-sensitive fluorescent reporter, and hydrazine as a labeling group. The results showed that the probe was stable in various solvents, had more prolonged emission at 550 nm, and led to maximum emission wavelength changes depending on solvent polarity only rather than pH and temperature. The probe was a powerful tool to firstly detect the N-terminal domain polarity in both heat-denatured and native R-lactalbumin.
In 2008, Ma's group75 synthesized a neutral red-based probe GGTDP (60) to label active-site arginine residues. The probe can detect the local polarity and conformational changes of the active site in rabbit muscle creatine kinase. In addition, in the same year, the authors79 devised another polarity-sensitive probe based on neutral red CMTDP (61) to study the local polarity and Cys121 domain of β-lactoglobulin.
Zhou et al.80 prepared an excellent biocompatibility probe based on neutral red for detecting Pt2+, Au3+, and Pd2+. The probe was used for bioimaging and biosensing of noble metal ions in vitro and in vivo. Pei et al.81 designed a specific light-up probe for i-motif DNA with neutral red (58). The results could provide a new method for researching the structures and functions of i-motifs, DNA-based anticancer drugs, and fluorescence sensing systems.
Nile red
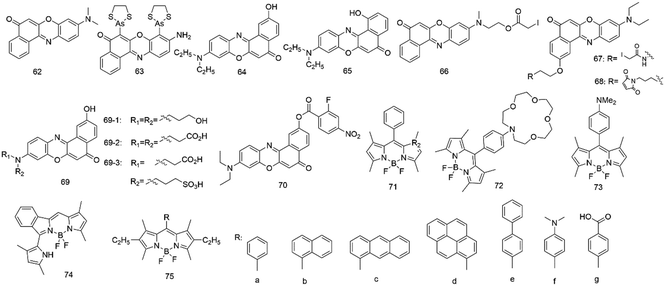
Nile red (62, Fig. 5) belongs to benzophenoxazine dyes, which show an intense color and have a lipophilic nature. However, Nile red has no formal charges and is photostable, but its fluorescence is highly susceptible to its environment polarity.82,83 The dye showed strong fluorescence and high quantum yield in apolar medium, whereas in water it was poorly soluble and shows almost no fluorescence. The dye also has a vast working wavelength range and is easy to remove from many biomolecules absorbed; in addition, its fluorescence is not influenced by pH (4.5 and 8.5).83,84 Therefore, it provides a powerful tool for lipid imaging and hydrophobic study sites on proteins.To study lipid droplets in normal cells and pathological states, Nile red (62) has been employed via fluorescence microscopy and flow cytofluorometry.85 Results showed that Nile red had better selectivity for cytoplasmic lipid droplets and emitted yellow-gold fluorescence, but not red fluorescence. Since then, Allen and colleagues86 used Nile red to detect cellular lipids in three species of Paramecium and Tetrahymena. The method was rapid and inexpensive due to the direct application of the dye to living or fixed cells.
In addition, Nile red (62) was also a valuable tool to probe hydrophobic sites on many native proteins.83 It can interact with many native proteins with hydrophobic binding domains such as β-lactoglobulin, κ-casein, and albumin, causing a wide range of spectral changes after interacting with different proteins. The dye can detect the exposure or formation of new hydrophobic surfaces upon binding to calmodulin, and oligomerization of melittin, or unfolding of ovalbumin during early thermal denaturation.
On the other hand, several Nile red-based derivatives have gradually emerged. In 2001, Tsien and colleagues87 employed a Nile red-based derivative (63) to probe the conformational change of proteins in live cells. This fluorescent probe can be attached to four cysteine residues in the recombinant protein. Upon response to its environment, the probe changed its fluorescence intensity, which provided monitoring of the conformational change for the proteins in live cells.
