Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microsensor systems for cell metabolism – from 2D culture to organ-on-chip
Jochen
Kieninger
 *,
Andreas
Weltin
*,
Andreas
Weltin
 ,
Hubert
Flamm
and
Gerald A.
Urban
,
Hubert
Flamm
and
Gerald A.
Urban
Laboratory for Sensors, IMTEK – Department of Microsystems Engineering, University of Freiburg, Germany. E-mail: kieninger@imtek.uni-freiburg.de; Tel: +49 761 203 7265
First published on 5th April 2018
Abstract
Microsensor systems for cell metabolism are essential tools for investigation and standardization in cell culture. Electrochemical and optical read-out schemes dominate, which enable the marker-free, continuous, online recording of transient effects and deliver information beyond microscopy and end-point tests. There has been much progress in microfluidics and microsensors, but the translation of both into standard cell culture procedures is still limited. Within this critical review, we discuss different cell culture formats ranging from standard culture vessels to dedicated microfluidic platforms. Key aspects are the appropriate supply of cells, mass transport of metabolites to the sensors and generation of stimuli. Microfluidics enable the transition from static to dynamic conditions in culture and measurement. We illustrate the parameters oxygen (respiration), pH (acidification), glucose and lactate (energy metabolism) as well as short-lived reactive species (ROS/RNS) from the perspective of microsensor integration in 2D and 3D cell culture. We discuss different sensor principles and types, along with their limitations, microfabrication technologies and materials. The state-of-the-art of microsensor platforms for cell culture is discussed with respect to sensor performance, the number of parameters and timescale of application. That includes the advances from 2D culture to the increasingly important 3D approaches, with specific requirements for organotypic microtissues, spheroids and solid matrix cultures. We conclude on the current progress, potential, benefits and limitations of cell culture monitoring systems from monolayer culture to organ-on-chip systems.
Introduction
The human body with its 1014 cells is a highly complex system often hindering the direct research on a specific organ function at the cellular level. Research on its physiology and pathophysiology asks for a sufficiently simple system of subunits. Classically, cells cultivated in monolayer culture (2D) are used for this purpose with both primary cells from donors or patients and immortal cell lines. On one hand, it became clear that cellular heterogeneity demands for the investigation on single cells, which themselves not always represent features of the whole tissue. On the other hand, the highly artificial situation with the absence of the third dimension and therefore typically also the absence of concentration gradients within the 2D cell population drove the demand for 3D in vitro models. Co-cultures of different cell types allow modeling of some basic interactions between different cell types. However, there is still a large gap between the classical 2D/3D cell culture systems and functional units in the human body. Therefore, organ-on-chip systems are developed combining culture of cells with the promise of organ-like functionality.1,2 Here, the term organ-on-chip is used for any chip-based 3D culture model with organotypic functions. Within the realm of in vitro models, from 2D culture to organ-on-chip, many aspects of human physiology and pathophysiology can be modeled and investigated. In particular, research can be conducted that would not at all be possible in humans from an ethical point of view. Besides the benefit from reduced complexity, in vitro experiments generally require much lower effort and cause substantially lower cost. While animal experiments are often seen as the link between in vitro models and humans – independent of ethical concerns and high cost – those results can only be transferred to a certain extent keeping in mind that all species-specific mechanisms cannot be seen. Here, in particular, organ-on-chip approaches could bring cell culture models closer to the human than animal models ever could be.In order to obtain relevant information from such in vitro models, it is important to acquire the actual state of the cells. Optical, continuous observation of cell morphology provides only basic information about the cellular state. Staining methods reveal more and also intracellular details. Typically, staining interferes with the cells, often needs fixation and thus can only provide end-point data. To address the cellular metabolism itself, observation of the living cells is needed, which is preferably carried out by accessing the small molecules involved in the metabolism (oxygen, glucose, lactate etc.) with microsensors. The usage of such sensors allows the continuous recording of transient mechanisms, which is often called metabolic monitoring. This is especially helpful in scenarios in which recovery or cyclic effects could be overseen if only end-point data is used.
Research using in vitro models along with the ability of metabolic monitoring can be driven by many different motivations. In fundamental research, the knowledge of metabolic functions can be of primary interest. In other fields, such as the study of gene expression or drug response, the metabolic state often determines the results, and thus, monitoring of metabolic parameters essentially contributes to the standardization of cell culture experiments.
Pharmacodynamics studies, drug testing and compound screening are all applications in which the metabolic monitoring provides indicating parameters for specific pharmacological interactions. Results from in vitro cultivation of patient material in personalized medicine can benefit from microsensor readings, as within the very limited time frame of typical clinical scenarios a large quantity of information can be obtained. In all these different fields of application, the in vitro models in combination with microfluidics and sensors provide a rather simple possibility to parallelize the experiments in a reproducible manner. This allows both a detailed parameter study and collection of sufficient data for good statistics. Many systems claim to provide easy scale-up for parallelization. However, it is rarely shown, and in most cases it would be technically challenging.
In our opinion, the available tools benefit a lot from great progress in microfluidic systems, while the application of microsensors therein still lags behind. Critically, this is particularly true for 3D cell cultures, although several promising approaches have been proposed.
Within this critical review, we discuss the different sensor principles for the most relevant metabolic parameters as well as microfabrication technologies and materials with respect to their applicability in mammalian cell culture monitoring systems. Then, we review the different systems described in the literature and commercial products. The key aspects of those systems are:
• How are the cells cultured?
• How do the metabolites come to the microsensors?
• Whether and how are stimuli generated?
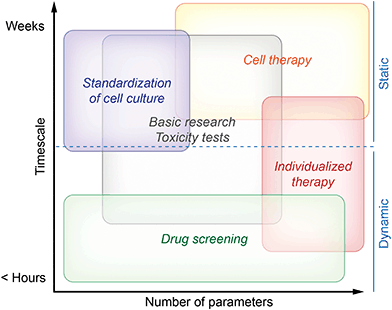
Monitoring systems can be separated into dynamic systems comprising microfluidics and static systems with stagnant medium following the routine procedure in cell culture. Depending on the field of application, either the culture system should provide results very fast, or microsensors should be able to measure for longer time, possibly without the need for recalibration. Another important aspect is the number of different sensing parameters provided by the system. In Fig. 1, we summarize the needs for the different fields of application. The fields are clustered depending on the desired measurement duration and on the number of measured parameters from single to multiparameter monitoring. Drug screening systems should be optimized for acquisition of results within minimal time, while standardization of cell culture typically addresses many cell cycles resulting in demands for stable sensors over weeks.
 | ||
| Fig. 1 Classification of cell culture monitoring systems by the needs of different fields of application. | ||
Fig. 1 describes both needs which are essential in order to obtain meaningful results and needs describing the optimal case. This is especially true for the field of cell therapy. While successful measurements are possible with less capable systems, we think that cell therapy will be the field with the highest demand for long-term stability and number of parameters in the future.
We do not comprehensively treat metabolic monitoring in bioreactors because of its different environment for sensors, resulting in completely different aspects for size, integration, stability and functionality. However, some works describing microsensors dedicated to bioreactors are applicable to mammalian cell culture and organ-on-chip systems as well and were therefore included. An overview of selected cell culture monitoring systems is given in Table 1.
| Name | Cell culture type | Material | Microfluidics | Parameters | Sensing principle | Notable features | Ref. |
|---|---|---|---|---|---|---|---|
| Cytosensor | 2D, dynamic | Silicon | Yes | pH | LAPS | Commercial | 108 |
| Oxygen | Amperometric (Pt) | 109, 110 | |||||
| Glucose, lactate | Amperometric biosensor (Pt) | 110 | |||||
| Sensing Cell Culture Flask | 2D, static | Glass | No | Oxygen | Amperometric (Pt) | Standard culture vessel | 28, 107 |
| pH | Potentiometric (iridium oxide) | 107 | |||||
| Glucose, lactate | Amperometric biosensor (Pt) | 107 | |||||
| Superoxide | Amperometric (Au) | 76 | |||||
| NO | Differential pulse voltammetry | 143 | |||||
| (Au) | |||||||
| Boero et al. | 2D, static | Silicon | Yes | Glucose, lactate | Amperometric biosensor (Au) | External biosensors | 102 |
| Presens OxoDish | 2D, static | Polymer | No | Oxygen | Luminescence | Commercial, standard format | 32 |
| Seahorse Bioscience | 2D/3D, static/dynamic | Polymer | No | Oxygen, pH | Fluorescence | Commercial, standard format, dip-in approach | 55 |
| Bionas SC1000 | 2D, dynamic | Silicon | Yes | pH | ISFET | Commercial | 56, 115 |
| Oxygen | Amperometric (Pd) | ||||||
| Adhesion | Impedance (IDES) | ||||||
| MetaScreen | 2D, dynamic | Glass | Yes | Oxygen | Amperometric | Downstream biosensors on-chip | 57 |
| pH | Potentiometric (iridium oxide) | ||||||
| Glucose, lactate | Amperometric biosensor | ||||||
| Misun et al. | 3D spheroid, dynamic | Glass | Yes | Glucose, lactate | Amperometric biosensor (Pt) | Hanging-drop network, in situ biosensors | 19 |
| Bavli et al. | 3D spheroid, dynamic | Glass/PDMS | Yes | Oxygen | Luminescence (tissue embedded) | Commercial, external downstream biosensors | 133 |
| Glucose, lactate | Amperometric biosensor | ||||||
| Zhang et al. | 3D spheroid, dynamic | Glass | Yes | pH | Optical absorption | Multiple sensor units, fluidic breadboard | 135 |
| Oxygen | Luminescence | ||||||
| Immunosensors | Cyclovoltammetric (Au) | ||||||
| Weltin et al. | 3D spheroid, static | Polyimide | No | Oxygen | Amperometric (Pt) | Standard culture vessel, in situ biosensors, dip-in approach | 18 |
| Lactate | Amperometric biosensor (Pt) | ||||||
| Domansky et al. | 3D, scaffold-based, dynamic | Polymer | Yes | Oxygen | Luminescence | On-chip pumps, dip-in approach | 144 |
Cell metabolism and cellular microenvironment
Metabolic pathways – energy metabolism and relevant parameters
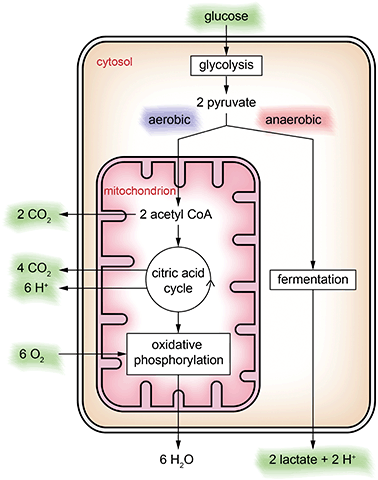
In general, metabolic pathways are quite complex and consist of catabolic pathways (breaking down molecules such as nutrients) and anabolic pathways (building up larger molecules from small building blocks such as proteins). In the context of metabolic cell culture monitoring, usually the focus is on the catabolic pathway of glucose only, which is the major energy source for cellular activity. In cell culture, glucose is often the only provided nutrient in the medium.Keeping in mind that this is a severe abstraction, we focus on the simplified energy metabolism of glucose as illustrated in Fig. 2 for the discussion of the cell culture monitoring systems. In the first step, glycolysis, glucose is broken down into pyruvate molecules. Depending on oxygenation around the cell, pyruvate is further catabolized following an aerobic or an anaerobic pathway. Because of its overall higher efficiency (the amount of energy generated per glucose molecule), the aerobic pathway is preferred.
