Open Access Article
Open Access ArticleA short, rigid linker between pyrene and guanidiniocarbonyl-pyrrole induced a new set of spectroscopic responses to the ds-DNA secondary structure†
Marijana
Radić Stojković
a,
Patryciusz
Piotrowski
b,
Carsten
Schmuck
*b and
Ivo
Piantanida
*a
aDivision of Organic Chemistry and Biochemistry, Ruđer Bošković Institute, P. O. Box 180, HR-10002 Zagreb, Croatia. E-mail: Ivo.Piantanida@irb.hr
bInstitute for Organic Chemistry, University of Duisburg-Essen, Universitässtrasse 7, 45141 Essen, Germany. E-mail: carsten.schmuck@uni-due.de; Fax: (+) 201 183 4259
First published on 2nd December 2014
Abstract
A novel pyrene-guanidiniocarbonyl-pyrrole dye, characterised by a short, rigid linker between the two chromophores, interacts strongly with ds-DNA but only negligibly with ds-RNA. Under neutral conditions the dye shows strong selectivity toward AT-DNA (with respect to GC-DNA). Binding is accompanied by a specific ICD band at 350 nm and fluorescence quenching for all DNAs/RNAs studied. At pH 5 the affinity of the dye is reversed, now favouring GC-DNA over AT-DNA. A strong emission increase for AT-DNA is observed but with quenching for GC-DNA.
Introduction
Versatile applications of small molecules targeting DNA/RNA in biochemical and biomedicinal applications have attracted enormous interest for more than 60 years.1,2 The research on DNA/RNA dyes was mostly focused on the impact on the DNA/RNA function or selective/specific DNA/RNA labelling.3,4 However, the huge complexity of DNA-coded processes, which do not depend only on the corresponding coding DNA basepair sequence but also include epigenetics, only recently attracted attention.5 To address such complex systems, during the last two decades researchers often combined two or more DNA/RNA binding modes in designing novel small molecules used for nucleic acid sensing. Especially specific 3-D binding-motifs are of interest to achieve such highly selective or specific interactions even with slightly different DNA/RNA structures.2,6 For the recognition reporting, fluorescence is the most popular method.7 However, upon increasing sensitivity many new techniques or even long established ones are faced with the challenge to circumvent fluorescence disadvantages. An alternative is circular dichroism (CD) spectropolarimetry, a useful method to study conformational changes in the secondary structure of polynucleotides.8 CD can also be used to study binding interactions as small achiral dyes can eventually acquire an induced CD spectrum (ICD) upon binding to polynucleotides.9 Thus, with currently increasing sensitivity, CD spectropolarimetry offers new possibilities of probing DNA/RNA structures.In previous work we already studied a series of DNA/RNA-binding compounds in which pyrene, as a fluorophore, was attached to a guanidiniocarbonyl-pyrrole (GCP) cation, an artificial tailor-made anion binding site, which is capable of interacting with the nucleic acid phosphate backbone compounds.10–13 These studies revealed several intriguing properties of such compounds: for instance 1 (Scheme 1, 1+ charge, pH dependent) showed dual spectroscopic recognition between ds-DNA and ds-RNA at pH 5, 2 (2+ charge, pH dependent) transferred the ICD band recognition pattern between ds-DNA and ds-RNA to neutral conditions, 3 and 4 (1+ charge, pH dependent and additional H-bonding sites in the linkers) finely tuned fluorescence and ICD sensing between various ds-polynucleotides based on minor groove recognition. Here we present a novel pyrene analogue 5 characterized by one pH dependent positive charge (GCP unit) and the shortest linker (with respect to 1–4) of increased rigidity, equipped with a neutral carboxylic-ester side chain. We hope that with a more rigid structure 5 should show increased selectivity in DNA/RNA binding, whereby the carboxylic-ester side chain (neutral to avoid electrostatic repulsion of the negative charges of the nucleic acid) could help to position 5 within the DNA/RNA binding site by a combination of steric hindrances and eventual H-bonding interactions. Moreover, 5 has a chiral centre in the main linker chain, which could introduce chiral control of the orientation of the two chromophores.