Since then, the fluorescence behaviors of the 2-hydroxy derivative (64) based on Nile red have been investigated in various solvents and micelles by Biczok et al.88 In apolar solvents, the extent of the vibronic coupling between the two lowest singlet excited states was suggested to govern the rate of the radiationless deactivation, whereas in polar medium, the hydrogen bonding with alcohols and the decrease of the energy gap between the singlet excited and ground states significantly accelerated the energy dissipation. It demonstrated that HONR was an outstanding probe for the study of micelles. In 2005, the same group89 studied the interaction of HONR with organic nitrogen compounds. They proved that HONR displayed high sensitivity to the basicity of hydrogen bond acceptors. The relative contribution of the competing reaction steps was very sensitive to solvent polarity and the basicity of the additive.
However, the fluorescence behavior of the 1-hydroxy Nile red derivative (65) was entirely different from that of the 2-hydroxy derivative.90 Deprotonation with a base was very difficult due to the strong hydrogen bonding between the 1-HO moiety and the heterocyclic nitrogen of the molecule, but the intramolecular proton transfer facilitated photoinduced tautomerization. The dye showed dual fluorescence in all solvents, and the relative intensity of the two bands was found to vary with the polarity and hydrogen bond donating power of the medium.
Two Nile red nucleosides were also synthesized by Okamoto's group in 2005 (ref. 91) and 2006.92 The absorption and fluorescence of these Nile red nucleosides both varied with the change of solvent polarity. The former was a β-C-2′-deoxyriboside based on Nile red as a powerful tool to detect the local polarity change and dynamics inside the DNA structure. However, the latter included a 2-hydroxylated Nile red derivative and 1,2-dideoxyglycan, which was used to detect the polarity change of the microenvironment surrounding DNA.
To obtain a long-wavelength fluorescent biosensor based on galactose and glucose binding protein (GGBP), Pitner and colleagues93 conjugated thiol-reactive Nile red derivatives (66–67) to GGBP proteins. Upon binding to the selected cysteine sites of GGBP, the Nile red derivatives showed fluorescence emission around 640–650 nm and changes in fluorescence intensity up to 50%.
In 2005, Moerner et al.94 employed a Nile red-based derivative (68) to study the sequence of local conformational changes caused by the bacterial chaperonin GroEL and its cofactor GroES which assist protein folding. The derivative was used to covalently label the single-cysteine mutant of GroEL (Cys261), locating its cysteine inside the central cavity at the apical region of the protein. This result showed that the emission intensity changes could help us to obtain insights into the sequence of conformational changes in GroEL-mediated protein folding.
It is well known that Nile red fluoresces with good quantum yields at about 530 nm in apolar solvents. Still, its fluorescence emission shifted to about 640 nm and its quantum yield dramatically decreased in polar solvents.84 However, Nile red has very poor solubility in aqueous media, limiting its use in labeling most biomolecules. Therefore, Burgess92 has designed and synthesized three Nile red derivatives (69) with water-solubilizing groups tending to show fluorescence with good quantum yields at around 640 nm in aqueous media. Compared to Nile red, derivative 69-1 with three hydroxyl groups was insoluble in aqueous media. However, the derivatives with dicarboxylic acid 69-2 and carboxylic/sulfonic acid 69-3 had excellent water solubility and fluoresced with good quantum yields in the 640 nm region in aqueous media.
In 2010, Klymchenko et al.95 developed a novel probe for cell plasma membranes with Nile red fluorophore. Unlike the parent Nile red, the new probe labels only the outer leaflet of cells and lipid vesicles through a unique switching property of Nile red. In 2014, a red fluorescent turn-on probe for GPCR imaging96 and a Nile red probe for SNAP-tagged plasma membrane proteins were developed.97 In 2019, Klymchenko's group98 developed probes based on Nile red for plasma membrane imaging. It combines ON/OFF switching, polarity sensing and target specificity, and the probe NR4A revealed the heterogeneity of lipid order at the cell surface and its connection with nanoscale membrane topology. In 2021, they also synthesized a series of Nile red probes for organelle imaging.99
Han et al.100 designed and synthesized a new fluorescent off–on probe (70) for detecting hydrogen polysulfides (H2Sn) selectively and sensitively with near-infrared (NIR) fluorescent Nile red dye. The probe responded to H2Sn in an aqueous solution with nanomolar LOD and without interference by relevant species. Endogenous H2Sn was imaged in living cells.