In the cytosol, glycolysis occurs, in which the six-carbon sugar glucose is broken down into two three-carbon sugars and converted further into two pyruvate ions. In the case of sufficient oxygen, pyruvate is oxidized within the mitochondria to acetyl-CoA releasing CO2. Acetyl-CoA is the starting point of the citric acid cycle, in which further energy is generated, with CO2 and protons as by-products. Downstream of the citric acid cycle, in the oxidative phosphorylation, further energy is harvested by reduction of oxygen to water. If oxygenation is not sufficient, oxidative phosphorylation cannot occur, and the citric acid cycle stops, resulting in accumulation of pyruvate ions in the cytosol. In this case, the anaerobic pathway, the fermentation of pyruvate into lactate and release of a proton, sets in.
Cell culture monitoring using microsensors per se can measure extracellular substances only. Except for cultivating isolated mitochondria, only the substances marked green in Fig. 2 can be accessed directly. The individual steps of the energy metabolism of glucose can only be seen indirectly.
Glucose as the major energy source in cell culture is available in the cell culture medium in high concentrations up to 10 mM or more. Physiological levels are typically lower (few mM), but glucose is not the sole energy source in vivo. In cell culture, glucose levels decrease depending on medium volume, cell density and metabolic activity and often reach values below the mM range before the medium is exchanged.
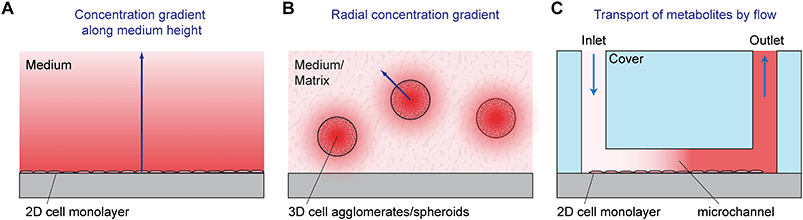
The concentration of dissolved oxygen is determinant for many different biological processes.3 Aerobic cell culture conditions (e.g. 5% CO2, 95% air) result in dissolved oxygen concentrations in the range of 200 μM. Depending on the type of culture, pericellular values are much lower because of the formation of diffusion gradients (see Fig. 3A). Typical oxygen concentrations in healthy tissue are between 20 and 150 μM. Pathologically low oxygen conditions (hypoxia) can be found, depending on the origin of the cells, at 25 μM or lower.4 In cell culture, it is important to establish a well-defined pericellular oxygen concentration in order to ensure the reproducibility of the desired cellular state. In situations with limited medium volume or the possibility of formation of gradients, cellular respiration rates can be accessed. Typical rates are in the range of 200 fmol h−1 per cell (T-47D breast cancer cell line).5 The aspect of oxygenation and oxygen control in cell culture has been discussed recently in more detail in this journal.6,7
Lactate ions are the end product of the anaerobic pathway. Initially, the cell culture medium contains no or, in case serum was used to prepare the medium, up to a few mM lactate. Under typical culture conditions, lactate concentrations can rise up to few mM, usually not higher than half of the initial glucose concentration in the medium. Taking into account the limitations of the simplified model described in Fig. 2, the fraction of twice the number of lactate molecules produced by the number of glucose molecules consumed can be used as a measure for the fraction of the anaerobic pathway. In a more detailed view, it should be considered that, depending on the cell type, lactate can be used as an energy source or even metabolized to glucose, such as when liver cells convert lactate produced in the muscles back to glucose (Cori cycle).
Dissolved CO2 gas and protons (pH) are interlinked by the dissolution of CO2 and formation of bicarbonate ions (eqn (1)). Therefore, monitoring of pH is often preferred over measuring dissolved CO2 gas in cell culture monitoring applications.
| CO2 + H2O ⇌ H2CO3 ⇌ HCO3− + H+ | (1) |
Additionally, the pH in a typical cell culture medium is stabilized by sodium bicarbonate and cultivating the cells in an incubator atmosphere containing CO2 gas (e.g. 5%), which results in a strong buffer system following eqn (1). The pH of the typical cell culture medium is adjusted to 7.4, decreasing over time due to cell metabolism to values not much below pH 7 under appropriate culture conditions. In case cellular acidification should be accessed and measured as acidification rates with microsensors, a small total medium volume along with a weakly buffered medium is required.
Short-lived, reactive species
Along with the stable products of and educts for cellular metabolism, short-lived and reactive species play a role in cell metabolism, namely reactive oxygen (ROS) and reactive nitrogen species (RNS). ROS include both oxygen radicals as superoxide (O2*−) or hydroxyl radicals (OH*) and non-radical derivatives such as hydrogen peroxide (H2O2). The reactive nitrogen family includes nitrogen-derived radicals like the initial nitric oxide (NO*) and related products nitrogen dioxide (NO2*) as well as the non-radical peroxynitrite (ONOO−). Among the ROS and RNS species, superoxide (O2*−) and nitric oxide (NO*) radicals are of primary interest. Besides their biological role, identification from cell/tissue release upon drug stimulation might be more straightforward, compared to follow-up reaction products, which makes the identification of cause–effect relationships more reliable.Cellular microenvironment in different culture formats
Most traditional cell cultures for adherent cells have been done in planar arrangements. In such 2D cell culture systems, cells are seeded as individual cells into a cell culture flask, dish or well plate. Coating of the surface with appropriate proteins can be performed prior to seeding in order to facilitate cell adhesion, which is often an important prerequisite for cell division and physiological behavior of the cells. After seeding, cells sediment and adhere to the bottom of the vessel. A stagnant layer of cell culture medium is present above the cells with a typical height in the mm to cm range. The media contain nutrients, can take up waste products and act as a diffusion barrier for dissolved gases provided by the incubator atmosphere. This situation results in gradients along the medium height (see Fig. 3A for an illustration).For parameters like glucose, the medium acts as a finite source; the cells are the sink. In the case of lactate and other waste products, the only source is the cells. For those parameters with a finite source or with just cells as the source, measurements at any position along the medium height of the 2D cell culture provide qualitatively meaningful results. However, to obtain quantitatively correct readings, a defined sensor position next to the cells is necessary. In contrast, for parameters like oxygen, the cells act as a sink, and the top surface of the medium (the incubator atmosphere) is an infinite supply. The established gradients and therefore the oxygen concentration next to the cells (pericellular) depend on the aerobic cell metabolism, which may be a function of the pericellular oxygen concentration itself.5 In this situation, it is essential to have sensors next to the cells in order to obtain meaningful sensor readings.
Three-dimensional (3D) cell cultures gain popularity because they resemble the in vivo situation more closely and show more organotypic properties than 2D cultures.8–10 Such properties make them promising models for drug and toxicity screening, where microphysiometry has been applied in 2D to measure cell metabolism, and also in tissue engineering, cancer research and multi-organ modeling. In 3D culture, cells are typically either mutually adherent but non-substrate adherent aggregates,11e.g. spheroids,11,12 in a scaffold-free medium or mutually adherent and embedded within a gel/solid matrix, such as Matrigel13 or other scaffolds. Today, spheroids are one of the most used 3D cell culture models, also in microfluidic systems. They can be formed in different ways which include:11,12 aggregation of cells by gravity in hanging droplets at the air/liquid interface; use of vessels, reactors or labware with non-adhesive/cell-repellent surfaces and shaking, spinning, rotating or centrifuging such vessels; employing microwells or molds made from non-adhesive materials; growth within or around a specific matrix/hydrogel, bead or emulsified droplet. The typical spheroid size is in the range of several hundred micrometers. Gradients for metabolic reagents and products can be found in the matrix outside the spheroid. Additionally, and usually dominant, there are gradients which occur within the spheroids themselves, as illustrated in Fig. 3B. Oxygen, for example, depletes along the radius towards the center. Assuming appropriate culture conditions, it is possible to achieve neighboring layers with normoxic and hypoxic states, which are an essential model system in tumor research.14 Spheroids can be understood as an inverse model of the tumor geometry: in tissue, oxygen and nutrients' concentrations drop radially along the distance from a blood vessel in the center, whereas in a spheroid, the lowest oxygen and nutrients' concentrations can be found in its center. Especially for the method of hanging droplets, microfluidic systems have been applied to enable parallelization and enhance reproducibility.15–17 Access to metabolic parameters using microsensors is often limited to the medium surrounding the spheroid.18,19 Measurement of values or even tracing of gradients inside the spheroids can hardly be achieved by microsensors as typical sensor geometries are in the same size as the diameter. Although the penetration of the spheroid with sensors in the format of a microneedle is feasible,20–22 the results would strongly be influenced by the physical presence of the sensor.
Other 3D cell culture formats comprise cells growing equally distributed in a 3D cell culture matrix in a tissue-like manner, sometimes also comprising vessel-like structures to mimic vascularization.23,24 In such systems, sensors can be used to provide concentration readings at distinct points, e.g. outside the matrix and in the middle.
Microsensors can also be used to monitor suspension cell culture, however these systems typically relate to the field of bioreactors, which is not covered by this review.
Sensors and relevant parameters
Different metabolic parameters can be classified by the lifetime of the species and the presence or absence of strong gradients in the cell culture system. Both species with short lifetime (e.g. ROS, RNS) and substances with strong gradients, i.e. the source and sink within the cell culture (e.g. oxygen), require sensors located next to the cells (pericellular monitoring). Other parameters which are stable and consumed or produced by the cells only (such as glucose and lactate) are less critical regarding the spatial position of the sensor in relation to the cells. In a microfluidic system, these parameters could also be monitored downstream resulting in the same information about the cellular metabolism compared to readings from pericellular sensors. A summary of commonly used sensor types and relevant ranges for the major metabolic parameters can be found in Table 2.| Parameter | Sensor types | Relevant range in cell culture | Analyte consumption |
|---|---|---|---|
| Oxygen | Direct amperometric sensor | 0–200 μM (0–20 kPa) | Yes |
| Clark-type sensor | Depends on the principle | ||
| Optical sensor (luminescence/fluorescence) | |||
| No | |||
| pH | Potentiometric sensor | 5–8 | No |
| Optical sensor (luminescence/fluorescence) | No | ||
| Lactate | Electrochemical biosensor | 0–15 mM | Yes |
| Optical biosensor | Yes | ||
| Glucose | Electrochemical biosensor | 0–25 mM | Yes |
| Optical biosensor | Yes |
Respiration, oxygen
It is important to know the absolute oxygen concentration because it is crucial for the aerobic or anaerobic pathways in cell metabolism. Additionally, the respiration rate (the change in oxygen concentration over time, typically referred to the cell number, e.g. in fmol h−1 per cell or in relative units) can be used as a measure for the activity of the aerobic cellular metabolism.Electrochemical oxygen sensors are based on the reduction of molecular oxygen at a noble metal electrode (usually platinum). Depending on whether the electrode is directly in the cell culture medium, those sensors are called direct amperometric sensors or, in case there is an independent sensor electrolyte separated by a gas-permeable membrane, they are called Clark-type sensors. It has to be noted that sometimes in the literature the term “Clark-type” is used for all types of electrochemical oxygen sensors. The advantage of the Clark-type arrangement is the independence of the electrode reactions from interfering or electrode poisoning substances from the cell culture medium, but at the cost of higher technological effort.25–27 Application of chronoamperometric protocols comprising cleaning steps enables stable measurements in cell culture medium including serum also with direct amperometric sensors.28 For both types of amperometric oxygen sensors, the transfer function is linear assuming sufficient diffusion limitation within the sensor membrane. Typically, the offset of such sensors is close to zero allowing one-point calibration at the beginning of the cell culture experiment, which suits the conventional cell culture routine much better than all other sensing methods requiring at least two different oxygen concentrations during the calibration procedure. A variant of the amperometric oxygen sensor is to use the local pH change caused by the oxygen reduction process to generate the measurement signal using pH-sensitive field effect transistors.29,30 A major advantage of this approach is the possibility to use the same readout circuitry as that used for pH sensing.
