Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mimicking arterial thrombosis in a 3D-printed microfluidic in vitro vascular model based on computed tomography angiography data†
Pedro F.
Costa‡
ab,
Hugo J.
Albers‡
 *cd,
John E. A.
Linssen‡
b,
Heleen H. T.
Middelkamp
d,
Linda
van der Hout
c,
Robert
Passier
d,
Albert
van den Berg
c,
Jos
Malda
abe and
Andries D.
van der Meer
*d
*cd,
John E. A.
Linssen‡
b,
Heleen H. T.
Middelkamp
d,
Linda
van der Hout
c,
Robert
Passier
d,
Albert
van den Berg
c,
Jos
Malda
abe and
Andries D.
van der Meer
*d
aUtrecht Biofabrication Facility, Utrecht University, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands
bDepartment of Orthopaedics, University Medical Center Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands
cBIOS Lab-on-a-Chip Group, MESA+ Institute for Nanotechnology, MIRA Institute for Biomedical Technology and Technical Medicine, University of Twente, 7500 AE Enschede, The Netherlands. E-mail: h.j.albers@utwente.nl
dApplied Stem Cell Technologies Group, MIRA Institute for Biomedical Technology and Technical Medicine, University of Twente, 7500 AE Enschede, The Netherlands. E-mail: andries.vandermeer@utwente.nl
eDepartment of Equine Sciences, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 112, 3584 CM Utrecht, The Netherlands
First published on 18th July 2017
Abstract
Arterial thrombosis is the main instigating factor of heart attacks and strokes, which result in over 14 million deaths worldwide every year. The mechanism of thrombosis involves factors from the blood and the vessel wall, and it also relies strongly on 3D vessel geometry and local blood flow patterns. Microfluidic chip-based vascular models allow controlled in vitro studies of the interaction between vessel wall and blood in thrombosis, but until now, they could not fully recapitulate the 3D geometry and blood flow patterns of real-life healthy or diseased arteries. Here we present a method for fabricating microfluidic chips containing miniaturized vascular structures that closely mimic architectures found in both healthy and stenotic blood vessels. By applying stereolithography (SLA) 3D printing of computed tomography angiography (CTA) data, 3D vessel constructs were produced with diameters of 400 μm, and resolution as low as 25 μm. The 3D-printed templates in turn were used as moulds for polydimethylsiloxane (PDMS)-based soft lithography to create microfluidic chips containing miniaturized replicates of in vivo vessel geometries. By applying computational fluid dynamics (CFD) modeling a correlation in terms of flow fields and local wall shear rate was found between the original and miniaturized artery. The walls of the microfluidic chips were coated with human umbilical vein endothelial cells (HUVECs) which formed a confluent monolayer as confirmed by confocal fluorescence microscopy. The endothelialised microfluidic devices, with healthy and stenotic geometries, were perfused with human whole blood with fluorescently labeled platelets at physiologically relevant shear rates. After 15 minutes of perfusion the healthy geometries showed no sign of thrombosis, while the stenotic geometries did induce thrombosis at and downstream of the stenotic area. Overall, the novel methodology reported here, overcomes important design limitations found in typical 2D wafer-based soft lithography microfabrication techniques and shows great potential for controlled studies of the role of 3D vessel geometries and blood flow patterns in arterial thrombosis.
Introduction
Arterial thrombosis is the main cause of heart attacks and strokes, which together cause over 14 million deaths worldwide every year.1 Thrombosis is a complex process, which is instigated by multiple factors, both genetic and acquired: blood-borne factors, vessel wall dysfunction and fluid dynamical effects. Animal models are the most popular experimental tool to study the pathophysiology of thrombosis in laboratory settings. While animal models mimic the mechanistic multifactorial complexity found in arterial thrombosis, they do not fully recapitulate human physiology or human disease. For example, rodent platelet biology2 and coagulation dynamics3 display inter-species differences, and mice arteries typically undergo wall shear stresses that are an order of magnitude higher4 than those in humans. Furthermore, thrombosis in mice can only be instigated in the largest arteries by using ferric chloride or by laser-induced injury,5–8 limiting their use in studying thrombosis in specific, well-controlled 3D vascular geometries. Therefore, engineered in vitro blood flow chambers, perfused with human blood, are often used to complement or confirm results from in vivo models.9,10In recent years, microfluidic organs-on-chips emerged as new engineered in vitro models that capture aspects of human physiology with an unprecedented level of realism.11–13 Microfluidic technology has also been used extensively to create new in vitro models for studying thrombosis.14–18 For example, when microfluidic channels with cultured endothelial cells and geometric constrictions mimicking stenotic vessels were perfused with human whole blood, platelet aggregates tend to form specifically