BODIPY
Boron-dipyrromethene (BODIPY, 71), a red-emitting fluorophore, has many advantages, including high fluorescence quantum yields (φf) values of 0.5–0.8 and a long excited emission wavelength of 500 nm. The fluorophore showed spectroscopically advantageous properties and high affinity, leading to fast and efficient charge transfer. Therefore, it has been used as a laser dye and labelled biomolecule. In addition, it has been incorporated in the radical ion pair of an electron-transfer probe leading to generated electric fields.101 BODIPY expressed high fluorescence, had sharp and narrow emission peaks and possessed reduced solvatochromic shifts in non-polar media.Daub's group101 synthesized a BODIPY dye with aza crown substituted (72) and its dimethylamino analogs (73). In a non-polar solvent, such as hexane, it was emitted from the locally excited (LE) state, however, from the charge-transfer (CT) state in more polar solvents. It was also found that the donor-substituted compounds indicate dual emission from the LE and CT states and had low fluorescence quantum yields.
In 2007, Nagano and co-workers102 synthesized a series of BODIPY derivatives with variously substituted at position 8 of benzene moieties and examined the solvent polarity dependence mechanism of the fluorescence ON/OFF and PET process for these fluorophores. In less polar media, the oxidation potential of the benzene moiety became more positive, and the reduction potential of the BODIPY fluorophore became more negative. Therefore, an enormous free energy change of PET from the benzene moiety could be observed in a non-polar environment. Based on these findings, these environment-sensitive probes were applied to label bovine serum albumin (BSA) and living cells, indicating that the polarity at the albumin surface was concluded to be similar to that of acetone, while the polarity at internal membranes of HeLa cells was identical to that of dichloromethane.
Another BODIPY-based fluorescent probe has been used to gain insight into the study of BSA. In 2014, Peng's group103 synthesized a boron-dipyrromethene (BODIPY) dye with a pyrrole ring (74) and studied its spectral properties in different solvents. It was found that the BODIPY-based dye was also sensitive to the solvent polarity and showed enhanced fluorescence emission in a less polar solvent. In an aqueous buffer, its fluorescence emission increased significantly at 622 nm but did not change at 575 nm after the addition of BSA. The BODIPY-based dye could serve as a BSA reporter due to the presence of suitable hydrophobic cavities for binding with the dye in BSA.
In 2018, the interaction between a series of BODIPY complexes (75) and BSA was investigated by Tomilova's group.104 They demonstrated that the fluorescence of these hydrophobic BODIPY-based dyes increases significantly in the presence of BSA. The reason is that these dyes bound with the subdomain IIA in the BSA hydrophobic cavities. Studies also showed that only the BODIPY dye with 8-phenyl substituted had the highest sensitivity and the fluorescence could be quantitated for BSA in an aqueous solution. Therefore, it could serve as a polarity sensor in the protein surface, and all the studied dyes with bulky aromatic groups could serve as hydrophobic molecule labeling agents for bioimaging.
Conclusions
The change and development of cellular polarity closely link with many cellular processes involved in membrane and protein structure and function, for example, cell differentiation and directional migration, membrane growth, molecule (e.g., drug) vectorial transportation, and immune response activation. The abnormal change of cellular polarity can be the feedback of function disorders and many related diseases. Consequently, an increasing number of polarity-sensitive (solvatochromic) probes have been developed and employed to rapidly monitor and detect the local polarity of a protein and the membranes in living cells.Herein, we summarized the fluorescence properties and advanced applications of several polarity-sensitive fluorescence probes. The ANS derivative dansyl amine and NBD and SBD are well-known fluorophores because of their small size, green-yellow emission wavelength, and significant Stokes shift. Nevertheless, their small fluorescence turn-on ratios and low selectivity also limited their application. Coumarin emits the blue-green region, and its derivatives have been widely used due to the remarkable Stokes shift and high quantum yield, but the short emission wavelength limited their use for animal imaging. ANS shows an emission maximum below the 500 nm region but can label proteins and image biological membranes. In addition, 4-MDN is also a yellow-green-emitting fluorophore and has better chemical stability; however, its PET effect may affect fluorescence emission. PRODAN is a hydrophobic fluorescent dye and shows a red shift from 401 nm to 531 nm with increasing local polarity. Neutral red shows an emission change from 540 nm to 637 nm with increasing local polarity, is actively absorbed by animal tissues and shows relatively nontoxic staining. Therefore, it has been widely used for staining cellular particles and in particular as a specific lysosomal polarity probe. Nile red is an uncharged hydrophobic and long-wavelength fluorophore and can show intense colors. Its solvatochromic properties are commonly used to examine the dynamics of membranes and quantitate lipid accumulation within cells. BODIPY dye is a red-emitting fluorophore and shows low background interference. However, a red-emitting fluorophore is difficult to synthesize and is unstable, thus limiting its use. It is worth noting that the different derivatives based on all the above fluorophores can show different fluorescence behaviors after conditions change.