Potentiometric sensors with their inherent logarithmic transfer function (following the Nernst equation) would often fit better to the possible oxygen concentration ranges.3 However, to our knowledge, no potentiometric oxygen sensor has been applied successfully to monitor dissolved oxygen concentration in cell culture.
Optical oxygen sensors are based on fluorescence quenching in an appropriate dye by molecular oxygen.31 Measurement principles include the evaluation of fluorescence amplitude, fluorescence lifetime or phase shift in the case of modulated excitation light. For all principles, the transfer function follows the non-linear Stern–Volmer relation. This relation states that the output of the sensors is inversely proportional to the concentration plus a constant. This means that there is no clear zero-point and therefore two-point calibration is needed. The fluorescent dye can be embedded in a polymer as a single sensor spot or spread in a membrane coating on the whole surface of the cell culture vessel in order to measure spatial changes in oxygen concentration.32,33 In both cases, fluorescence excitation and readout can be carried out directly using LEDs or coupled through an optical fiber outside transparent cell culture vessels. This is a big advantage with respect to easy handling and sterilization of the cell culture vessels and allows separation of the single-use sensor spot from the reusable optical setup. Similar sensor spots can be read out with an inverted microscope.34
Acidification, pH
Potentiometric pH sensors, namely the pH glass electrode, are the most widely used sensors for proton activity. However, for application in cell culture, their size limits their usage during medium preparation. Only in bioreactors such electrodes are commonly applied. For cell culture monitoring, either light-addressable potentiometric sensors (LAPS), ion-selective field effect transistors (ISFET), metal oxide-based potentiometric sensors or optical sensors are used to monitor pH or the acidification rate (decrease of pH over time). The LAPS principle comprises a pH-sensitive insulation material like silicon oxynitride or Ta2O5 as the sensing element exposed to the electrolyte. Activation of charge carriers by LED or laser causes a photocurrent transducing the pH to an electrical quantity.35 While early cell culture monitoring devices36 measured acidification only using LAPS, this sensor principle was employed in recent multiparameter systems as well.37,38 ISFETs with a pH-sensitive gate material are among the smallest possible pH sensors employed in cell culture.39,40 Both LAPS and ISFET are limited to the availability of silicon as the substrate including CMOS circuits (see also the section on microfabrication and materials). Fanigliulo et al. compared LAPS- and ISFET-based sensors to access cellular metabolism.41Potentiometric pH sensors based on metal oxides, such as iridium oxide,42–46 tungsten oxide47–49 or ruthenium oxide,50–52 can also be used on transparent substrates and are often preferred because of their simple integration. However, the overall accuracy is also dependent on the stability of the reference electrode. Optical pH sensors are based on fluorescence quenching at alkaline pH, but usually need, in contrast to optical oxygen sensors, a second reference dye (dual lifetime referencing).53
LAPSs, ISFETs and potentiometric sensors have a logarithmic transfer function referring to the proton concentration (i.e. linear with pH), while optical pH sensors follow a more complex non-linear function.54 Both transfer functions are well suited for larger pH changes. The case of typical changes within cell culture of less than one pH unit requires very stable sensor signals over days. In order to stimulate faster changes as response to a stimulus, a medium with low buffer capacity can be used.55–57
Biosensors: glucose, lactate, glutamate, pyruvate
Electrochemical biosensors dominate the field of enzyme-based biosensors. Among them are widely used first-generation biosensors with an enzyme immobilized in a membrane or matrix directly on the electrode. The enzyme converts the analyte by generating a by-product, which is oxidized or reduced at an appropriately polarized electrode. Often, oxidase enzymes (such as glucose oxidase, lactate oxidase, glutamate oxidase or pyruvate oxidase)57–59 are used resulting in the by-product H2O2, which is oxidized at a noble metal electrode (usually platinum). Common to all those sensors is the consumption of oxygen during the enzymatic conversion in equimolar concentration as the analyte. This is especially critical in the case of glucose (concentrations in the mM range) in hypoxic cell culture (oxygen concentration can drop down to 10 μM or below). This limitation can be overcome by taking advantage of the faster diffusion of oxygen compared to glucose and usage of a membrane enabling strong diffusion limitation but resulting in a lower sensitivity of the sensor. The use of a blank electrode with the same membranes but without the enzyme is advisable in order to exclude any background signal from species oxidized at the potential chosen for the oxidation of the by-product of the enzymatic reaction. Sensors based on oxidase enzymes release H2O2, also into the cell culture. For analytes with high concentrations (glucose, lactate), this can lead to adverse effects of H2O2 on the cells. In case spatial separation of the sensors from the cells as in microfluidic systems is not possible, an additional membrane containing catalase, which decomposes H2O2 to oxygen and water, can be applied on top of the enzymatic sensor.58 Recent works also describe the application of optical biosensors for glucose and lactate for cell culture applications.60 Compared to electrochemical biosensors, until now, they have not played an equally important role in metabolic monitoring in cell culture.Sensors for short-lived species
The measurement of reactive oxygen (ROS) and reactive nitrogen species (RNS) in a cellular environment remains a challenge, even with the utilization of microsensor approaches in combination with electrochemical sensing techniques. General considerations on sensor/measurement design should include the complex nature of these molecules. Inherently, ROS and RNS are often molecules acting on short distance with a short lifetime and low concentrations. Their concentrations and temporal distribution strongly depend on the cellular state and microenvironment.The physiological concentration of NO is in the range of only pM to μM during burst events.61,62 NO radicals show a typical diffusion of around 150–300 μm within a 4–15 s range in biological media,63–67 whereas other sources report a diffusion of 10 μm within 15 ms.68
Under physiological conditions, the half-life of free superoxide radicals is very limited in the range of milliseconds to some few seconds. This short lifetime results in a low cellular basal concentration of some pM (ref. 69) and a free diffusion path length of around 40 μm under in vivo conditions.70 The spontaneous decomposition rate of superoxide is strongly dependent on the local pH.
The short half-life and diffusion length of ROS and RNS arise from spontaneous or enzymatic decomposition and high reactivity with a bundle of different biomolecules, including membrane lipids, proteins, nucleic acids, other reactive species and transition metal ions. One example for a fast conversion is the reaction of NO* with O2*− to ONOO− with an almost diffusion-controlled rate (the rate constant is 4–16 × 109 mol−1 L s−1).71 As a consequence, the positioning of a sensor in the diffusion zone of ROS/RNS and the cellular microenvironment has tremendous influence on the signal height,66e.g. a complete signal loss at a position 1 mm away from single cell for NO measurement.68 Also, Brovkovych et al. showed the direct influence of sensor tip distance on the NO signal in a single endothelial cell experiment.72 Lee et al. demonstrated the impact of electrode and sealing size at a needle-shaped NO microsensor setup.65 It becomes obvious that a closer electrode cell distance (artificial synapse arrangement) increases the collection efficiency of the sensor. On the other hand, the diffusion of potential reaction partners like oxygen or arginine may be hindered by the sensor body, which leads to analyte accumulation.
In particular, in planar, chip-based sensor approaches with randomly growing cells, the complex hemispherical diffusion profiles for ROS and RNS can hardly be predicted. Further, the relationship between the diffusion profile/electrode overlap and electrode size may lead to a misinterpretation of the measured extracellular concentration when compared to an external calibration setup with homogeneously distributed analyte standards, even with correct sensor readings.
Microfabrication techniques allow for sensor design to measure within the expected area of occurrence and enable, along with appropriate electrochemical techniques, label-free measurement with sufficient spatial and time resolution. One approach to address the low concentration profiles of short-lived species might be the utilization of ultra-microelectrode (UME) arrays. A higher mass transfer rate, compared to millimetric electrodes, in terms of studying fast kinetics, and a better signal/noise ratio are the main aspects advocating their use, whereas smaller overall signals from lower electrode areas challenge the readout electronics' performance. Zhang et al. described an array of 2500 ultra-microelectrodes (carbon, 2 μm diameter) exploiting the effect of UME arrays for NO measurement.73 This report demonstrated the occurrence of steady-state regimes and the additivity of electrode currents. Later, the electrodes were coated with permselective membranes (Nafion and a commercial membrane from World Precision Instruments, USA) to enhance the selectivity towards NO, which can be detected by amperometry at 0.86 V vs. Ag/AgCl.
Parallel measurement of both ROS and RNS in stimulated adherent cell culture under static conditions was successfully demonstrated by Chang et al.74 They used planar sputtered gold/cytochrome c electrodes for superoxide and screen-printed carbon electrodes with electrodeposited tetrasulfonated nickel phthalocyanine (NiTSPc) for nitric oxide detection. The system consists of a full 24-well format sensor array and makes use of standard transwell inserts for cell culture pre-cultivation. The transwell is inserted into the measurement setup and avoids direct electrode/cell contact by the transfer membrane.
Another interesting approach by Isik et al. is the use of pyramid-shaped Pt tip electrodes on chip, to avoid direct cell–electrode contact and possible cell death upon electrical polarization.75 The electrode tips are located between/above the cells, which enables a measurement of NO within its diffusion zone. The tips were modified with NiTSPc to improve their NO sensitivity and measurement with HUVEC cells was performed amperometrically at 0.75 V vs. Ag/AgCl. The study focused on adherent cell line cultivation under static conditions, but the 3D geometry of the electrodes might pave the way for measurement in tissue slices or 3D cell culture models.
Electrochemical measurement of ROS and RNS molecules makes use of specific redox potentials to discriminate between the main interfering substances, whereas the high oxidation potential especially for NO (>700 mV vs. Ag/AgCl) is not suitably specific. If high time resolution is required, fast-scan voltammetry and amperometric measurements are the methods of choice, but also with limited selectivity. Therefore, techniques like differential pulse voltammetry are a good compromise between acceptable time resolution and increased selectivity.
Alternative approaches to reduce cross-sensitivities use permselective membranes to cover the sensing electrodes, allowing only the analytes of interest to reach the electrode surface. Typical membrane materials in use are hydrogels, Nafion, polypyrrole, o-phenylenediamine or polymethylcellulose. In addition, we demonstrated selective and robust in vitro measurement of the superoxide radical by direct oxidation on a planar gold microelectrode covered with a polyethylenimine membrane.76
Microfabrication and materials
Technology and substrate material
Early integrated cell culture monitoring systems used silicon as a substrate and were fabricated using (enhanced) CMOS technology.36,39,77–79 Silicon is essential in the case of LAPS and ISFETs, but also convenient for electrochemical sensors. An important possibility is to use the CMOS functionality to integrate analog circuitry for amplification and multiplexing or even circuitry for digitalization directly into the sensor chip. Additionally, optical functionality like cameras80–82 could be integrated.However, none of the described monitoring systems used integrated circuits to enhance signal performance or even eliminate external instrumentation. The sensor chip which is in direct contact with the cells should ideally be used once and therefore chips with integrated circuits generate unreasonably high costs. A critical disadvantage of silicon as a substrate is its opaqueness for visible light. Especially in the case of 2D cell culture, standard routines include optical inspection of the cells with through-light microscopy. Also, for 3D systems, optical access is beneficial. In the case of microfluidic systems, observation of bubbles simplifies successful measurements.