in the outlet zone of the constriction.14,15,19 Mimicking arterial physiology also encompasses applying an arterial shear rate to the endothelial cells and blood. It has been shown that shear rate is very important in von Willebrand factor activation and thus influences platelet adhesion and aggregation.14,15,20,21 However, due to inherent limitations posed by 2D wafer-based microfabrication techniques of microfluidic chips, the channels in almost all on-chip thrombosis models inevitably possess planar configurations with rectangular cross-sections instead of the typical 3D, round cross-section blood vessel structures found in vivo.11,22 A number of strategies have been employed to produce chips that contain microfluidic channels with circular cross-sections. For example, acrylic optical fibres in combination with standard soft lithography can be used to fabricate stylized geometries reminiscent of vessel stenoses and aneurysms.19 Moreover, microfabrication techniques, such as 3D bioprinting and gelatin-based soft lithography, have also been used to engineer microfluidic channels with circular cross-sections, which were then perfused with human blood to study thrombosis and thrombolysis.23,24
So far, none of these studies yielded microfluidic chips in which thrombosis can be studied in geometries that are truly representative of human vessel geometries. This type of 3D complexity is essential to accurately mimic the in vivo flow patterns which are fundamental in the mechanism of thrombosis.14,25,26
Three-dimensional printing has the potential to revolutionize the field of microfluidics by allowing to overcome the limitations posed by conventional 2D wafer-based microfabrication techniques.22 Furthermore, it allows the fabrication of fully 3D structures, which closely mimic the natural shape of complex human tissue structures and could, therefore, also be useful in replicating the architecture of human blood vessels.27–29 Stereolithography (SLA) in particular could be a suitable technique to fabricate architecturally complex vessel models, since it can reach very high printing resolutions and has already been successfully used in manufacturing microfluidic chips.30–32 In SLA-based technologies, 3D constructs are manufactured in a layer-by-layer fashion from liquid photolabile polymers (resins), which are cured at specific locations through exposure to light.32,33
In this study, we describe an advanced method to produce microfluidic chips that replicate the exact 3D architecture of both healthy and stenotic coronary arteries. Our method relies on SLA-based fabrication of miniaturized models of a coronary artery based on computed tomography angiography (CTA) imaging data. The printed models are used as moulds in polydimethylsiloxane (PDMS)-based soft lithography to produce microfluidic chips that contain exact replicas of the arterial architecture, as well as inlets and outlets to allow perfusion of the chips. By replicating the geometries found in vivo it becomes possible to overcome the limitations usually encountered when using square channel geometries, e.g. unrealistic flow profiles and fibrin formation in low-flow zones in channel corners. Moreover, we demonstrate that microfluidic chips produced with our method are compatible with cell culture and high-resolution confocal microscopy by seeding human umbilical vein endothelial cells (HUVECs) followed by image analysis. Finally, as a proof-of-principle, we assessed the effect of vessel geometries on platelet aggregation by perfusing healthy and stenotic vessel designs with human whole blood under real arterial shear rates.
Materials & methods
Coronary artery 3D models
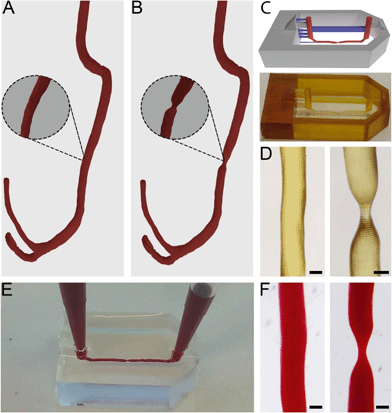
Anonymised thoracic CTA scans were acquired from a free online digital imaging and communication in medicine (DICOM) library.34 DICOM files consist of stacks of 2D tissue image sections which, when combined, can be used to generate 3D models. The utilized DICOM files were read and segmented using 3D Slicer (http://www.slicer.org).35 In short, by applying thresholding functions, all irrelevant tissue regions were excluded in order to only highlight vascular tissue. The result from this threshold was then converted to a 3D model by using the “model making” option in 3D Slicer.Directly after segmentation, many artefact volumes remained from surrounding tissue. The 3D model was, therefore, further processed in computer-aided design (CAD) software Blender (Blender foundation, Amsterdam, The Netherlands, version 2.72) to remove artefacts and smoothen the surface. Through specific selection of meshes within Edit Mode, the residual surrounding tissue geometries and artefacts were removed. The model was subsequently smoothened within Sculpt Mode using the ‘F smooth function’. After downsizing, this resulted in a 3D vessel model with a channel diameter of 400 μm (Fig. 1A).
Stenosis models with 55% and 67% constrictions were then further created by narrowing a straight segment of the channel to diameters of 180 μm and 140 μm respectively (Fig. 1B, 55% not shown). This step was performed in Blender with the Sculpt Mode by selecting the ‘F-inflate/deflate’ and ‘F-smooth’ functions.