It should be highlighted that a significant number of fluorophores and hybrid systems have been reported to develop better fluorescent probes for monitoring local polarity in the membranes and to detect conformational changes in proteins. There is no doubt that these fluorescent probes play a vital role in studying biochemical processes that occur during cell proliferation, embryonic development, and even diseases. Therefore, shortly, outstanding achievements in studying various function proteins and membranes using these polarity-sensitive fluorescent probes to further study related diseases are expected. These studies would provide insights into biological progress in cell biology, drug discovery, and therapeutic imaging.
Author contributions
All authors contributed equally to this work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The present work was supported by grants from the National Natural Science Foundation of China (No. 81673393 and 81874308), the Taishan Scholar Program at Shandong Province and the Shandong Natural Science Foundation (No. ZR2018ZC0233).Notes and references
- K. Kavallieratos, J. M. Rosenberg, W. Z. Chen and T. Ren, J. Am. Chem. Soc., 2005, 127, 6514–6515 CrossRef CAS PubMed.
- E. P. Petrov and P. Schwille, Springer Ser. Fluoresc., 2008, 6, 145–197 CAS.
- J. Y. Kwon, Y. J. Jand, Y. J. Lee, K. M. Kim, M. S. Seo, W. Nam and J. Y. Yoon, J. Am. Chem. Soc., 2005, 127, 10107–10111 CrossRef CAS PubMed.
- Z. Yang, J. Cao, Y. He, J. H. Yang, T. Kim, X. Peng and J. S. Kim, Chem. Soc. Rev., 2014, 43, 4563–4601 RSC.
- P. A. Russell, R. H. Pottier and D. P. Valenzeno, Photochem. Photobiol., 1994, 59, 309–313 CrossRef PubMed.
- M. A. Haidekker and E. A. Theodorakis, Org. Biomol. Chem., 2007, 5, 1669–1678 RSC.
- M. Monti, L. Brandt, J. Ikomi-Kumm and H. Olsson, Scand. J. Haematol., 1986, 36, 353–357 CrossRef CAS PubMed.
- Y. Saito, R. R. Desai and S. K. Muthuswamy, Biochim. Biophys. Acta, 2018, 1869, 103–116 CAS.
- D. G. Drubin and W. J. Nelson, Cell, 1996, 84, 335–344 CrossRef CAS.
- M. T. Butler and J. B. Wallingford, Nat. Rev. Mol. Cell Biol., 2017, 18, 375–388 CrossRef CAS PubMed.
- M. Li, J. Fan, H. Li, J. Du, S. Long and X. Peng, Biomaterials, 2018, 164, 98–105 CrossRef CAS PubMed.
- M. Simons and M. Mlodzik, Annu. Rev. Genet., 2008, 42, 517–540 CrossRef CAS PubMed.
- H. R. Pires and M. Boxem, J. Mol. Biol., 2018, 3521–3544 CrossRef CAS PubMed.
- G. Weber and F. J. Farris, Biochemistry, 2002, 18, 3075–3078 CrossRef PubMed.
- G. Signore, R. Nifosì, L. Albertazzi and R. Bizzarri, J. Biomed. Nanotechnol., 2009, 5, 722–729 CrossRef CAS PubMed.