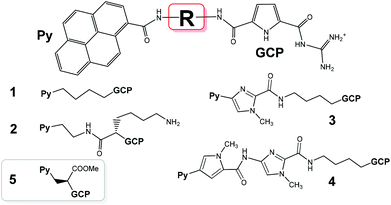 | ||
| Scheme 1 Previously studied pyrene–guanidiniocarbonyl–pyrrole conjugates 1–4 and the novel derivative studied here (5), which is characterized by the shortest linker studied so far. | ||
Results and discussion
Synthesis of compound 5 was performed according to Scheme 2. First a pyrene functionalized amino acid 12 was synthesized, which was then coupled to a Boc-protected guanidinio-carbonylpyrrole derivative 14. Details of the synthesis and characterization of new compounds are given in ESI.†Physico-chemical and spectroscopic properties of aqueous solutions of 5
Due to the low solubility of 5 a stock solution was prepared in DMSO (0.01 M) and solutions for the subsequent studies were prepared by adding small aliquots of this stock solution to buffer solutions (0.05 M Na cacodylate, DMSO content of the final solutions <0.01%). Clear solutions of 5 in the 10−5 M range were obtained in this way. At pH 7 compound 5 is neutral, while at pH 5 the GCP group is protonated, so that 5 bears one positive charge.10,13 The concentration dependence of the UV/vis spectrum was linear up to 2 × 10−5 M and the shape of the fluorimetric spectrum showed a well-defined pyrene emission maximum at λ = 402 nm confirming that under these conditions 5 is not aggregated but present as individual molecules in solution. Absorption maxima and the corresponding molar extinction coefficients (ε) are given in ESI.† Heating of the aqueous solutions of 5 up to 90 °C did not cause any significant changes in the UV/Vis spectra and reproducibility upon cooling to room temperature verified the chemical stability of the compound.Interactions with DNA/RNA in aqueous medium
Because of the significantly different protonation states of 5 at different pH values, binding studies were performed at pH 7 and pH 5. At pH 7 compound 5 is mostly neutral whereas at pH 5 it is mostly protonated (pKs of the GCP unit is around 6). In thermal denaturation experiments at pH 5 and 7, 5 stabilized exclusively AT-DNA (pH 7/pH 5, ΔTm = 3.0/9.0 °C, respectively) but not ct-DNA which has a mixed sequence with 42% GC-basepairs. This is significantly different from the previously studied analogues 1–4. Moreover, 5 did not stabilize ds-RNA. The increased stabilisation effect of 5 at pH 5 can be attributed to increased electrostatic interactions of the now protonated form of 5 with the negatively charged backbone of ds-DNA. In line with this interpretation is the observation that compound 2 which is positively charged even at neutral pH (due to the lysine side chain) shows a significantly stronger binding than 5 at pH 7.The UV/vis titrations of 5 with ds-DNA are characterized by a moderate hypochromic of the absorption band at λ > 300 nm, which suggests aromatic stacking interactions. In contrast, the addition of ds-RNA resulted in only negligible changes (ESI Table S2†).
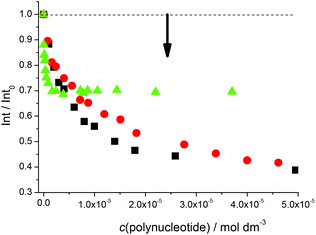
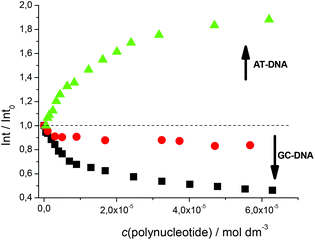
Fluorimetric titrations revealed emission changes which depend on the pH, the type of nucleic acid (DNA vs. RNA) and the basepair composition of the polynucleotide added. Namely, at pH 7 the emission of 5 was quenched by all added ds-DNA/RNA studied (Fig. 1). However, at pH 5 (Fig. 2) upon the addition of AT-DNA the emission considerably increased while the addition of GC-DNA strongly quenched the fluorescence. Again ds-RNA only negligibly influenced the emission of 5.