Microfluidic device design and fabrication
Microfluidic devices were fabricated by common PDMS-based soft lithography using moulds created by SLA.22,31The full model for 3D printing contained the 3D vessel models, as well as inlets, outlets and a box-like container and was designed in Blender CAD software. The generated 3D vessel models were further extended with cylindrical sections so they would fit the length (15 mm) of the box-like container. The container (width 10 mm) was composed of six 4 mm thick walls which were designed to restrict the PDMS into a confined volume upon casting and moulding in later steps. Furthermore, the full 3D model contained inlets and outlets (diameter 1 mm, height 5 mm), and support structures (width 300 × 500 μm, height 1.8 mm) connected to the inlets, outlets and vessel models to allow accurate printing of the suspended section of the 3D artery model. The maximum amount of overhang in an SLA model is roughly 45°, and any structures positioned below this angle regularly fail to print. The final curvature of the printed model can exceed 45° as long as each subsequent layer does not exceed the 45° overhang compared to the previous layer. The inlets and outlets were positioned at a horizontal angle (i.e. 0/180°) and therefore support structures were needed to facilitate printing. The vessel structures were positioned at the centre of the moulding area of the device and placed at only 100 μm distance from the mould's bottom plane to ensure that after moulding, the resulting chips would be compatible with high magnification microscopy. The arrangement of the vessel within the full 3D model was defined in such a way that the channel would be suspended and horizontally levelled to the Z-axis along the whole length of the device (Fig. 1C). Any residual artefacts and mesh errors present in the 3D models were removed with Magics software (Materialise NV, Leuven, Belgium).
The full 3D models were then printed from PIC100 resin in a Perfactory 3 mini-multi lens SLA printer (EnvisionTEC GmbH, Gladbeck, Germany) using a resolution of 25 μm (Fig. 1C). The resulting devices were flashed for 3500 cycles in an Otoflash G171 apparatus (EnvisionTEC) and rinsed in 100% ethanol at 37 °C for 7 hours, refreshing ethanol roughly every 2 hours. Using Fiji image analysis software,36 the resulting construct dimensions were determined. PDMS (Sylgard 184, Dow Corning, Midland MI, USA) pre-polymer was then mixed with the crosslinking agent in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, poured into the moulds and set to cure overnight at 60 °C.
1 ratio, poured into the moulds and set to cure overnight at 60 °C.
The chips were finished by first breaking open the moulds manually and then gently removing the vessel segments of the moulds by pulling with forceps and leaving an open channel inside the PDMS (Fig. S1†).
Finally, the chips were sterilized by means of plasma treatment (60 s at 100 W, 600 mTorr, Harrick Plasma) and bonded to a microscope cover glass.
Endothelial cell culture and seeding
After the plasma treatment, the microfluidic channels in the chips were coated with a solution of 0.1 mg ml−1 rat tail collagen-I (Cultrex-Trevigen) in 1× phosphate buffered saline (PBS; Sigma Aldrich). The collagen solution was introduced in the microfluidic device and incubated for 30 minutes followed by a PBS wash. The microfluidic device was endothelialised by introducing 40 μl of 4 × 106 cells per ml HUVEC (Lonza) suspension in EGM-2 (Lonza) culture medium and incubating for 30 minutes on a platform rocker (Thermolyne Vari-mix) set to 6 rpm. The introduction of cells was repeated and the microfluidic device was incubated upside down for another 30 minutes. The endothelialised microfluidic device was cultured for a period of two days to ensure a confluent monolayer, during which the EGM-2 culture medium was refreshed twice per day.Staining, fluorescence microscopy and confocal microscopy
A live/dead staining was performed using the live/dead viability/cytotoxicity kit for mammalian cells according to the manufacturer's protocol (ThermoFisher Scientific – calcein AM, ethidium homodimer-1). Alternatively, to visualize cell morphology, the cells were fixated using 4% paraformaldehyde (Sigma Aldrich) solution and permeated using a 0.3% Triton X-100 (Sigma Aldrich) solution in PBS. The nuclei and F-actin filaments were stained using two molecular probes: NucBlue (ThermoFisher Scientific) and ActinGreen (ThermoFisher Scientific), respectively. The stained cells were imaged using an EVOS FL cell imaging system using DAPI, CY5 and GFP filter cubes. Confocal fluorescent microscopy was performed on a Nikon confocal A1 microscope with excitation lasers of wavelength 405 and 488 nm.Blood perfusion