- S. Uchiyama, K. Kimura, C. Gota, K. Okabe, K. Kawamoto, N. Inada, T. Yoshihara and S. Tobita, Chemistry, 2012, 18, 9552–9563 CrossRef CAS PubMed.
- A. S. Klymchenko, Acc. Chem. Res., 2016, 50, 366–375 CrossRef PubMed.
- A. P. Demchenko, K. C. Tang and P. T. Chou, Chem. Soc. Rev., 2012, 42, 1379–1408 RSC.
- Z. R. Grabowski and A. K. Rotkiewicz, Chem. Rev., 2003, 103, 3899–4031 CrossRef PubMed.
- S. Uchiyama, T. Santa, N. Okiyama, T. Fukushima and K. Imai, Biomed. Chromatogr., 2001, 15, 295–318 CrossRef CAS PubMed.
- S. Uchiyama, T. Santa and K. Imai, J. Chem. Soc., Perkin Trans. 2, 1999, 2, 2525–2532 RSC.
- A. J. Schadock-Hewitt, T. F. Bruce and R. K. Marcus, Langmuir, 2015, 31, 10418–10425 CrossRef CAS PubMed.
- Y. D. Zhuang, P. Y. Chiang, C. W. Wang and K. T. Tan, Angew. Chem., Int. Ed., 2013, 52, 8124–8128 CrossRef CAS PubMed.
- Z. Liu, T. Jiang, B. Wang, B. Ke, Y. Zhou, L. Du and M. Li, Anal. Chem., 2016, 88, 1511–1515 CrossRef CAS PubMed.
- F. Liu, H. J. Liu, X. J. Liu, W. Chen, F. Wang, R. Q. Yu and J. H. Jiang, Anal. Chem., 2017, 89, 11203–11207 CrossRef CAS PubMed.
- P. S. Song and W. H. Gordon, J. Phys. Chem., 1970, 74, 4234–4240 CrossRef CAS PubMed.
- B. D. Wagner, Molecules, 2009, 14, 210–237 CrossRef CAS PubMed.
- H. Pal, S. Nad and M. Kumbhakar, J. Chem. Phys., 2003, 119, 443–452 CrossRef CAS.
- R. F. Epand, R. Kraayenhof, G. J. Sterk, H. W. Wong Fong Sang and R. M. Epand, Biochim. Biophys. Acta, 1996, 1284, 191–195 CrossRef.
- G. Signore, R. Nifosi, L. Albertazzi, B. Storti and R. Bizzarri, J. Am. Chem. Soc., 2010, 132, 1276–1288 CrossRef CAS PubMed.
- A. S. Abreu, B. F. C. Hermenegildo, P. M. T. Ferreira, M. J. R. P. Queiroz and E. M. S. Castanheira, RSC Adv., 2016, 6, 72141–72148 RSC.
- N. Scholz, A. Jadhav, M. Shreykar, T. Behnke, N. Nirmalananthan, U. Resch-Genger and N. Sekar, J. Fluoresc., 2017, 27, 1949–1956 CrossRef CAS PubMed.
- A. Ghose, M. Amaro, P. Kovaricek, M. Hof and J. Sykora, Methods Appl. Fluoresc., 2018, 025005 CrossRef PubMed.
- O. B. Ptitsyn, R. H. Pain, G. V. Semisotnov, E. Zerovnik and O. I. Razgulyaev, FEBS Lett., 1990, 262, 20–24 CrossRef CAS PubMed.
- G. V. Semisotnov, N. A. Rodionova, O. I. Razgulyaev, V. N. Uversky, A. F. Gripas and R. I. Gilmanshin, Biopolymers, 1991, 31, 119–128 CrossRef CAS PubMed.
- R. Golbik, R. Zahn, S. E. Harding and A. R. Fersht, J. Mol. Biol., 1998, 276, 505–515 CrossRef CAS PubMed.
- D. Sarkar, RSC Adv., 2013, 3, 24389 RSC.
- S. C. Tu and J. W. Hastings, Biochemistry, 1975, 14, 4310–4316 CrossRef CAS PubMed.
- J. Bino, R. D. S. Patrick and K. L. Anil, Curr. Sci., 2011, 80, 287–290 Search PubMed.