Fluorimetric data were processed using the Scatchard equation14 to calculate binding parameters (Table 1). The binding affinity of 5 changed considerably between pH 7 and pH 5 (Table 1). Under neutral conditions 5 showed two orders of magnitude higher binding constant for poly(dA-dT)2 in comparison with other ds-polynucleotides. Intriguingly, the absolute change in fluorescence quenching (Fig. 1, I/I0) was the smallest upon addition of poly(dA-dT)2 with respect to the other DNA/RNA. This suggests that the fluorophore (pyrene) emission response is not proportionally related to the DNA/RNA binding affinity. Most intriguingly, at pH 5 the selectivity of 5 for a different ds-DNA is reversed (Table 1). For ds-RNA no quantitative binding data could be obtained as negligible fluorimetric changes hampered data processing.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ks), ratios n = [bound compound]/[polynucleotide] of 5 and 2 with ds-polynucleotides calculated from fluorimetric titrations at pH 7 and pH 5 (buffer sodium cacodylate, I = 0.05 M).a
Ks), ratios n = [bound compound]/[polynucleotide] of 5 and 2 with ds-polynucleotides calculated from fluorimetric titrations at pH 7 and pH 5 (buffer sodium cacodylate, I = 0.05 M).a
dAdT–dAdT log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ks (n) Ks (n) |
dGdC–dGdC log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ks (n) Ks (n) |
rA-rU log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ks (n) Ks (n) |
||
|---|---|---|---|---|
a Titration data were processed using the Scatchard equation,14 accuracy of the obtained n ± 10 − 30%, consequently log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ks values vary in the same order of magnitude.
b Aggregation occurred.
c Published results.11,12
d Too small changes for accurate data calculation. Ks values vary in the same order of magnitude.
b Aggregation occurred.
c Published results.11,12
d Too small changes for accurate data calculation.
|
||||
| pH 7 | 5 | 7.3 (0.3) | 5.4 (0.9) | 5.0 (0.9) |
| 2 | 6.6 (0.2) | 5.7 (0.2) | ||
| pH 5 | 5 | 5.3 (0.2) | 6.4 (0.1) | |
| 2 | 6.5 (0.3) | 6.3 (0.2) | 5.4 (0.2) | |
Comparison of the binding parameters with those of the previously studied analogue 2 revealed several distinct differences in pH dependent affinity as well as in the fluorimetric response. However, fluorimetric titrations showed a very complex pattern of interactions between 5 and DNA/RNA, with no straightforward correlations with the structure–activity relationship. Thus for a more detailed structural analysis of the complexes formed, CD spectroscopy was applied.8,9 It should be noted that due to its weak intensity the CD spectrum of 5 (ESI†) could be accurately subtracted from CD spectra of 5/DNA complexes. The CD spectrum of ds-RNA upon titration with 5 did not change significantly (ESI†). The absence of any ICD bands was attributed to the lack of one dominant and well-structured binding mode of 5 with respect to the chiral axis of the RNA double helix.
In contrast to ds-RNA, the addition of 5 to any of the ds-DNA studied at either pH 5 or pH 7 resulted in pronounced changes in the CD spectra. Common for all studied DNA was a moderate decrease of the CD band of the DNA itself at λ = 245–250 nm, which is characteristic for a distortion of the double stranded helix. For both GC- and AT-DNA isoelliptic points λ < 265 nm supports one dominant type of DNA structure in the complex. However, at λ > 265 nm the observed changes in the CD spectra were strongly influenced by the DNA basepair composition. Spectral changes at λ = 265–300 nm arise from several spectroscopically active species (free and bound DNA, ICD effects of bound 5). The individual contributions of each species could not be differentiated from one another.
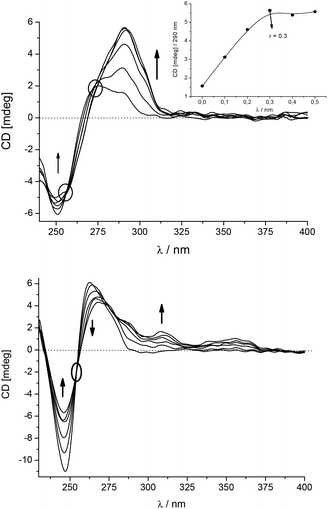
A more detailed analysis revealed that the CD spectrum of poly dGdC–poly dGdC significantly changed at λ > 275 nm upon addition of 5 (Fig. 3, up) due to the appearance of a strong ICD band at 290–313 nm. As this is the absorption band of the GCP moiety, this ICD band can be attributed to the positioning of the GCP chromophore along one of the DNA grooves.9,10,12 Furthermore, the absence of any ICD band > 330 nm suggests that the pyrene chromophore is not bound in the minor groove but rather outside of the GC-DNA. Most likely steric hindrance between the amino groups of guanine in the minor groove and the sterically demanding linker prevents alignment of the pyrene within the groove. A strongly non-linear dependence of the ICD intensity at 290 nm pointed to a saturation of the dominant binding site of the GCP moiety at r[5]/[dGdC–dGdC] = 0.3.