Microfluidic devices were connected to a syringe pump (Harvard Apparatus PhD 2000) using custom connectors and Tygon tubing. The Tygon tubing was coated with a washing buffer of HEPES, 1% bovine serum albumin (Sigma Aldrich), 1% glucose (Merck Millipore), 200 mM CaCl2 (Sigma Aldrich) and 0.1 μl ml−1 heparin (Sigma Aldrich) to avoid clotting. Human citrated whole blood was used in perfusion experiments. The blood was provided by the Experimental Centre for Technical Medicine (ECTM, MIRA Institute, University of Twente) and used within a maximum of 4 hours after donation. The citrated blood was re-calcified using a re-calcification buffer of 63.2 mM CaCl2 and 31.6 mM MgCl2 (ThermoFisher Scientific) and stained with 2 μg ml−1 DiOC6 (Life Technologies). Blood was pulled through the microfluidic device using the syringe pump at a flow rate of 0.29 ml min−1 yielding a shear rate of approximately 1000 s−1. The chosen shear rate would recapitulate the shear rate experienced by endothelium in large arteries.37–39 Platelet aggregation dynamics were quantified in Fiji. An image stack was generated with a resolution of 1 frame per second, capturing the full length of the perfusion. This stack was then converted to black/white images showing either the presence or absence of platelets, through specific thresholding. Finally, the platelets were quantified by measuring the white intensity (platelets) inside a region of interest (ROI) interpolated over the full image stack (Fig. S8†).Computational fluid dynamics
COMSOL 5.1 Multiphysics CFD module was used to predict the fluid flow profile, streamlines and shear rate distribution inside the 3D vessel geometries. The laminar flow module, that assumes incompressible flow, no turbulence and does not neglect the inertial term in Navier–Stokes' law, was used. The CAD 3D vessel model that was incorporated in the chip design was also used in COMSOL using a straight section modelled with a vessel diameter of 400 μm, which is compared to the original CTA vessel geometry with a vessel diameter of 4 mm diameter. A flow rate of 4.83 × 10−9 m3 s−1 was set on the inlet for the miniaturized model, and a flow rate of 1.0 × 10−6 m3 s−1 was set for the original model, both resulting in a wall shear rate of 1000 s−1. An entrance length of 1 m was chosen to provide a fully developed flow on entry of the geometry. The outlet pressure was set to atmospheric pressure with no back flow. On the outer boundary of the geometries, excluding the inlets and outlets, a no-slip boundary condition was imposed.Results & discussion
Fabrication of microfluidic chips with 3D blood vessel geometries
The flow properties of vessels are greatly determined by their 3D shape. Therefore, to accurately mimic in vivo flow conditions in our model, it is crucial to closely replicate natural shapes. To attain exact in vivo shapes, human thoracic CTA scans were segmented and converted to 3D models, which were further isometrically reduced in size by a factor of 10 before being used to fabricate chips (Fig. 1A and B).It was found that only vessels with a diameter of approximately 100 μm would still 3D print successfully; by further reducing the diameter, the printed vessels would become fragile and collapse during printing. Given this lower limit, we always printed the model vessels with a non-stenosis diameter of 400 μm, so that even a severe stenosis of 67% (stenotic diameter of approximately 130 μm) would still print successfully (Fig. 1C and D). Even more severe occlusions could be printed by increasing the vessel diameter while keeping the stenotic diameter constant. Constructs with healthy vessel geometries showed a printing success rate of roughly 80%, while stenotic constructs showed a printing success of 50%. Printing defects consisted mainly of small offsets perpendicular to the vessel direction (Fig. S2†). In later experiments, such offsets would potentially alter flow patterns within the microfluidic chips and therefore the constructs possessing these defects were discarded. However, this productivity loss could be compensated by the fast production method and the number of chips that could be printed at once. All printed constructs also showed artefacts consisting of ring-like indentations (approximately 2 to 5 μm in size) that were the result of the partial overlap of consecutively printed layers. These artefacts are inherent to SLA at these resolutions and they were also found in the chips after moulding (Fig. S3†). Although these artefacts cannot be avoided, we did not see any indication of negative effects caused by these ridges (see sections below).
It was possible to observe that the successfully printed constructs closely matched the designed 3D model dimensions (Fig. 1C). By using Fiji image analysis software, actual diameters of printed constructs were found to be 10 to 30 μm smaller than the design, with diameters of 370 μm in the straight channel segments, and 180 μm (55%) and 120 μm (67%) for the stenosis geometries.
When using the moulds in PDMS-based soft lithography, chips containing healthy and stenotic geometries could be successfully created. Both the stenotic and cylindrical geometries were easily removable from within the cured PDMS and did not damage the integrity of the chips whatsoever (Fig. 1F). Although the vessel segments used in our studies were relatively straight, we also successfully used our method to produce chips with vessel segments containing bends with angles of up to 80° (Fig. S4†). The success rate of manufacturing chips with such dramatic curves is substantially lower due to entrapment of parts of the 3D-printed constructs inside the cross-linked PDMS. In order to produce chips with complex 3D curves, that more closely mimic in vivo shapes, success rates may be increased by using sacrificial resins40 or modular chips.41
The produced PDMS chips could be perfused with fluids, without any observation of leakage (Fig. 1E). The created inlets and outlets allowed tight sealing of pipette tips and tube connectors and facilitated perfusion and hydrostatic pressure-driven flow between two pipette tips for cell culturing.
Unlike what is usually seen in conventional soft lithography techniques, within the proposed method, alignment of PDMS layers or bonding to glass is no longer needed to create channels.14,22,42 Moreover, it was possible to create perfusable free-form 3D channels, rather than the typical square-section or stylized circular channels observed in standard PDMS microfluidic chips.
Endothelialisation of microfluidic chips with 3D model vessels
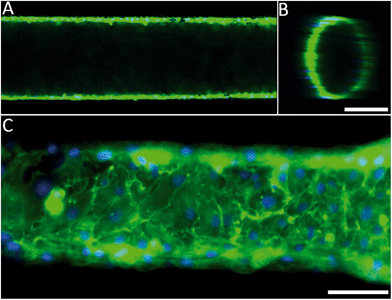
The engineered 3D microfluidic devices were endothelialised by culturing human umbilical vein endothelial cells (HUVECs) in the channels for two days. Since the PDMS could potentially still contain cytotoxic photo-crosslinker residues,43 which originated from the PIC100, a viability assay was performed on the microfluidic device. A live/dead staining of the endothelial monolayer showed that the number of dead cells was negligible (Fig. S5†). Confocal fluorescence microscopy and epifluorescence microscopy combined with nuclei and actin filament staining (Fig. 2A and B) was used to analyse the morphology of the endothelial cells for a 1250 μm section of a microfluidic device. The results (Fig. 2C) confirmed that the endothelial cells had indeed formed a confluent monolayer and that the HUVECs possessed a typical cobblestone morphology. Furthermore, the inset (Fig. 2C) demonstrated that the HUVECs were not affected by the microtopographical artefacts (Fig. S3†) that result from the SLA.Computational fluid dynamics on the 3D vessel geometries
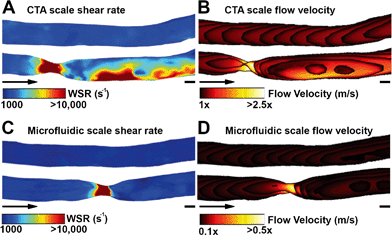
Shear rate distributions and fluid flow profiles in both the microfluidic 3D geometries and the original CTA scale 3D vessel geometries were analysed by CFD. A comparison between healthy and stenotic geometries showed a difference in and near the stenotic area (Fig. 3). Typical shear rates in the non-stenotic sections of the 3D vessel were in the range of 1000 s−1. Because of the 67% occlusion, higher velocities (Fig. 3B and D) and consequently shear rates (Fig. 3A and C) were measured in stenotic sections with shear rates higher than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s−1. A comparison between the CTA scale geometry (Fig. 3A and B) and the smaller microfluidic scale model (Fig. 3C and D), showed that, with the chosen volumetric flow rate, the shear rate distribution in the stenosis is very similar, and this is also the case for higher shear rates. Shear rates higher than 5000 s−1 are considered as pathological and shear rates higher than 8000–10
000 s−1. A comparison between the CTA scale geometry (Fig. 3A and B) and the smaller microfluidic scale model (Fig. 3C and D), showed that, with the chosen volumetric flow rate, the shear rate distribution in the stenosis is very similar, and this is also the case for higher shear rates. Shear rates higher than 5000 s−1 are considered as pathological and shear rates higher than 8000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s−1 activate von Willebrand Factor.14,21 Therefore, all shear rates above 10
000 s−1 activate von Willebrand Factor.14,21 Therefore, all shear rates above 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s−1 are considered as relevant. The CTA scale computational model showed an elevated shear rate downstream of the stenosis, which was a result of inertial forces in the curved section after the stenosis. Recirculation might have contributed to the different shear rate distributions found in the two stenotic models. The stenotic CTA scale model (Fig. S6d†) showed severe recirculation with elevated backflow velocities. The stenotic microfluidic scale model (Fig. S6c†) also showed recirculation, but the affected area and backflow velocities are smaller. The two healthy models (Fig. S6a and b†) did not show any sign of recirculation. The microfluidic scale model showed a different shear rate pattern because the Reynolds number in the CTA scale model is significantly higher in comparison to the microfluidic scale model, resulting in an altered shear rate distribution and recirculation.
000 s−1 are considered as relevant. The CTA scale computational model showed an elevated shear rate downstream of the stenosis, which was a result of inertial forces in the curved section after the stenosis. Recirculation might have contributed to the different shear rate distributions found in the two stenotic models. The stenotic CTA scale model (Fig. S6d†) showed severe recirculation with elevated backflow velocities. The stenotic microfluidic scale model (Fig. S6c†) also showed recirculation, but the affected area and backflow velocities are smaller. The two healthy models (Fig. S6a and b†) did not show any sign of recirculation. The microfluidic scale model showed a different shear rate pattern because the Reynolds number in the CTA scale model is significantly higher in comparison to the microfluidic scale model, resulting in an altered shear rate distribution and recirculation.
To further investigate the effects of the aforementioned ring-like indentations, a smooth channel and a channel with 4 μm ridges, both with a diameter of 400 μm, were compared using a CFD model. No significant differences in flow velocity profiles were found and shear rate distributions did show minor differences (Fig. S7†).
Thrombosis-on-a-chip
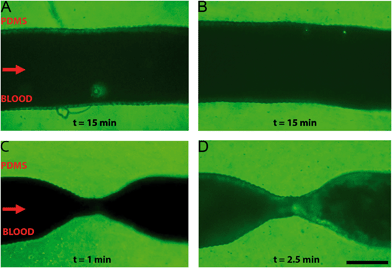
A comparison between healthy and stenotic geometries was made by perfusion of re-calcified citrated human whole blood through a series of endothelialised microfluidic devices with varying degrees of occlusion: 0%, 55% and 67%, respectively. Healthy and stenotic microfluidic devices were perfused at the same flow rates. The healthy geometries did not show any sign of thrombosis along the entire length of the microfluidic device, after at least 15 minutes of blood perfusion (Fig. 4A and B). Furthermore, this demonstrated that the ring-like indentations did not influence platelet aggregation. The stenotic geometries did induce thrombosis at and downstream of the stenotic apices as demonstrated by clear fluorescent platelet aggregation (Fig. 4C and D). The first signs of platelet aggregation in the stenosis region could be observed as soon as two minutes after the start of whole blood perfusion.Minor recirculation was expected based on the CFDs in the supplementary figures (Fig. S6†). Indeed, platelet aggregation was also observed in this recirculation zone distal to the stenotic geometries. Recirculation plays an important role in thrombus formation, as shear rate and exposure time increases.44 Recirculation also results in a longer exposure time to paracrine activation factors and allows for easier deposition of fibrin in low flow zones because of a longer retention time.45,46 Interestingly, the formation of a thrombus was not limited merely to the recirculation area, as platelet aggregation also occurred near the onset of the stenosis and at the apex of the stenosis where no recirculation is to be expected. The local increase in shear rate in these areas is likely to also contribute to the observed thrombosis, for example by the activation of von Willebrand factor resulting in platelet adhesion.14,15,47 Additionally, by using re-calcified whole-blood, the intrinsic coagulation pathway will become active over the course of the experiment. This contact activation of coagulation will likely also contribute to the observed platelet aggregation because of the known cross-talk between activated coagulation factors and platelets.48 Whole blood perfusion through healthy and stenotic geometries showed that thrombosis only occurred in microfluidic devices with a stenotic geometry, which suggests that – in line with previous reports14,15 – fluid flow velocity gradients, minor recirculation, intrinsic coagulation pathways and increased shear rates are likely to be the determining factors of thrombogenesis in our microfluidic chips.
Although the constant flow rate used in this experiment is not completely representative of physiological flow patterns, it can be suitable to investigate the onset of thrombosis in a well-controlled setting. In future studies, a pressure-driven flow could be used instead, as this better mimics the dynamic changes in blood flow during vessel occlusion.
The thrombus size over time was determined by a semi-quantitative analysis in several microfluidic devices (Fig. S8†). Thrombus formation in 3 stenotic geometries (Fig. S8b–d†) was compared with a healthy geometry (Fig. S8a†). Different aggregation mechanics are observed in the stenotic geometries. First, continuous aggregate growth which is characterized by a steady increase in thrombus size and intensity. Second, stick and slip growth which involves small aggregates to grow and dislodge, leading to an increase in intensity followed by a sharp drop in intensity after thrombus dislodgement. The stenotic geometries showed a combination of the aggregation mechanics although each experiment showed a different predominant mechanism.
Conclusions
By combining CTA data, SLA 3D printing and microfluidics, we have developed a new approach for manufacturing microfluidic in vitro cell culture models which closely mimic the architectures found in both healthy and stenotic arteries with high precision. With this approach thrombosis can be recapitulated in 3D vessel geometries in a way that is not possible in microfluidic chips fabricated with typical 2D wafer-based soft lithography, or even in microfluidic chips produced with acrylic fibres or by advanced biofabrication methods.14,15,49 Furthermore, because it is based on 3D models, this methodology allows for easy correlation and comparison of in vitro studies with in silico fluid flow simulations. In the future, the devices and method described herein could be used to employ medical CTA imaging data to produce patient-specific microfluidic chips, and as such, the method and proof-of-principle can be considered to be an important stepping stone towards patient-specific arterial thrombosis studies. Overall, our method represents an important step towards making microfluidic organ-on-chip-based technology increasingly relevant in experimental studies of arterial thrombosis.Acknowledgements
The authors acknowledge the funding received from Utrecht Life Sciences, the European Research Council (ERC) under the Advanced Grant ‘VESCEL’ Program (Grant no. 669768) of prof. Van den Berg, and the University of Twente Strategic Research Orientation ‘Organs-on-Chips’.Notes and references
- United Nations World Health Organization, Fact sheet No 310 http://www.who.int/mediacentre/factsheets/fs310/en/ 310.
- M. Jirouskova, A. S. Shet and G. J. Johnson, A guide to murine platelet structure, function, assays, and genetic alterations, J. Thromb. Haemostasis, 2007, 5, 661–669 CrossRef CAS PubMed.
- J. Suo, et al., Hemodynamic shear stresses in mouse aortas: Implications for atherogenesis, Arterioscler., Thromb., Vasc. Biol., 2007, 27, 346–351 CrossRef CAS PubMed.
- C. Lemini, R. Jaimez and Y. Franco, Gender and inter-species influence on coagulation tests of rats and mice, Thromb. Res., 2007, 120, 415–419 CrossRef CAS PubMed.
- P. Cullen, et al., Rupture of the atherosclerotic plaque: Does a good animal model exist?, Arterioscler., Thromb., Vasc. Biol., 2003, 23, 535–542 CrossRef CAS PubMed.
- L. Badimon, G. Vilahur and T. Padro, Atherosclerosis and thrombosis: Insights from large animal models, J. Biomed. Biotechnol., 2011, 2011, 1–12 Search PubMed.
- B. B. C. Furie and B. B. C. Furie, Review series Thrombus formation in vivo, J. Clin. Invest., 2005, 115, 3355–3362 CrossRef CAS PubMed.
- P. Pérez, M. Alarcón, E. Fuentes and I. Palomo, Thrombus formation induced by laser in a mouse model, Exp. Ther. Med., 2014, 8, 64–68 Search PubMed.
- R. Van Kruchten, J. M. E. M. Cosemans and J. W. M. Heemskerk, Measurement of whole blood thrombus formation using parallel-plate flow chambers - a practical guide, Platelets, 2012, 23, 229–242 CrossRef CAS PubMed.
- K. H. Benam, et al., Engineered In Vitro Disease Models, Annu. Rev. Phytopathol., 2015, 10, 195–262 CrossRef CAS PubMed.
- S. N. Bhatia and D. E. Ingber, Microfluidic organs-on-chips, Nat. Biotechnol., 2014, 32, 760–772 CrossRef CAS PubMed.
- A. D. Van Der Meer and A. Van Den Berg, Organs-on-chips: breaking the in vitro impasse, Integr. Biol., 2012, 4, 461 RSC.
- A. D. van der Meer, V. V. Orlova, P. ten Dijke, A. van den Berg and C. L. Mummery, Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device, Lab Chip, 2013, 13, 3562–3568 RSC.
- E. Westein, et al., Atherosclerotic geometries exacerbate pathological thrombus formation poststenosis in a von Willebrand factor-dependent manner, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 1357–1362 CrossRef CAS PubMed.
- W. S. Nesbitt, et al., A shear gradient-dependent platelet aggregation mechanism drives thrombus formation, Nat. Med., 2009, 15, 665–673 CrossRef CAS PubMed.
- E. Westein, S. de Witt, M. Lamers, J. M. E. M. Cosemans and J. W. M. Heemskerk, Monitoring in vitro thrombus formation with novel microfluidic devices, Platelets, 2012, 23, 501–509 CrossRef CAS PubMed.
- S. Zhu, et al., In microfluidico: Recreating in vivo hemodynamics using miniaturized devices, Biorheology, 2015, 52, 303–318 CrossRef CAS PubMed.
- M. Tsai, et al., In vitro modeling of the microvascular occlusion and thrombosis that occur in hematologic diseases using microfluidic technology, J. Clin. Invest., 2012, 122, 408–418 CAS.
- R. G. Mannino, et al., ‘Do-it-yourself in vitro vasculature that recapitulates in vivo geometries for investigating endothelial-blood cell interactions’, Sci. Rep., 2015, 5, 12401 CrossRef PubMed.
- D. L. Bark and D. N. Ku, Platelet transport rates and binding kinetics at high shear over a thrombus, Biophys. J., 2013, 105, 502–511 CrossRef CAS PubMed.
- L. D. C. Casa, D. H. Deaton and D. N. Ku, Role of high shear rate in thrombosis, J. Vasc. Surg., 2015, 61, 1068–1080 CrossRef PubMed.
- Y. Xia and G. M. Whitesides, Soft Lithography, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- Y. Zheng, et al., In vitro microvessels for the study of angiogenesis and thrombosis, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9342–9347 CrossRef CAS PubMed.
- Y. S. Zhang, et al., Bioprinted thrombosis-on-a-chip, Lab Chip, 2016, 16, 4097–4105 RSC.
- S. Han, et al., A versatile assay for monitoring in vivo-like transendothelial migration of neutrophils, Lab Chip, 2012, 12, 3861 RSC.
- R. Estrada, G. A. Giridharan, M. D. Nguyen, S. D. Prabhu and P. Sethu, Microfluidic endothelial cell culture model to replicate disturbed flow conditions seen in atherosclerosis susceptible regions, Biomicrofluidics, 2011, 5, 1–11 CrossRef PubMed.
- J. Visser, et al., Biofabrication of multi-material anatomically shaped tissue constructs, Biofabrication, 2013, 5, 35007 CrossRef PubMed.
- J. Malda, et al., 25th anniversary article: Engineering hydrogels for biofabrication, Adv. Mater., 2013, 25, 5011–5028 CrossRef CAS PubMed.
- P. S. D'urso, et al., Stereolithographic biomodelling in cranio-maxillofacial surgery: a prospective trial, J. Craniomaxillofac. Surg., 1999, 27, 30–37 CrossRef.
- A. K. Au, W. Lee and A. Folch, Mail-order microfluidics: evaluation of stereolithography for the production of microfluidic devices, Lab Chip, 2014, 14, 1294–1301 RSC.
- S. Knowlton, et al., 3D-printed microfluidic chips with patterned, cell-laden hydrogel constructs, Biofabrication, 2016, 8, 25019 CrossRef PubMed.
- P. Zorlutuna, et al., Microfabricated biomaterials for engineering 3D tissues, Adv. Mater., 2012, 24, 1782–1804 CrossRef CAS PubMed.
- C. W. Hull, Apparatus for production of three-dimensional objects by stereolithography, US Pat., 4575330, 1984 Search PubMed.
- AGECANONIX: Cardiac and coronary artery CT from http://www.osirix-viewer.com/resources/dicom-image-library/ (last checked 23-05-17).
- A. Fedorov, et al., 3D Slicer as an image computing platform for the Quantitative Imaging Network, Magn. Reson. Imaging, 2012, 30, 1323–1341 CrossRef PubMed.
- J. Schindelin, et al., Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- R. Dammers, et al., Shear stress depends on vascular territory: comparison between common carotid and brachial artery, J. Appl. Physiol., 2003, 94, 485–489 CrossRef PubMed.
- S. P. Wu, et al., Wall Shear Rates Differ between the Normal Carotid, Femoral, and Brachial Arteries: An In Vivo MRI Study, J. Magn. Reson. Imaging, 2004, 19, 188–193 CrossRef PubMed.
- A. Schmidt-Trucksass, et al., Arterial properties of the carotid and femoral artery in endurance-trained and paraplegic subjects, J. Appl. Physiol., 2000, 89, 1956–1963 CAS.
- R. Liska, F. Schwager, C. Maier, R. Cano-Vives and J. Stampfl, Water-soluble photopolymers for rapid prototyping of cellular materials, J. Appl. Polym. Sci., 2005, 97, 2286–2298 CrossRef CAS.
- K. Vittayarukskul and A. P. Lee, A truly Lego ® -like modular microfluidics platform, J. Micromech. Microeng., 2017, 27, 35004 CrossRef.
- D. Huh, et al., Reconstituting organ level lung functions on a chip, Science, 2010, 328, 1662–1668 CrossRef CAS PubMed.
- R. J. Narayan, Rapid Prototyping of Biomaterials: Principles and Applications, Woodhead Publishing, 2014 Search PubMed.
- T. L. Poepping, R. N. Rankin and D. W. Holdsworth, Flow patterns in carotid bifurcation models using pulsed doppler ultrasound: Effect of concentric vs. eccentric stenosis on turbulence and recirculation, Ultrasound Med. Biol., 2010, 36, 1125–1134 CrossRef PubMed.
- K. Rana and K. B. Neeves, Blood flow and mass transfer regulation of coagulation, Blood Rev., 2016, 30, 357–368 CrossRef CAS PubMed.
- S. P. Jackson, Arterial thrombosis—insidious, unpredictable and deadly, Nat. Med., 2011, 17, 1423–1436 CrossRef CAS PubMed.
- Z. M. Ruggeri, The role of von Willebrand factor in thrombus formation, Thromb. Res., 2007, 120, 5–9 CrossRef PubMed.
- J. W. M. Heemskerk, E. M. Bevers and T. Lindhout, Platelet activation and blood coagulation, Thromb. Haemostasis, 2002, 88, 186–193 CAS.
- A. Jain, et al., Assessment of whole blood thrombosis in a microfluidic device lined by fixed human endothelium, Biomed. Microdevices, 2016, 18, 1–7 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7lc00202e |
| ‡ Shared first-authorship. |
| This journal is © The Royal Society of Chemistry 2017 |