- S. Mukhopadhyay, Ira, G. Krishnamoorthy and U. Maitra, J. Phys. Chem. B, 2003, 107, 2189–2192 CrossRef CAS.
- S. Basu, M. Dandapat, D. Ghosh and D. Mandal, Colloids Surf., A, 2014, 457, 196–202 CrossRef CAS.
- M. Kordts, A. Kerth, S. Drescher, M. Ott and A. Blume, J. Colloid Interface Sci., 2017, 501, 294–303 CrossRef CAS PubMed.
- J. G. Benito, A. Aznar and J. Baselga, J. Fluoresc., 2001, 11, 307–314 CrossRef.
- K. J. Shea, Y. Okahata and T. K. Dougherty, Macromolecules, 1983, 17, 296–300 CrossRef.
- V. Janout, M. Lanier and S. L. Regen, J. Am. Chem. Soc., 1997, 119, 640–647 CrossRef CAS.
- C. M. Cardona, J. Alvarez, A. E. Kaifer, T. D. McCarley, S. Pandey, G. A. Baker, N. J. Bonzagni and F. V. Bright, J. Am. Chem. Soc., 2000, 122, 6139–6144 CrossRef CAS.
- P. Ceroni, I. Laghi, M. Maestri, V. Balzani, S. Gestermann, M. Gorka and F. Vögtle, New J. Chem., 2002, 26, 66–75 RSC.
- D. Summerer, S. Chen, N. Wu, A. Deiters, J. W. Chin and P. G. Schultz, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9785–9789 CrossRef CAS PubMed.
- E. C. Buruiana, M. Zamfir and T. Buruiana, Rev. Roum. Chim., 2009, 54, 993–999 CAS.
- R. Patil, A. Laguerre, J. Wielens, S. J. Headey, M. L. Williams, M. L. Hughes, B. Mohanty, C. J. Porter and M. J. Scanlon, ACS Chem. Biol., 2014, 9, 2526–2534 CrossRef CAS PubMed.
- T. Liu, Y. Gao, X. Zhang, Y. Wan, L. Du, H. Fang and M. Li, Anal. Chem., 2017, 89, 11173–11177 CrossRef CAS PubMed.
- E. Wagner-Rousset, M. C. Janin-Bussat, O. Colas, M. Excoffier, D. Ayoub, J. F. Haeuw, I. Rilatt, M. Perez, N. Corvaia and A. Beck, mAbs, 2014, 6, 273–285 CrossRef PubMed.
- A. P. de Silva, A. Goligher, H. Q. Nimal Gunaratne and T. E. Rice, ARKIVOC, 2003, 229–243 Search PubMed.
- G. Loving and B. Imperiali, J. Am. Chem. Soc., 2008, 130, 13630–13638 CrossRef CAS PubMed.
- G. Loving and B. Imperiali, Bioconjugate Chem., 2009, 20, 2133–2141 CrossRef CAS PubMed.
- C. Huang, Q. Yin, W. Zhu, Y. Yang, X. Wang, X. Qian and Y. Xu, Angew. Chem., Int. Ed., 2011, 50, 7551–7556 CrossRef CAS PubMed.
- A. A. Fuller, F. J. Seidl, P. A. Bruno, M. A. Plescia and K. S. Palla, Biopolymers, 2011, 96, 627–638 CrossRef CAS PubMed.
- N. Tamai, T. Izumikawa, S. Fukui, M. Uemura, M. Goto, H. Matsuki and S. Kaneshina, Biochim. Biophys. Acta, 2013, 1828, 2513–2523 CrossRef CAS PubMed.
- M. Patra, M. Mitra, A. Chakrabarti and C. Mukhopadhyay, J. Biomol. Struct. Dyn., 2014, 32, 852–865 CrossRef CAS PubMed.
- F. G. Prendergast, M. Meyer, G. L. Carlson, S. Iida and J. D. Potter, J. Biol. Chem., 1983, 258, 7541–7544 CrossRef CAS.
- T. Hiratsuka, J. Biol. Chem., 1999, 274, 29156–29163 CrossRef CAS PubMed.