In contrast to GC-DNA, the addition of 5 to poly dAdT–poly dAdT (Fig. 3, down) resulted in a decrease and a bathochromic shift of the CD band of the DNA at λ = 263 nm. Two positive ICD bands appeared at 308 and 350 nm, which agreed nicely with UV/vis maxima of 5 bound to the poly dAdT–poly dAdT complex (ESI,† Table S2). The ICD band at 350 nm is specific for the 5/AT-DNA complex as this band is not observed in the 5/GT-DNA complex. This band can be attributed to the uniform positioning of pyrene within the DNA double helix.10,12 Furthermore, the positive sign of this ICD band suggests that the pyrene is either positioned along the minor groove9 or partially intercalated with its long axis oriented perpendicularly to the long axes of the adjacent DNA basepairs.9 For the accurate differentiation between these two binding modes NMR analysis of 5/oligonucleotide complexes would be necessary, which was however hampered by the insufficient solubility of 5/DNA complexes at concentrations needed for such NMR studies. Therefore, we performed simple molecular docking studies to probe the insertion of 5 into AT-DNA as we did previously for the intercalating analogue 110 and the minor groove binder 4.12 The obtained structures (ESI†) show that the rigid linker of 5 hampers the efficient insertion of the GCP moiety into the minor groove when the pyrene is intercalated. Thus the insertion of the whole molecule 5 into the minor groove is the most likely binding mode.
The most intriguing observation was that the addition of 5 to ct-DNA (58% AT; 42% GC) resulted in much smaller changes of the CD spectrum (only weak ICD bands at 290–310 nm, ESI†) in comparison with synthetic AT- or GC-DNA. Obviously a homogeneous basepair composition favours the specific binding mode, which is characteristic for the observed ICD signature.
Conclusions
In contrast to the previously studied compound 1, which interacted significantly with DNA/RNA only at pH 5 when the GCP moiety is protonated, 2 and 5 showed significant interactions also at pH 7. However, there are several distinct differences between 5 and previously studied 1–4. Firstly, only 5 revealed high selectivity toward AT-DNA at pH 7 as confirmed by the two orders of magnitude higher binding constant (Table 1), the exclusive thermal stabilisation of only AT-DNA but of no other DNA/RNA, and the specific positive ICD band at 350 nm, supporting a uniform positioning of the pyrene within the DNA double helix. Secondly, this selectivity of 5 was reversed at pH 5, now favouring GC-DNA over AT-DNA and with no ICD band showing up at 350 nm. Hence, the pH change induced a different binding mode in which the pyrene is removed from the DNA-double helix. Furthermore, 5 showed only at pH 5 a fluorescence increase exclusively for AT-DNA while GC-DNA yielded emission quenching, for similar reasons as for analogue 2.11 Intriguingly, 2 interacted also strongly with ds-RNA, in contrast to the negligibly weak interaction of 5. The reason for this is not clear yet.Most likely these distinct spectrophotometric responses of 5, which are significantly different from the previously studied analogues 1–4, can be attributed to its short, rigid linker. This linker only allows two chromophores to adopt very limited orientations upon interaction with the different DNA binding sites. Especially interesting is the strong interaction of neutral 5 with DNA at pH 7, since only very few neutral compounds show biorelevant DNA interactions (>90% DNA dyes are positively charged).2
However, a major disadvantage is the low solubility of 5, which will be addressed in future research to allow more detailed structural studies of DNA complexes formed (e.g. by NMR studies). Furthermore, future work will also focus on the enantiomer of 5, which could show different patterns of DNA/RNA interactions, similarly to recently reported chiral perylene bisimide derivatives.15 Also, biological studies of 5 and comparison with the activity of its close analogues11,12 are of the highest interest, to characterize its antiproliferative activity, cellular uptake and distribution. Nonetheless, 5 is particularly convenient for further synthetic modifications and the design of second generation derivatives since the carboxy-group allows easy functionalization to structurally even more complicated analogues by simple peptide bond formation, for instance based on the peptide backbone recombination.16
Acknowledgements
Financial supports from the Croatian Science Foundation (grant no. 1477) and FP7-REGPOT-2012-2013-1, grant agreement number 316289 – InnoMol are gratefully acknowledged. I. P. and C. S. are grateful to COST Action CM1005 for the support in networking and mutual cooperation.Notes and references
-
R. B. Silverman, The Organic Chemistry of Drug Design and Drug Action, Elsevier Academic Press, New York, 2004 Search PubMed
.