Approaches to introduce transparency by integration of a transparent window into the silicon chip79 or small silicon chips into glass substrates39 were mentioned. From today's perspective, the benefit of such approaches compared to the technological costs does not seem justifiable.
Glass substrates allow optical inspection of the cells as well as the possibility to use optical markers in the cell culture parallel to the microsensors. Thin-film processing for electrochemical sensors is possible using metals like platinum or gold along with silicon nitride or silicon oxide for passivation. Electrodes can be modified by electrodeposition of other metals onto the thin-film layers or coating with polymers similar to backend processes employed after CMOS fabrication in the case of silicon substrates.28,57
Polymer-based systems can be flexible, such as dip-in microsensors based on polyimide,18,83 or used as stiff materials comparable to other cell culture labware, such as well plates with integrated optical sensors.32 Gas permeability is excellent in silicones, which can be employed for gas perfusion into the cell culture. Polydimethylsiloxane (PDMS) replicas are often used to form channels on top of glass chips in order to combine sensor chips with microfluidics.
Sensor functionalization
Electrochemical microsensors need (thin-film) electrodes based on metals or carbon. Depending on the sensor type, fully inert electrodes (e.g. glassy carbon), catalytically active but inert electrodes (e.g. platinum, gold) or redox-active electrodes (e.g. silver, copper) are employed. Often, the electrodes are covered with a diffusion-limiting membrane (e.g. pHEMA), which also prevents cells from sticking to the electrode surface. In the case of biosensors, the enzymes need to be immobilized directly on the electrode or dispersed in a membrane. All electrochemical sensors need a reference electrode, which is often silver/silver chloride with an appropriate membrane or coating, to avoid the adverse effect of dissolved silver ions on the cells.Sensor functionalization for optical sensors is performed by application of the fluorescent dye dissolved in a membrane material, typically made of silicone or similar gas-permeable polymeric material.
Material selection
All used materials need to be non-cytotoxic or be fully encapsulated. Additionally, the materials in direct contact with the cells should be optimized for the cells to adhere or, depending on the application, consist of cell-repelling material. Passivation layers from thin-film technology like silicon oxide and silicon nitride have shown to be well suited for culture cells, in the case of tumor cells even without additional coating.28 Among the thick-film materials, the cytocompatibility of the epoxy-based polymer SU-8 is discussed controversially for different processing and cells.84–87 Coatings containing polyethylene glycol domains were suggested to render the material cell-repellent.88 Because of the large variety of different cells used in culture, it is important to test all materials with respect to cytotoxicity and cytocompatibility for the specific cell type.Sterilization
An important aspect is the selection of the appropriate sterilization method. Both the integrity of the material and the functionality of sensors have to be taken into account for choosing the procedure and dose. While for clinical application the employed sterilization method has to comply with regulatory demands, in research the importance of the topic is often neglected. In the case of microfabricated devices, the microbiological burden is typically low because of the clean room environment. This often allows applying disinfection using ethanol and culture media comprising antibiotics instead of sterilization.For sensor systems fabricated in CMOS technology, gamma sterilization is often not preferred because of altered characteristics mainly due to generation of defects in the gate oxide. Elevated temperature (autoclaving) is often avoided because mechanical stress can cause failure in hybrid mounted sensor chips. Biosensors comprising enzymes rule out higher temperature at all, while ethanol should not come into contact with the enzyme membranes. For biosensors, gamma sterilization is often a good choice if enzyme stability is considered.
Systems for 2D cell culture – static conditions
The class of systems to monitor one or several parameters in 2D cell culture under static conditions typically replicates or enhances conventional culture vessel formats, like Petri dishes, well plates or tissue culture flasks. Depending on the desired parameter, fixed integration of the sensor points next to the cell layer is essential. Alternatively, dip-in approaches with sensors at defined positions using a motorized micro-stage can be applied.5The majority of monitoring systems focusing on oxygen as the sole parameter employ optical oxygen sensors based on fluorescence quenching (Fig. 4A). Those works cover toxicity studies,32 evaluation of oxygen consumption rates,89 investigation of hypoxia for cancer research90 or in non-tumor cells,91–93 optimization of culture conditions,94 stem cell differentiation,95 and endocrinology.96 Also, electrochemical oxygen sensors were used, mainly Clark-type27 or direct amperometric sensors.28 In our opinion, electrochemical sensors are only reasonable in the case of systems for multi-parameter monitoring, especially in combination with biosensors, or if very low oxygen concentrations need to be measured.
 | ||
| Fig. 4 Sensor systems for metabolic monitoring in 2D static culture. A: Optical monitoring system to measure oxygen at the bottom of cell culture wells32 (reprinted with permission from Elsevier). B: Microdialysis system to monitor glucose and lactate102 (reprinted with permission from Elsevier). C: Multiparameter monitoring on a silicon chip with oxygen, pH, glucose and lactate sensors105 (reprinted from Pemberton et al., 2016, DOI 10.3390/s141120519, under CC BY 4.0). D: Sensing Cell Culture Flask: a transparent sensor chip with different electrochemical sensors is embedded in the bottom of a tissue culture flask.107 | ||
Instead of integrating oxygen sensors into the bottom of the cell culture area, an approach was described using integrated sensor strips enabling measurements at different heights above the bottom of the cell culture.83 This system is intended for determining oxygen gradients in stagnant medium above the cells in 2D culture, but would be an interesting approach to access oxygenation in 3D cell culture as well.
Another group of systems focuses on oxygen and pH as measurement parameters, for which mainly optical sensors are used.97,98 Further steps towards comprehensive multiparameter monitoring of cellular metabolism is the integration of biosensors into the sensor systems for static culture conditions. Systems were proposed using biosensors combined with microdialysis to measure glucose and lactate in static cell cultures (Fig. 4B).99–102 Glucose was also measured directly using a sensor chip dipped into a static cell culture.103,104
A multiparameter system measuring oxygen, pH, glucose and lactate using silicon chips was described for static cell culture in wells (Fig. 4C).105,106 The Sensing Cell Culture Flask (Fig. 4D) was introduced as an electrochemical sensor platform on transparent glass chips, embedded into the bottom of conventional tissue culture flasks, with sensors for oxygen, pH, glucose and lactate.28,76,83,107
Systems for 2D cell culture – dynamic conditions
In traditional 2D cell cultures, cells grow as an adherent monolayer at the bottom of relatively large vessels, e.g. flasks or microtiter plates. Total media volumes are in the range of 100 μL to several mL. With typical handling, i.e. medium change every few days, strong gradients of metabolites develop slowly because diffusion is comparably slow (Fig. 3A). The aim of dynamic cell culture monitoring is to reduce liquid volume to a few microliters and therefore to generate fast concentration changes. To maintain appropriate culture conditions, this approach requires periodic exchange of medium to keep cells alive (Fig. 3C). By such periodic exchange through microfluidics in stop/flow protocols, metabolic rates can be measured repetitively within minutes by microsensors placed directly underneath the cells in the culture chamber or in the microfluidic outlet channels. Such systems for the label-free, non-invasive monitoring of cell metabolism are called microphysiometers. Metabolic rates are accessible online, and changes upon substance exposure can be quantified. Suspended cells cannot attach properly to the chip surface under flow conditions to form 2D monolayers. Therefore, such systems are typically operated in an open configuration similar to a microtiter plate during a culture phase over up to a few days with a larger volume above the cells, during which the cells form adherent layers. For measurement, the systems are sealed with plugs, and the volume is reduced drastically for microfluidic operation.The first microphysiometer (Fig. 5A) was based on a silicon chip with an integrated light-addressable potentiometric sensor (LAPS) for pH measurement.35,77 A sample of 104 to 106 cells was inserted onto a membrane. A plug with vertical in- and outflow channels was inserted for operation, reducing the chamber volume to 2.8 μL for measurement,108 and a peristaltic pump was connected. The flow was stopped for around 30 s, during which extracellular acidification changed the pH in the range of 0.1 units. The system was later commercialized as the Cytosensor36 and enhanced by other sensor types. By insertion of platinum wires into the sealing plug and subsequent polishing, microelectrodes were formed above the cell layer. Electrochemical biosensors for glucose and lactate and dissolved oxygen sensors were integrated.109–111 Newer implementations of the LAPS microphysiometry principle exist.37,38,112 With similar protocols, pH was also measured by means of ion-selective field effect transistors (ISFET) using CMOS technology on silicon chips.78,79,113 ISFETs also allowed the determination of dissolved oxygen by local pH changes.30 More common is the direct amperometric measurement of oxygen by reduction at noble metal electrodes.57,110,114
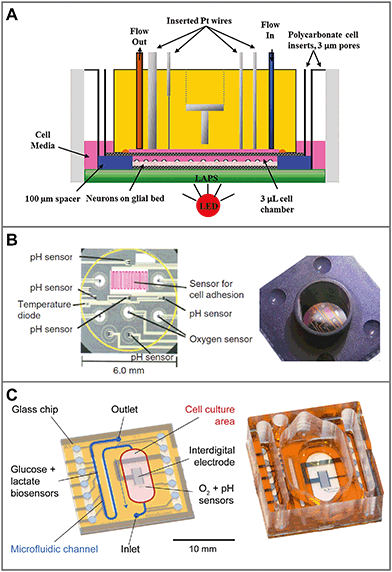 | ||
| Fig. 5 Sensor systems for metabolic monitoring in 2D dynamic culture (microphysiometers). Cells are cultured directly on microsensor chips. For measurement, the medium volume is reduced to a few microliters and periodically exchanged by microfluidics. A: The Cytosensor microphysiometer based on a silicon chip, in which pH is measured with a light-addressable potentiometric sensor (LAPS). Amperometric microsensors are included by integrating wires in the cover111 (reprinted with permission, Copyright 2012 American Chemical Society). B: Silicon-based system with planar sensor integration for pH (ISFET), oxygen (amperometric) and cellular adhesion (interdigital electrode)56,115 (reprinted with permission from Elsevier). C: Glass-based system for better microscopy with in situ amperometric oxygen sensors (platinum), potentiometric pH sensors (iridium oxide) and an interdigital electrode, as well as amperometric glucose and lactate biosensors integrated in the downstream microfluidics.57,107 | ||
Another parameter of interest in such systems is cellular adhesion, which is measured by impedance analysis.78,79 Attachment and spreading of cells on interdigital electrode structures (IDES) is determined by capacitance changes. Adhesion can be monitored continuously during all phases of cell growth, and is mostly independent of flow. The combination of IDES-based impedance monitoring with ISFET-based pH sensing and amperometric oxygen measurement for drug and toxicology screening on a silicon chip was commercialized by Bionas (Fig. 5B).56,115 The key feature of microphysiometers, in comparison to traditional end-point tests, is the rapid determination of metabolic rates in a continuous, online measurement (Fig. 6A), which reveals transient recovery effects of cellular metabolism and allows the investigation of pharmacodynamics in drug screening (Fig. 6B) and cancer research.56,57,116
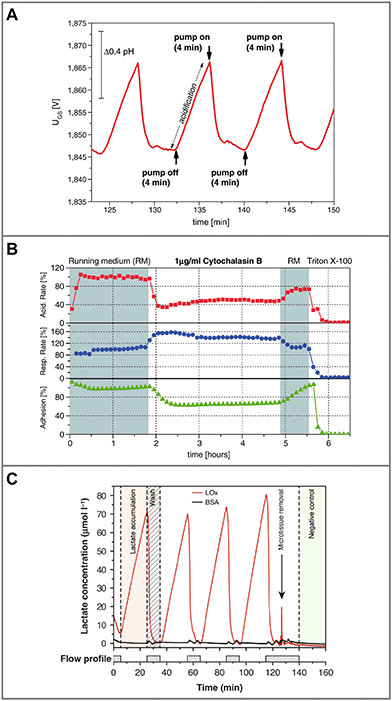 | ||
| Fig. 6 Typical measurements from dynamic cell culture monitoring with microsensors. A: Extracellular acidification of 2D cells during the stop phase (pump off) in a microphysiometer system and return to the baseline during the flow phase (pump on)56 (reprinted with permission from Elsevier). B: Metabolic rates for acidification, respiration and adhesion extracted from such an experiment. Rates change upon substance exposure, and continuous, online monitoring allows the measurement of pharmacodynamics and recovery effects in the drug screening application56 (reprinted with permission from Elsevier). C: Biosensor measurement of lactate production from 3D microtissue spheroids in hanging droplets, including the blank signal, which shows lactate production and return to baseline depending on the flow profile19 (reprinted from Misun et al., 2016, DOI 10.1038/micronano.2016.22, under CC BY 4.0). | ||
Particularly for enzyme-based biosensors which produce hydrogen peroxide as an intermediary product, sensor placement outside the cell culture chamber is desirable to reduce oxidative stress. We demonstrated the placement of glucose and lactate biosensors in the outlet channel (Fig. 5C).57,117 Undiluted medium from the cell chamber was transported to the biosensor array and measured during the flow phase. Amperometric oxygen sensors based on platinum electrodes and potentiometric pH sensors based on iridium oxide electrodes were included in the cell culture chamber. In contrast to silicon-based devices, we integrated these electrochemical, multiparameteric sensors on a transparent glass chip, which facilitates microscopy. Other groups focused on reduction of the volume to the sub-microliter range for glucose118 and lactate119 measurement or alternative fabrication methods such as screen- and inkjet-printing.120
The method and systems commercialized by Seahorse Bioscience fall between static and dynamic monitoring.55 Cells are cultured in largely standard microtiter plates, into which a sensor probe is lowered, temporarily reducing the volume above the cells to around 2 μL. Oxygen and pH are measured using optical sensors. Pneumatic reservoirs above the medium enable the addition of test compounds, and the movement of the sensor tip mixes the medium. Metabolic rate determination within minutes is possible, but options for microfluidics and sensor placement are limited. The system allows parallel experiments in the entire plate with automated sequential readout. From an engineering standpoint, using microphysiometer systems seems straightforward. However, their operation with a combination of fluidics, sensors and cell handling is more complex than standard procedures in cell biology. The amount of data acquired is high compared to end-point tests and requires at least some basic data processing to obtain meaningful results. Valuable efforts to automate operation121 and parameterize data processing122 have been made. However, few novel concepts or parameters have emerged within the last decade, and many existing systems have faded away. Other systems never left academia and might have suffered from reliability problems. Their complexity and their limited compatibility with true high-throughput screening are drawbacks which still persist even among the novel approaches for 3D cell cultures.