- A. Jacobson, A. Petric, D. Hogenkamp, A. Sinur and J. R. Barrio, J. Am. Chem. Soc., 1996, 118, 5572–5579 CrossRef CAS.
- S. J. L. Zhikuan Lu, H. Wang, W. E. Moerner and R. J. Twieg, J. Org. Chem., 2006, 71, 9651–9657 CrossRef PubMed.
- A. Okamoto, K. Tainaka, T. Unzai and I. Saito, Tetrahedron, 2007, 63, 3465–3470 CrossRef CAS.
- O. A. Kucherak, P. Didier, Y. Mely and A. S. Klymchenko, J. Phys. Chem. Lett., 2010, 1, 616–620 CrossRef CAS.
- D. Dziuba, P. Pospisil, J. Matyasovsky, J. Brynda, D. Nachtigallova, L. Rulisek, R. Pohl, M. Hof and M. Hocek, Chem. Sci., 2016, 7, 5775–5785 RSC.
- Y. O. Niko, S. Kawauchi and G. I. Konishi, Chem. – Eur. J., 2013, 19, 9760–9765 CrossRef CAS PubMed.
- P. D. Yosuke Niko, Y. Mely, G.-i. Konishi and A. S. Klymchenko, Sci. Rep., 2016, 6, 18870 CrossRef PubMed.
- E. K. J. Valanciunaite, H. Seki, D. Danylchuk, N. Peyriéras, Y. Niko and A. Klymchenko, Anal. Chem., 2020, 92, 6512–6520 CrossRef PubMed.
- A. P. A. Ashoka, Y. P. Kovtun and A. Klymchenko, J. Phys. Chem. Lett., 2019, 10, 2414–2421 CrossRef CAS PubMed.
- T. Parasassi, E. K. Krasnowska, L. Bagatolli and E. Gratton, J. Fluoresc., 1998, 8, 365–373 CrossRef CAS.
- D. M. Owen, C. Rentero, A. Magenau, A. Abu-Siniyeh and K. Gaus, Nat. Protoc., 2011, 7, 24–35 CrossRef PubMed.
- C. Sousa, T. S. e. Melo, M. Geze, J. M. Gaullier, J. C. Maziere and R. Santus, Photochem. Photobiol., 1996, 63, 601–607 CrossRef CAS PubMed.
- S. Y. Dong, H. M. Ma, X. J. Duan, X. Q. Chen and J. Li, J. Proteome Res., 2005, 1, 161–166 CrossRef PubMed.
- S. Wang, X. Wang, W. Shi, K. Wang and H. Ma, Biochim. Biophys. Acta, Proteins Proteomics, 2008, 1784, 415–422 CrossRef CAS PubMed.
- M. K. Singh, H. Pal, A. C. Bhasikuttan and A. V. Sapre, Photochem. Photobiol., 1998, 68, 32–38 CrossRef CAS.
- J. G. Dubrovsky, M. Guttenberger, A. Saralegui, S. Napsucialy-Mendivil, B. Voigt, F. Baluska and D. Menzel, Ann. Bot., 2006, 97, 1127–1138 CrossRef PubMed.
- S. Y. Dong, H. M. Ma, X. J. Duan, X. Q. Chen and J. Li, J. Proteome Res., 2005, 4, 161–166 CrossRef CAS PubMed.
- X. Wang, S. Wang and H. Ma, Analyst, 2008, 133, 478 RSC.
- W. Gao, H. Song, X. Wang, X. Liu, X. Pang, Y. Zhou, B. Gao and X. Peng, ACS Appl. Mater. Interfaces, 2018, 10, 1147–1154 CrossRef CAS PubMed.
- L. Xu, J. Wang, N. Sun, M. Liu, Y. Cao, Z. Wang and R. Pei, Chem. Commun., 2016, 52, 14330–14333 RSC.
- A. K. Dutta, K. Kamada and K. Ohta, J. Photochem. Photobiol., A, 1996, 93, 57–64 CrossRef CAS.