-
DNA and RNA Binders, ed. M. Demeunynck, C. Bailly and W. D. Wilson, Wiley-VCH, Weinheim, 2002 Search PubMed
.
- E. Trinquet and G. Mathis, Mol. Biosyst., 2006, 2, 381 RSC
.
- Y. N. Teo and E. T. Kool, Chem. Rev., 2012, 112, 4221 CrossRef CAS PubMed
.
-
(a) S. B. Baylin and K. E. Schuebel, Nature, 2007, 448, 548 CrossRef CAS PubMed
; (b) K. K. Li, C. Luo, D. X. Wang, H. L. Jiang and Y. G. Zheng, Med. Res. Rev., 2012, 32, 815 CrossRef CAS PubMed
.
- B. Willis and D. P. Arya, Bioorg. Med. Chem. Lett., 2009, 19, 4974 CrossRef CAS PubMed
.
-
(a) R. J. Lipshutz, S. P. A. Fodor, T. R. Gingeras and D. J. Lockhart, Nat. Genet., 1999, 21, 20 CrossRef CAS PubMed
; (b) L. M. Smith, J. Z. Sanders, R. J. Kaiser, P. Hughes, C. Dodd, C. R. Connell, C. Heiner, S. B. H. Kent and L. E. Hood, Nature, 1986, 321, 674 CrossRef CAS PubMed
; (c) T. Oida, Y. Sako and A. Kusumi, Biophys. J., 1993, 64, 676 CrossRef CAS
; (d) A. Hillisch, M. Lorenz and S. Diekmann, Curr. Opin. Struct. Biol., 2001, 11, 201 CrossRef CAS
.
-
A. Rodger and B. Nordén, Circular Dichroism and Linear Dichroism, Oxford University Press, New York, 1997, Ch. 2 Search PubMed
.
- M. Eriksson and B. Nordén, Methods Enzymol., 2001, 340, 68–98 CAS
.
- L. Hernandez-Folgado, C. Schmuck, S. Tomić and I. Piantanida, Bioorg. Med. Chem. Lett., 2008, 18, 2977 CrossRef CAS PubMed
.
- L. Hernandez-Folgado, D. Baretić, I. Piantanida, M. Marjanović, M. Kralj, T. Rehm and C. Schmuck, Chem. – Eur. J., 2010, 16, 3036 CrossRef CAS PubMed
.
- K. Gröger, D. Baretić, I. Piantanida, M. Marjanović, M. Kralj, M. Grabar, S. Tomić and C. Schmuck, Org. Biomol. Chem., 2011, 9, 198 Search PubMed
.
- K. Klemm, M. Radić Stojković, G. Horvat, V. Tomišić, I. Piantanida and C. Schmuck, Chem. – Eur. J., 2012, 18, 1352 CrossRef CAS PubMed
.
- J. D. McGhee and P. H. von Hippel, J. Mol. Biol., 1976, 103, 679 CrossRef
.
- T. H. Rehm, M. R. Stojković, S. Rehm, M. Škugor, I. Piantanida and F. Wurthner, Chem. Sci., 2012, 3, 3393 RSC
.
- M. Dukši, D. Baretić, V. Čaplar and I. Piantanida, Eur. J. Med. Chem., 2010, 45, 2671 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, synthesis, spectrophotometric data, additional DNA/RNA binding data. See DOI: 10.1039/c4ob02169j |
| This journal is © The Royal Society of Chemistry 2015 |