Systems for 3D cell culture and organ-on-chip
Regarding the access to cell metabolites, there are two main differences between 3D and monolayer cultures. First, cell numbers are usually lower in 3D culture12 (103 in microtissues or spheroids vs. 104–106 in standard 2D monolayers), and cell aggregates are often distributed heterogeneously in space. Thus, concentration changes are smaller. Gradients are less homogeneous and multidirectional (Fig. 3B). Second, if cells are non-adherent, microfluidic protocols are not as easily applicable, because cells may be washed away. Measurements by impedance, which often rely on cell attachment and are used extensively in 2D, are also more difficult. If cells are in a solid matrix, there is not even direct fluidic access, and metabolite exchange relies on diffusion of metabolites through the matrix. Fluidic channels must be formed by microstructuring of the matrix.Many microfluidic systems have been developed for the formation, trapping and culture of non-adherent microtissues, but very few include sensors to measure cell metabolites. Systems are often microfluidic channel networks fabricated from molded or thermoformed polymers with small wells to trap the microtissues, while the medium flows over or alongside the well.123–127 Paper has also been used for both fluidic compartments and cell scaffold.128,129 Supernatants and perfusion media are then transferred to external readers or analysed in situ. Measurement is often limited to viability testing by optical methods such as fluorimetric live/dead staining.16,130 As these systems often require periodic or constant perfusion, a strict differentiation between dynamic and static measurement is more difficult.
The straightforward, static dip-in approach is to measure metabolites directly in the stagnant supernatant in standard culture vessels, i.e. microtiter plates. Here, two difficulties arise: due to the low cell number, concentration changes are small, and medium exchange happens only every few days. This requires highly sensitive and long-term stable sensors since the fluid volume cannot be reduced indefinitely. We showed the direct measurement of lactate and oxygen using electrochemical microsensors in standard 96-well plates containing a single microtissue spheroid.18,131 Needle-type sensor strips132 with microelectrodes at the tip were dipped into and remained in the well over days. The integration of the sensor system into the standardized setup enables compatibility with standard handling procedures. Lactate production rates from 2000-cell spheroids in the range of 5 μM h−1 were measured over up to three days. Oxygen measurement in the direct vicinity of the spheroids also revealed no hypoxic conditions in the well even though assumingly anaerobic metabolic pathways were active. By measuring altered metabolic rates under drug exposure, we demonstrated that metabolic access yields valuable information in drug screening, which complements and enhances microscopy or viability tests. Generally, electrochemical systems with electrode sizes in the micrometer range allow a highly localized measurement.
As in 2D systems, microfluidics can be utilized to reduce total media volumes and thus increase concentration changes. Collection and undiluted transport of media to downstream sensors enables the integral measurement over numerous microtissues. Such a system was realized for a multiwell bioreactor for hepatic spheroids (Fig. 7A).133 Nine microtissues were kept in approx. 1.2 μL wells each, within a gas-tight bioreactor with constant perfusion. Sensors included in situ oxygen sensors composed of tissue-embedded phosphorescent beads and commercial amperometric biosensors for glucose and lactate off-chip, in a combined outlet channel.134 Oxygen was measured continuously over 28 d and consumption rates were determined. After 4 d of adaptation, distinctly lower concentrations than air saturation were measured in the microwells. Glucose and lactate metabolism from all microtissues combined was measured from 40 μL medium extracted from the bioreactor. Microfluidic switching allowed frequent calibration of the biosensors with external solutions to account for drift of the sensors. A comparable concept for multi-organoid drug screening has been shown recently,135 in which cardiac and liver organoids within a microfluidic network were combined with optical pH and oxygen sensors,136 as well as electrochemical immunobiosensors.137 Even though these systems are designed to work largely automated, they require considerable technological effort and instrumentation to fabricate and operate both microfluidic networks and sensor periphery.
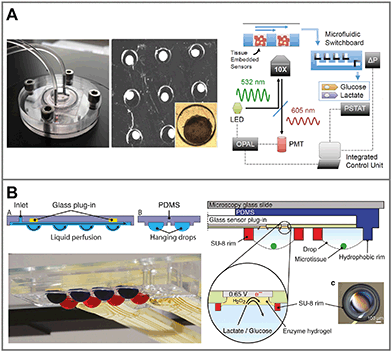 | ||
| Fig. 7 Sensor systems for metabolic monitoring in 3D cell culture. A: 3D microtissue spheroids trapped in microwells within a perfused microbioreactor. Oxygen and pH are measured optically through nanoparticles embedded in the microtissues. Glucose and lactate are measured using downstream amperometric biosensors connected by a microfluidic switchboard133 (copyright 2016 National Academy of Sciences). B: 3D microtissue spheroids cultured in interconnected hanging droplet networks. A glass-based sensor chip and microstructures allow fluidic connection between the drops and integration of in situ microsensors for, e.g. lactate, glucose or impedance19 (reprinted from Misun et al., 2016, DOI 10.1038/micronano.2016.22, under CC BY 4.0). | ||
Hanging droplets are another method for scaffold-free culture of non-adherent microtissues. Cells agglomerate and form spheroids at the bottom of the hanging drop, driven by gravity and convection forces, where only a liquid/gas interface exists.16 By balancing inflow and outflow through active pumping17,138 or surface tension-driven flow,139 droplet size can be maintained constant. In the same way, medium can be transferred between drops, while spheroids remain trapped within the same droplet. Interconnected hanging droplet networks up to 96 wells138 allow intertissue contact and various options for periodic medium exchange and controlled substance exposure, while metabolites remain confined in very small drop volumes around 10 μL.17 This allowed the direct integration of enzyme-based amperometric glucose and lactate biosensors into the droplets by combination with a glass-based sensor chip (Fig. 7B).19 In short-term experiments, within a 20 min static phase, lactate accumulation and glucose production could be measured in parallel (Fig. 6C). Size-dependent metabolic rates from 300–500 μm spheroids in the range of nmol h−1 could be measured. Impedance measurements are also possible. Small tissue size and its position away from the electrodes, which can only be at the ceiling of the droplet, make them challenging however.140 Reducing the droplet volume to bring the spheroid closer to the electrodes for measurement was necessary. Overall, the drawbacks of the hanging droplet approach are the complex liquid handling, necessary prevention of evaporation and limited robustness against mechanical shocks of the systems. Also, the open nature of the principle makes the measurement of dissolved gases, e.g. oxygen, difficult.
Although a number of systems exist with promising new approaches, to date, in 3D cultures, the exact metabolic state and immediate microenvironment, i.e. nutrient and oxygen supply, of the microtissues have still been largely unknown and relied mainly on the controlled exchange of media and diffusion of metabolites. Especially for oxygen, measurement within the microtissue would be desirable, but current microsensors are typically not small enough to be inserted with sufficiently low tissue interference.
Conclusion
Potential and benefits of metabolic monitoring
Microsensors can be integrated in different cell culture vessels or dedicated microfluidic platforms to access parameters of cellular metabolism in the extracellular space (culture medium) in close vicinity to the cells (pericellular). Microelectrode sizes can be as small as cellular dimensions, and arrays can be realized to cover larger areas. Electrochemical and optical read-out techniques dominate, especially for the classical metabolic parameters oxygen (respiration), pH (acidification), glucose (energy supply) and lactate (anaerobic waste product). Electrical read-out, typically in the form of impedance, is limited to mechano-physical properties, often cell attachment. Particularly, electrochemical techniques allow a low detection limit with a defined zero-point, combined with a high temporal resolution to resolve fast dynamics also of short-lived substances. Microsensors typically aim at continuous online measurements, which not only allow observations during the experiment, but also reveal transient (e.g. recovery) effects that are not detectable in end-point tests.The primary advantages of microfluidic systems are the reduction of media volume and thus faster concentration changes, the transport of undiluted medium to downstream sensors, as well as the possibility to repeatedly and efficiently generate a broad range of stimuli. The interplay between the state of the cells, microenvironment and sensing method is complex. Both the choice of a suitable measurement system and the obtained experimental findings must be critically evaluated for the aspects how cells are cultivated and supplied, how metabolites are transported to the sensors and how stimuli are generated. From the perspective of metabolic sensing, it is evident that even in simple, standard cell culture experiments, in which sometimes complex markers or pathways are investigated, basic metabolic parameters at the pericellular level are often unknown. Diffusion of metabolites in stagnant media or even solid matrices is comparably slow. If there is no flow, transport of metabolites to and from the cells will be governed by diffusion and thermal convection only. Therefore, nutrient or oxygen depletion can occur near the cells, even if those parameters seem globally well controlled. In 3D, the state and measurement of metabolites can be even more challenging. As a consequence, this may negatively affect the reproducibility or validity of findings in such experiments. At the moment, microsensors can therefore most likely still contribute more to standardization of cell culture experiments or help in basic physiological investigations, rather than replace classical high-throughput screening approaches.
Limitations in metabolic monitoring
Current dimensions of microsensors limit the access to extracellular readings only. It is not expected that in the near future sensors could be significantly scaled down in order to penetrate cells and provide meaningful sensor readings (besides counting nanoparticles along with optical readout as sensors). Thus, by principle, metabolic monitoring provides indirect information about the metabolic state of the cells only.Bringing sensors close to cells is important in order to obtain pericellular readings. In contrast, the physical presence of a sensor hinders a directly neighboring cell at the position of the sensor and slightly changes the diffusion profile next to the cell. In the case of single cell monitoring, this effect is crucial, and a trade-off has to be made. In 2D or 3D cell cultures, the presence of the microsensor typically does not have much influence, compared to other boundary conditions of the culture itself.
In general, sensors without analyte consumption are preferred to minimize influence on the cellular microenvironment. An exception is the case of 2D cell culture with oxygen sensors positioned between the cells. Typically, cells cannot grow at the very same position of the sensor, thus causing spots in the cell layer without cellular respiration. Therefore, it is beneficial that such a microsensor shows analyte consumption in the range of the cellular respiration to avoid an anomaly in the diffusion profile next to the sensor.
The electrochemical measurement of short-lived reactive species concentration (ROS/RNS) and its spatial distribution is only possible in close vicinity to the releasing cell under stagnant conditions, which excludes downstream detection in microfluidic devices. Planar microdisc sensors in direct vicinity to adherent 2D cell cultures and needle-shaped electrodes, especially suited for 3D culture models, are the only way to address the fleeting existence of these substances. Needle-shaped sensors suffer from bad reproducibility, vibration sensitivity, fragile handling and low throughput due to (motorized) manual positioning under the microscope. Measurement results from planar microsensor platforms with randomly growing cells should be evaluated critically with respect to a particular system design/setup, whose influence on the findings might be underestimated.
Electrochemical, enzyme-based biosensors require frequent calibration because of sensitivity drift, and the enzymatic reaction may release harmful by-products. Placement outside the actual cell culture area addresses both aspects. For long-term application, schemes for recalibration should be considered.
The sometimes considerable complexity of fabrication technology and operation, as well as incompatibility with standard procedures, limits the widespread application of more advanced microfluidic systems. A simplification of microfluidic handling would often be desirable, which could mean the elimination of pumps and reliance on pipetting or passive flow for media exchange. Instrumentation for electrochemical sensors and optical sensors appears to be an expensive investment at first glance. However, upon closer look, the costs for instrumentation equipment are low compared to the typical installation costs for a cell laboratory. Still, there is a need for cost-effective parallelization in order to meet high-throughput screening demands.
Once standardization is established, it is unclear whether the measurement of basic metabolic parameters (O2, pH, glucose, lactate) will be equally important as other indicating parameters like gene expression etc. For 3D cell culture, in which major gradients occur within the microtissues, it is desirable to measure inside such structures with minimal invasion. Instead of sensors, embedded particles which could be read out from the outside are promising alternatives for some parameters and can also address the overall heterogeneity of such cultures.
Impact of microsensor systems on organ-on-chip and human-on-chip systems
In organ-on-chip systems, the cells as sub-units are linked together to model the functionality of a specific organ. Similarly, such organ-on-chip systems as sub-units can be linked together in order to model partial functionality of the human body and are therefore sometimes visionarily called body-on-chip or human-on-chip systems.141,142The basic requirements and challenges for microsensors are the same, independent of whether it is a 2D cell culture or an organ-on-chip system. Notably, the complexity increases from 2D to 3D. Once a system can be used in 3D cell culture models, it can be readily applied to organ-on-chip systems. From the point of view of cell culture monitoring, there is no difference between a 3D culture of cells without organotypic function and an organ-on-chip system. However, the impact of these systems changes from just reporting the metabolic state of the cells to providing insight into cell–cell interactions enabling the functionality of the organ by observing the metabolism of some representative cells within the model. Here, sensor readings can be linked to and provide input for computer models (“in silico”), which become more and more important with the increasing complexity from organ-on-chip to human-on-chip. In our understanding, a meaningful human-on-chip cellular model can hardly be described and accessed without microsensors reading the metabolic state at characteristic points in the system.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank all collaborators in the European Union projects EUROXY (FP6-LIFESCIHEALTH) and METOXIA (FP7-HEALTH-2007-B), namely Peter Ebbesen, Erik O. Pettersen, Joe A. Sandvik and Jan Villadsen, who shaped the authors' views on the relevance and limitation of microsensors for cell culture monitoring and its implications on cancer research.References
- D. Huh, Y. Torisawa, G. A. Hamilton, H. J. Kim and D. E. Ingber, Lab Chip, 2012, 12, 2156–2164 RSC
.
- B. Zhang and M. Radisic, Lab Chip, 2017, 17, 2395–2420 RSC
.
- P. Ebbesen, K.-U. Eckardt, F. Ciampor and E. O. Pettersen, Acta Oncol., 2004, 43, 598–600 CrossRef CAS PubMed
.
- P. Ebbesen, E. O. Pettersen, J. Denekamp, B. Littband, J. Keski-Oja, A. Schousboe, U. Sonnewald, Ø. Åmellem and V. Zachar, Acta Oncol., 2000, 39, 247–248 CrossRef CAS PubMed
.
- E. O. Pettersen, L. H. Larsen, N. B. Ramsing and P. Ebbesen, Cell Proliferation, 2005, 38, 257–267 CrossRef PubMed
.
- M. D. Brennan, M. L. Rexius-Hall, L. J. Elgass and D. T. Eddington, Lab Chip, 2014, 14, 4305–4318 RSC
.
- P. E. Oomen, M. D. Skolimowski and E. Verpoorte, Lab Chip, 2016, 16, 3394–3414 RSC
.
- L. G. Griffith and M. A. Swartz, Nat. Rev. Mol. Cell Biol., 2006, 7, 211–224 CrossRef CAS PubMed
.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed
.
- M. Simian and M. J. Bissell, J. Cell Biol., 2017, 216, 31–40 CrossRef CAS PubMed
.
- E. Fennema, N. Rivron, J. Rouwkema, C. van Blitterswijk and J. De Boer, Trends Biotechnol., 2013, 31, 108–115 CrossRef CAS PubMed
.
- R. Z. Lin and H. Y. Chang, Biotechnol. J., 2008, 3, 1172–1184 CrossRef CAS PubMed
.
- H. K. Kleinman and G. R. Martin, Semin. Cancer Biol., 2005, 15, 378–386 CrossRef CAS PubMed
.
- D. R. Grimes, C. Kelly, K. Bloch and M. Partridge, J. R. Soc., Interface, 2014, 11, 20131124–20131124 CrossRef PubMed
.
- S. Messner, I. Agarkova, W. Moritz and J. M. Kelm, Arch. Toxicol., 2013, 87, 209–213 CrossRef CAS PubMed
.
- Y.-C. Tung, A. Y. Hsiao, S. G. Allen, Y. Torisawa, M. Ho and S. Takayama, Analyst, 2011, 136, 473–478 RSC
.
- O. Frey, P. M. Misun, D. A. Fluri, J. G. Hengstler and A. Hierlemann, Nat. Commun., 2014, 5, 4250 CAS
.
- A. Weltin, S. Hammer, F. Noor, Y. Kaminski, J. Kieninger and G. A. Urban, Biosens. Bioelectron., 2017, 87, 941–948 CrossRef CAS PubMed
.
- P. M. Misun, J. Rothe, Y. R. F. Schmid, A. Hierlemann and O. Frey, Microsyst. Nanoeng., 2016, 2, 16022 CrossRef CAS
.
- J. Carlsson, C. G. Stålnacke, H. Acker, M. Haji-Karim, S. Nilsson and B. Larsson, Int. J. Radiat. Oncol., Biol., Phys., 1979, 5, 2011–2020 CrossRef CAS
.
- R. M. Sutherland and W. F. Mueller-Klieser, Cancer Res., 1982, 42, 237–242 Search PubMed
.
- W. F. Mueller-Klieser and R. M. Sutherland, Br. J. Cancer, 1982, 45, 256–264 CrossRef CAS PubMed
.
- H. Lee, M. Chung and N. L. Jeon, MRS Bull., 2014, 39, 51–59 CrossRef
.
- D. T. T. Phan, X. Wang, B. M. Craver, A. Sobrino, D. Zhao, J. C. Chen, L. Y. N. Lee, S. C. George, A. P. Lee and C. C. W. Hughes, Lab Chip, 2017, 17, 511–520 RSC
.
- H. Suzuki, A. Sugama and N. Kojima, Sens. Actuators, B, 1990, 2, 297–303 CrossRef CAS
.
- G. Jobst, G. Urban, A. Jachimowicz and F. Kohl, Biosens. Bioelectron., 1993, 8, 123–128 CrossRef CAS
.
- C. C. Wu, H. N. Luk, Y. T. T. Lin and C. Y. Yuan, Talanta, 2010, 81, 228–234 CrossRef CAS PubMed
.
- J. Kieninger, K. Aravindalochanan, J. A. Sandvik, E. O. Pettersen and G. A. Urban, Cell Proliferation, 2014, 47, 180–188 CrossRef CAS PubMed
.
- B.-K. Sohn and C.-S. Kim, Sens. Actuators, B, 1996, 34, 435–440 CrossRef CAS
.
- M. Lehmann, W. Baumann, M. Brischwein, H. Gahle, I. Freund, R. Ehret, S. Drechsler, H. Palzer, M. Kleintges, U. Sieben and B. Wolf, Biosens. Bioelectron., 2001, 16, 195–203 CrossRef CAS PubMed
.
- I. Klimant and O. S. Wolfbeis, Anal. Chem., 1995, 67, 3160–3166 CrossRef CAS
.
- S. Beckers, F. Noor, U. Müller-Vieira, M. Mayer, A. Strigun and E. Heinzle, Toxicol. In Vitro, 2010, 24, 686–694 CrossRef CAS PubMed
.
- C. J. Ochs, J. Kasuya, A. Pavesi and R. D. Kamm, Lab Chip, 2014, 14, 459–462 RSC
.
- P. C. Thomas, M. Halter, A. Tona, S. R. Raghavan, A. L. Plant and S. P. Forry, Anal. Chem., 2009, 81, 9239–9246 CrossRef CAS PubMed
.
- D. Hafeman, J. Parce and H. McConnell, Science, 1988, 1182–1185 CAS
.
- H. McConnell, J. Owicki, J. Parce, D. Miller, G. Baxter, H. Wada and S. Pitchford, Science, 1992, 257, 1906–1912 CAS
.
- C. Wu, J. Zhou, N. Hu, D. Ha, X. Miao and P. Wang, Sens. Actuators, A, 2013, 199, 136–142 CrossRef CAS
.
- K. Su, J. Zhou, L. Zou, T. Wang, L. Zhuang, N. Hu and P. Wang, Sens. Actuators, A, 2014, 220, 144–152 CrossRef CAS
.
- W. H. Baumann, M. Lehmann, A. Schwinde, R. Ehret, M. Brischwein and B. Wolf, Sens. Actuators, B, 1999, 55, 77–89 CrossRef CAS
.
- P. Bergveld, Sens. Actuators, B, 2003, 88, 1–20 CrossRef CAS
.
- A. Fanigliulo, P. Accossato, M. Adami, M. Lanzi, S. Martinoia, S. Paddeu, M. T. Parodi, A. Rossi, M. Sartore, M. Grattarola and C. Nicolini, Sens. Actuators, B, 1996, 32, 41–48 CrossRef CAS
.
- W. Olthuis, M. A. M. Robben, P. Bergveld, M. Bos and W. E. van der Linden, Sens. Actuators, B, 1990, 2, 247–256 CrossRef CAS
.
- S. A. M. Marzouk, Anal. Chem., 2003, 75, 1258–1266 CrossRef CAS PubMed
.
- I. A. Ges, B. L. Ivanov, D.
K. Schaffer, E. A. Lima, A. A. Werdich and F. J. Baudenbacher, Biosens. Bioelectron., 2005, 21, 248–256 CrossRef CAS PubMed
.
- I. A. Ges, B. L. Ivanov, A. A. Werdich and F. J. Baudenbacher, Biosens. Bioelectron., 2007, 22, 1303–1310 CrossRef CAS PubMed
.
- T. Y. Kim and S. Yang, Sens. Actuators, B, 2014, 196, 31–38 CrossRef CAS
.
- K. Yamamoto, G. Shi, T. Zhou, F. Xu, M. Zhu, M. Liu, T. Kato, J. Y. Jin and L. Jin, Anal. Chim. Acta, 2003, 480, 109–117 CrossRef CAS
.
- P. Salazar, F. J. Garcia-Garcia, F. Yubero, J. Gil-Rostra and A. R. González-Elipe, Electrochim. Acta, 2016, 193, 24–31 CrossRef CAS
.
- Q. Guo, X. Wu, E. H. Han and W. Ke, J. Electroanal. Chem., 2016, 782, 91–97 CrossRef CAS
.
- Y. H. Liao and J. C. Chou, Sens. Actuators, B, 2008, 128, 603–612 CrossRef CAS
.
- M. Brischwein, H. Grothe, J. Wiest, M. Zottmann, J. Ressler and B. Wolf, Chem. Anal., 2009, 54, 1193–1201 CAS
.
- A. Sardarinejad, D. K. Maurya and K. Alameh, Sens. Actuators, A, 2014, 214, 15–19 CrossRef CAS
.
- G. Liebsch, I. Klimant, C. Krause and O. S. Wolfbeis, Anal. Chem., 2001, 73, 4354–4363 CrossRef CAS PubMed
.
- Y. Tian, B. R. Shumway, A. Cody Youngbull, Y. Li, A. K. Y. Jen, R. H. Johnson and D. R. Meldrum, Sens. Actuators, B, 2010, 147, 714–722 CrossRef CAS PubMed
.
- M. Wu, A. Neilson, A. L. Swift, R. Moran, J. Tamagnine, D. Parslow, S. Armistead, K. Lemire, J. Orrell, J. Teich, S. Chomicz and D. A. Ferrick, Am. J. Physiol., 2006, 292, C125–C136 CrossRef PubMed
.
- E. Thedinga, A. Kob, H. Holst, A. Keuer, S. Drechsler, R. Niendorf, W. Baumann, I. Freund, M. Lehmann and R. Ehret, Toxicol. Appl. Pharmacol., 2007, 220, 33–44 CrossRef CAS PubMed
.
- A. Weltin, K. Slotwinski, J. Kieninger, I. Moser, G. Jobst, M. Wego, R. Ehret and G. A. Urban, Lab Chip, 2014, 14, 138–146 RSC
.
- G. Jobst, I. Moser, M. Varahram, P. Svasek, E. Aschauer, Z. Trajanoski, P. Wach, P. Kotanko, F. Skrabal and G. Urban, Anal. Chem., 1996, 68, 3173–3179 CrossRef CAS PubMed
.
- A. Weltin, J. Kieninger and G. A. Urban, Anal. Bioanal. Chem., 2016, 408, 4503–4521 CrossRef CAS PubMed
.
- V. B. Koman, C. Santschi and O. J. F. Martin, Biomed. Opt. Express, 2015, 6, 2353 CrossRef CAS PubMed
.
- J. C. Toledo and O. Augusto, Chem. Res. Toxicol., 2012, 25, 975–989 CrossRef CAS PubMed
.
- J. S. Stamler, D. I. Simon, J. A. Osborne, M. E. Mullins, O. Jaraki, T. Michel, D. J. Singel and J. Loscalzo, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 444–448 CrossRef CAS
.
- C. Amatore, S. Arbault, M. Guille and F. Lemaître, Chem. Rev., 2008, 108, 2585–2621 CrossRef CAS PubMed
.
- J. R. Lancaster, Nitric Oxide, 1997, 1, 18–30 CrossRef CAS PubMed
.
- Y. Lee, J. Yang, S. M. Rudich, R. J. Schreiner and M. E. Meyerhoff, Anal. Chem., 2004, 76, 545–551 CrossRef CAS PubMed
.
- S. Isik, M. Etienne, J. Oni, A. Blöchl, S. Reiter and W. Schuhmann, Anal. Chem., 2004, 76, 6389–6394 CrossRef CAS PubMed
.
- S. Isik and W. Schuhmann, Angew. Chem., Int. Ed., 2006, 45, 7451–7454 CrossRef CAS PubMed
.
- D. M. Porterfield, J. D. Laskin, S. K. Jung, R. P. Malchow, B. Billack, P. J. Smith and D. E. Heck, Am. J. Physiol., 2001, 281, L904–L912 CAS
.
- X. J. Chen, A. C. West, D. M. Cropek and S. Banta, Anal. Chem., 2008, 80, 9622–9629 CrossRef CAS PubMed
.
- M. Saran and W. Bors, Chem.-Biol. Interact., 1994, 90, 35–45 CrossRef CAS PubMed
.
- G. Ferrer-Sueta and R. Radi, ACS Chem. Biol., 2009, 4, 161–177 CrossRef CAS PubMed
.
- V. Brovkovych, E. Stolarczyk, J. Oman, P. Tomboulian and T. Malinski, J. Pharm. Biomed. Anal., 1999, 19, 135–143 CrossRef CAS PubMed
.
- X. Zhang, J. Lin, L. Cardoso, M. Broderick and V. Darley-Usmar, Electroanalysis, 2002, 14, 697–703 CrossRef CAS
.
- S.-C. Chang, N. Pereira-Rodrigues, J. R. Henderson, A. Cole, F. Bedioui and C. J. McNeil, Biosens. Bioelectron., 2005, 21, 917–922 CrossRef CAS PubMed
.
- S. Isik, L. Berdondini, J. Oni, A. Blöchl, M. Koudelka-Hep and W. Schuhmann, Biosens. Bioelectron., 2005, 20, 1566–1572 CrossRef CAS PubMed
.
- H. Flamm, J. Kieninger, A. Weltin and G. A. Urban, Biosens. Bioelectron., 2015, 65, 354–359 CrossRef CAS PubMed
.
- J. Owicki and J. W. Parce, Biosens. Bioelectron., 1992, 7, 255–272 CrossRef CAS PubMed
.
- R. Ehret, W. Baumann, M. Brischwein, A. Schwinde, K. Stegbauer and B. Wolf, Biosens. Bioelectron., 1997, 12, 29–41 CrossRef CAS PubMed
.
- B. Wolf, M. Brischwein, W. Baumann, R. Ehret and M. Kraus, Biosens. Bioelectron., 1998, 13, 501–509 CrossRef CAS PubMed
.
- A. Ozcan and U. Demirci, Lab Chip, 2008, 8, 98–106 RSC
.
- G. Zheng, S. A. Lee, Y. Antebi, M. B. Elowitz and C. Yang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16889–16894 CrossRef CAS PubMed
.
- A. Greenbaum, W. Luo, T.-W. Su, Z. Göröcs, L. Xue, S. O. Isikman, A. F. Coskun, O. Mudanyali and A. Ozcan, Nat. Methods, 2012, 9, 889–895 CrossRef CAS PubMed
.
- P. Ebbesen, E. O. Pettersen, T. A. Gorr, G. Jobst, K. Williams, J. Kieninger, R. H. Wenger, S. Pastorekova, L. Dubois, P. Lambin, B. G. Wouters, T. Van Den Beucken, C. T. Supuran, L. Poellinger, P. Ratcliffe, A. Kanopka, A. Görlach, M. Gasmann, A. L. Harris, P. Maxwell and A. Scozzafava, J. Enzyme Inhib. Med. Chem., 2009, 24, 1–39 CrossRef CAS PubMed
.
- Z.-Z. Wu, Y. Zhao and W. S. Kisaalita, Colloids Surf., B, 2006, 52, 14–21 CrossRef CAS PubMed
.
- V. N. Vernekar, D. K. Cullen, N. Fogleman, Y. Choi, A. J. García, M. G. Allen, G. J. Brewer and M. C. LaPlaca, J. Biomed. Mater. Res., Part A, 2008, 89A, 138–151 Search PubMed
.
- K. V. Nemani, K. L. Moodie, J. B. Brennick, A. Su and B. Gimi, Mater. Sci. Eng., C, 2013, 33, 4453–4459 CrossRef CAS PubMed
.
- A. Ajetunmobi, D. McAllister, N. Jain, O. Brazil, A. Corvin, Y. Volkov, D. Tropea and A. Prina-Mello, J. Biomed. Mater. Res., Part A, 2017, 105, 2129–2138 CrossRef CAS PubMed
.
- D. J. Kenan, E. B. Walsh, S. R. Meyers, G. A. O'Toole, E. G. Carruthers, W. K. Lee, S. Zauscher, C. A. H. Prata and M. W. Grinstaff, Chem. Biol., 2006, 13, 695–700 CrossRef CAS PubMed
.
- R. R. Deshpande and E. Heinzle, Biotechnol. Lett., 2004, 26, 763–767 CrossRef CAS PubMed
.
- A. Wouters, B. Pauwels, H. A. J. Lambrechts, G. G. O. Pattyn, J. Ides, M. Baay, P. Meijnders, S. Dewilde, J. B. Vermorken and F. Lardon, Cancer Lett., 2009, 286, 180–188 CrossRef CAS PubMed
.
- N. Dehne, G. Hintereder and B. Brüne, Exp. Cell Res., 2010, 316, 1179–1189 CrossRef CAS PubMed
.
- T. G. Fernandes, M. M. Diogo, A. Fernandes-Platzgummer, C. L. da Silva and J. M. S. Cabral, Stem Cell Res., 2010, 5, 76–89 CrossRef CAS PubMed
.
- H. E. Abaci, R. Truitt, E. Luong, G. Drazer and S. Gerecht, Am. J. Physiol., 2010, 298, C1527–C1537 CrossRef CAS PubMed
.
- R. R. Deshpande, C. Wittmann and E. Heinzle, Cytotechnology, 2004, 46, 1–8 CrossRef CAS PubMed
.
- S. M. Browne, H. Daud, W. G. Murphy and M. Al-Rubeai, J. Biotechnol., 2014, 187, 135–138 CrossRef CAS PubMed
.
- W. Wang, L. Upshaw, D. M. Strong, R. P. Robertson and J. Reems, J. Endocrinol., 2005, 185, 445–455 CrossRef CAS PubMed
.
- M. Naciri, D. Kuystermans and M. Al-Rubeai, Cytotechnology, 2008, 57, 245–250 CrossRef CAS PubMed
.
- S. Lee, B. L. Ibey, G. L. Coté and M. V. Pishko, Sens. Actuators, B, 2008, 128, 388–398 CrossRef CAS
.
- C. Boero, S. Carrara, G. Del Vecchio, L. Calzà and G. De Micheli, IEEE Trans. Nanobiosci., 2011, 10, 59–67 CrossRef PubMed
.
- J. Olivo, L. Foglia, M. A. Casulli, C. Boero, S. Carrara and G. De Micheli, IEEE Trans. Biomed. Circuits Syst., 2014, 400–403 Search PubMed
.
- C. Boero, J. Olivo, S. Carrara and G. De Micheli, IEEE J. Emerg. Sel. Topic Circuits Syst., 2012, 2, 658–671 CrossRef
.
- C. Boero, M. A. Casulli, J. Olivo, L. Foglia, E. Orso, M. Mazza, S. Carrara and G. De Micheli, Biosens. Bioelectron., 2014, 61, 251–259 CrossRef CAS PubMed
.
- R. M. Pemberton, J. Xu, R. Pittson, N. Biddle, G. A. Drago, S. K. Jackson and J. P. Hart, Anal. Biochem., 2009, 385, 334–341 CrossRef CAS PubMed
.
- R. M. Pemberton, J. Xu, R. Pittson, G. A. Drago, J. Griffiths, S. K. Jackson and J. P. Hart, Biosens. Bioelectron., 2011, 26, 2448–2453 CrossRef CAS PubMed
.
- R. M. Pemberton, T. Cox, R. Tuffin, G. A. Drago, J. Griffiths, R. Pittson, G. Johnson, J. Xu, I. C. Sage, R. Davies, S. K. Jackson, G. Kenna, R. Luxton and J. P. Hart, Sensors, 2014, 14, 20519–20532 CrossRef PubMed
.
- S. Mross, T. Zimmermann, N. Winkin, M. Kraft and H. Vogt, Sens. Actuators, B, 2016, 236, 937–946 CrossRef CAS
.
- E. O. Pettersen, P. Ebbesen, R. G. Gieling, K. J. Williams, L. Dubois, P. Lambin, C. Ward, J. Meehan, I. H. Kunkler, S. P. Langdon, A. H. Ree, K. Flatmark, H. Lyng, M. J. Calzada, L. Del Peso, M. O. Landazuri, A. Görlach, H. Flamm, J. Kieninger, G. Urban, A. Weltin, D. C. Singleton, S. Haider, F. M. Buffa, A. L. Harris, A. Scozzafava, C. T. Supuran, I. Moser, G. Jobst, M. Busk, K. Toustrup, J. Overgaard, J. Alsner, J. Pouyssegur, J. Chiche, N. Mazure, I. Marchiq, S. Parks, A. Ahmed, M. Ashcroft, S. Pastorekova, Y. Cao, K. M. Rouschop, B. G. Wouters, M. Koritzinsky, H. Mujcic and D. Cojocari, J. Enzyme Inhib. Med. Chem., 2015, 30, 689–721 CrossRef CAS PubMed
.
- F. Hafner, Biosens. Bioelectron., 2000, 15, 149–158 CrossRef CAS PubMed
.
- S. E. Eklund, D. E. Cliffel, E. Kozlov, A. Prokop, J. Wikswo and F. Baudenbacher, Anal. Chim. Acta, 2003, 496, 93–101 CrossRef CAS
.
- S. E. Eklund, D. Taylor, E. Kozlov, A. Prokop and D. E. Cliffel, Anal. Chem., 2004, 76, 519–527 CrossRef CAS PubMed
.
- J. R. McKenzie, A. M. Palubinsky, J. E. Brown, B. McLaughlin and D. E. Cliffel, ACS Chem. Neurosci., 2012, 3, 510–518 CrossRef CAS PubMed
.
- N. Hu, C. Wu, D. Ha, T. Wang, Q. Liu and P. Wang, Biosens. Bioelectron., 2013, 40, 167–173 CrossRef CAS PubMed
.
- M. Lehmann, W. Baumann, M. Brischwein, R. Ehret, M. Kraus, A. Schwinde, M. Bitzenhofer, I. Freund and B. Wolf, Biosens. Bioelectron., 2000, 15, 117–124 CrossRef CAS PubMed
.
- M. Brischwein, E. R. Motrescu, E. Cabala, A. M. Otto, H. Grothe and B. Wolf, Lab Chip, 2003, 3, 234–240 RSC
.
- P. Mestres and A. Morguet, Expert Opin. Drug Discovery, 2009, 4, 785–797 CrossRef CAS PubMed
.
- H. Alborzinia, S. Can, P. Holenya, C. Scholl, E. Lederer, I. Kitanovic and S. Wölfl, PLoS One, 2011, 6, e19714 CAS
.
- A. Weltin, J. Kieninger, G. Urban, I. Moser, G. Jobst, M. Wego and R. Ehret, Proc. IEEE Sens., 2010, 2113–2116 Search PubMed
.
- I. A. Ges and F. Baudenbacher, Biosens. Bioelectron., 2010, 25, 1019–1024 CrossRef CAS PubMed
.
- I. A. Ges and F. Baudenbacher, Biosens. Bioelectron., 2010, 26, 828–833 CrossRef CAS PubMed
.
- J. R. McKenzie, A. Cognata, A. N. Davis, J. P. Wikswo and D. E. Cliffel, Anal. Chem., 2015, 87, 7857–7864 CrossRef CAS PubMed
.
- P. Wolf, M. Brischwein, R. Kleinhans, F. Demmel, T. Schwarzenberger, C. Pfister and B. Wolf, Biosens. Bioelectron., 2013, 50, 111–117 CrossRef CAS PubMed
.
- J. Wiest, A. Namias, C. Pfister, P. Wolf, F. Demmel and M. Brischwein, IEEE Trans. Biomed. Eng., 2016, 63, 2368–2375 CrossRef PubMed
.
- M. H. Wu, S. B. Huang, Z. Cui, Z. Cui and G. B. Lee, Sens. Actuators, B, 2008, 129, 231–240 CrossRef CAS
.
- Y. Chen, D. Gao, H. Liu, S. Lin and Y. Jiang, Anal. Chim. Acta, 2015, 898, 85–92 CrossRef CAS PubMed
.
- C. Kim, J. H. Bang, Y. E. Kim, S. H. Lee and J. Y. Kang, Lab Chip, 2012, 12, 4135–4142 RSC
.
- J. Ruppen, F. D. Wildhaber, C. Strub, S. Hall, T. Geiser, R. A. Schmid and O. T. Guenat, Lab Chip, 2015, 15, 3076–3085 RSC
.
- E. J. Vrij, S. Espinoza, M. Heilig, A. Kolew, M. Schneider, C. A. van Blitterswijk, R. K. Truckenmüller and N. C. Rivron, Lab Chip, 2016, 16, 734–742 RSC
.
- A. Mazzeo, E. Hong, D. E. Ingber and R. Derda, Anal. Chem., 2013, 85, 8085–8094 CrossRef PubMed
.
- K. A. Simon, B. Mosadegh, K. T. Minn, M. R. Lockett, M. R. Mohammady, D. M. Boucher, A. B. Hall, S. M. Hillier, T. Udagawa, B. K. Eustace and G. M. Whitesides, Biomaterials, 2016, 95, 47–59 CrossRef CAS PubMed
.
- K. Kwapiszewska, A. Michalczuk, M. Rybka, R. Kwapiszewski and Z. Brzózka, Lab Chip, 2014, 14, 2096–2104 RSC
.
- S. Hammer, A. Weltin, Y. Kaminski, F. Noor, J. Kieninger and G. A. Urban, Procedia Eng., 2015, 120, 961–964 CrossRef CAS
.
- A. Weltin, J. Kieninger, B. Enderle, A.-K. Gellner, B. Fritsch and G. A. Urban, Biosens. Bioelectron., 2014, 61, 192–199 CrossRef CAS PubMed
.
- D. Bavli, S. Prill, E. Ezra, G. Levy, M. Cohen, M. Vinken, J. Vanfleteren, M. Jaeger and Y. Nahmias, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2231–E2240 CrossRef CAS PubMed
.
- S. Prill, M. S. Jaeger and C. Duschl, Biomicrofluidics, 2014, 8, 34102 CrossRef PubMed
.
- Y. S. Zhang, J. Aleman, S. R. Shin, T. Kilic, D. Kim, S. A. Mousavi Shaegh, S. Massa, R. Riahi, S. Chae, N. Hu, H. Avci, W. Zhang, A. Silvestri, A. Sanati Nezhad, A. Manbohi, F. De Ferrari, A. Polini, G. Calzone, N. Shaikh, P. Alerasool, E. Budina, J. Kang, N. Bhise, J. Ribas, A. Pourmand, A. Skardal, T. Shupe, C. E. Bishop, M. R. Dokmeci, A. Atala and A. Khademhosseini, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E2293–E2302 CrossRef CAS PubMed
.
- S. A. M. Shaegh, F. De Ferrari, Y. S. Zhang, M. Nabavinia, N. B. Mohammad, J. Ryan, A. Pourmand, E. Laukaitis, R. B. Sadeghian, A. Nadhman, S. R. Shin, A. S. Nezhad, A. Khademhosseini and M. R. Dokmeci, Biomicrofluidics, 2016, 10, 44111 CrossRef PubMed
.
- R. Riahi, S. A. M. Shaegh, M. Ghaderi, Y. S. Zhang, S. R. Shin, J. Aleman, S. Massa, D. Kim, M. R. Dokmeci and A. Khademhosseini, Sci. Rep., 2016, 6, 24598 CrossRef CAS PubMed
.
- J.-Y. Kim, D. A. Fluri, J. M. Kelm, A. Hierlemann and O. Frey, J. Lab. Autom., 2015, 20, 274–282 CrossRef CAS PubMed
.
- T. E. de Groot, K. S. Veserat, E. Berthier, D. J. Beebe and A. B. Theberge, Lab Chip, 2016, 16, 334–344 RSC
.
- Y. R. F. Schmid, S. C. Bürgel, P. M. Misun, A. Hierlemann and O. Frey, ACS Sens., 2016, 1, 1028–1035 CrossRef CAS
.
- C. Luni, E. Serena and N. Elvassore, Curr. Opin. Biotechnol., 2014, 25, 45–50 CrossRef CAS PubMed
.
- C. Zhang, Z. Zhao, N. A. Abdul Rahim, D. van Noort and H. Yu, Lab Chip, 2009, 9, 3185–3192 RSC
.
- K. Aravindalochanan, J. Kieninger, J. A. Sandvik, E. O. Pettersen and G. A. Urban, in TRANSDUCERS 2009 - 15th International Conference on Solid-State Sensors, Actuators and Microsystems, 2009, pp. 1003–1006 Search PubMed.
- K. Domansky, W. Inman, J. Serdy, A. Dash, M. H. M. Lim and L. G. Griffith, Lab Chip, 2010, 10, 51–58 RSC
.
| This journal is © The Royal Society of Chemistry 2018 |