- D. L. Sackett and J. Wolff, Anal. Biochem., 1987, 167, 228–234 CrossRef CAS PubMed.
- C. M. Golini, B. W. Williams and J. B. Foresman, J. Fluoresc., 1998, 8, 395–404 CrossRef CAS.
- P. Greenspan, E. P. Mayer and S. D. Fowler, J. Cell Biol., 1985, 100, 965–973 CrossRef CAS PubMed.
- T. A. Cole, A. K. Fok, M. S. Ueno and R. D. Allen, Eur. J. Protistol., 1990, 25, 361–368 CrossRef CAS PubMed.
- S. R. Adams, R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis and R. Y. Tsien, J. Am. Chem. Soc., 2002, 124, 6063–6076 CrossRef CAS PubMed.
- K. Nagy, S. Goktiirk and L. Biczok, J. Phys. Chem. A, 2003, 107, 8784–8790 CrossRef CAS.
- K. Sebok-Nagy, Z. Miskolczy and L. Biczok, Photochem. Photobiol., 2005, 81, 1212–1218 CrossRef CAS PubMed.
- Z. Miskolczy, L. Biczók and I. Jablonkai, Chem. Phys. Lett., 2007, 440, 92–97 CrossRef CAS.
- K. Tainaka, Y. Fujiwara and A. Okamoto, Nucleic Acids Symp. Ser., 2005, 155–156 CrossRef CAS PubMed.
- A. Okamoto, K. Tainaka and Y. Fujiwara, J. Org. Chem., 2006, 71, 3592–3598 CrossRef CAS PubMed.
- K. J. Thomas, D. B. Sherman, T. J. Amiss, S. A. Andaluz and J. B. Pitner, Diabetes Technol. Ther., 2006, 8, 261–268 CrossRef CAS PubMed.
- S. Y. Kim, A. N. Semyonov, R. J. Twieg, A. L. Horwich, J. Frydman and W. E. Moerner, J. Phys. Chem. B, 2005, 109, 24517–24525 CrossRef CAS PubMed.
- O. A. Kucherak, S. Oncul, Z. Darwich, D. A. Yushchenko, Y. Arntz, P. Didier, Y. Mély and A. S. Klymchenko, J. Am. Chem. Soc., 2010, 132, 4907–4916 CrossRef CAS PubMed.
- L. A. Karpenko, R. Kreder, C. Valencia, P. Villa, C. Mendre, B. Mouillac, Y. Mély, M. Hibert, D. Bonnet and A. S. Klymchenko, ChemBioChem, 2014, 15, 359–363 CrossRef PubMed.
- L. R. E. Prifti, M. Umebayashi, R. Hovius, H. Riezman and K. Johnsson, ACS Chem. Biol., 2014, 9, 606–612 CrossRef PubMed.
- D. I. Danylchuk, S. Moon, K. Xu and A. S. Klymchenko, Angew. Chem., Int. Ed., 2019, 14920–14924 CrossRef CAS PubMed.
- D. I. Danylchuk, P. H. Jouard and A. S. Klymchenko, J. Am. Chem. Soc., 2021, 143, 912–924 CrossRef CAS PubMed.
- K.-B. Li, F.-Z. Chen, S. Zhang, W. Shi, D.-M. Han, C. Cai and C.-X. Chen, Anal. Methods, 2017, 9, 6443–6447 RSC.
- M. Kollmannsberger, K. Rurack, U. R. Genger and J. Daub, J. Phys. Chem. A, 1998, 102, 10211–10220 CrossRef CAS.
- H. Sunahara, Y. Urano, H. Kojima and T. Nagano, J. Am. Chem. Soc., 2007, 129, 5597–5604 CrossRef CAS PubMed.
- F. Song, Y. Xue, X. Wang, J. Wang, X. Xiong and X. Peng, Chem. Res. Chin. Univ., 2014, 30, 738–742 CrossRef CAS.
- O. S. Vodyanova, B. A. Kochergin, S. D. Usoltsev, Y. S. Marfin, E. V. Rumyantsev, E. L. Aleksakhina and I. K. Tomilova, J. Photochem. Photobiol., A, 2018, 350, 44–51 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |