Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
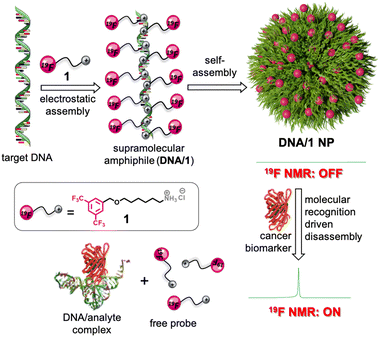
19F NMR ON/OFF nanoparticles: a universal approach for the specific detection of DNA-binding cancer biomarkers†
Devanathan
Perumal
,
Jithu
Krishna
,
Kaloor S.
Harikrishnan
,
Gowtham
Raj
,
Jemshiya
Kalathil
,
Minu
Saji
,
Kavyasree
M.
and
Reji
Varghese
 *
*
School of Chemistry, Indian Institute of Science Education and Research (IISER) Thiruvananthapuram, Trivandrum 695541, Kerala, India. E-mail: reji@iisertvm.ac.in
First published on 19th April 2023
Abstract
A supramolecular approach for the design of assembly–disassembly-driven 19F ON/OFF nanoparticles, triggered by specific molecular recognition, for the detection of DNA binding cancer biomarkers is reported. The key to our design strategy is the characteristic 19F NMR signal of the probe, which completely vanishes in the aggregated state due to the shortening of T2 relaxation. However, molecular recognition of DNA by the cancer biomarkers through specific molecular recognition results in the disassembly of the nanoparticles, which causes the restoration of the characteristic 19F signal of the probe. The universal nature of the approach is demonstrated through the selective detection of various cancer biomarkers including miRNA, ATP, thrombin, and telomerase.
Early-stage and precise diagnosis of cancer is extremely important for its successful treatment.1 Biomarker-targeted in vitro detection of cancer is undoubtedly one of the most promising non-invasive approaches for cancer diagnosis.2 Recent years have witnessed substantial growth in this field and different detection strategies based on colorimetry,3 fluorescence,4 electrochemical,5 enzyme-linked immunosorbent assay (ELISA),6 polymerase chain reaction (PCR),7 and surface enhanced Raman spectroscopy (SERS)8 have been developed. Although robust and highly efficient, most of them still suffer from a lack of sensitivity and specificity. Hence, there is always a rising demand for the development of in vitro biosensors that allow cancer diagnosis with extremely high precision and sensitivity. Among the various non-invasive diagnostic tools available for cancer diagnosis, 19F magnetic resonance imaging (19F MRI) has attracted enormous attention. This is mainly due to the following reasons: (i) high sensitivity (83% relative to 1H), (ii) 100% natural abundance, and (iii) essentially no 19F in animal bodies and hence no background signals.9–11 Though few strategies have been reported for the design of 19F-based biosensors, this area is still in its infancy.
One of the remarkable features of 19F is the large chemical shift distribution and hence even a small perturbation in the chemical shift produces distinct 19F signals. By exploring this, a large number of 19F containing biosensors have been reported for the detection of enzymes,12,13 reactive oxygen species,14,15 metal ions,16 and small organic analytes.17,18 Another strategy through which 19F-based biosensors were designed is by modulating the transverse relaxation time (T2) of 19F through the paramagnetic relaxation enhancement (PRE) effect. Several 19F ON/OFF probes have been reported for the detection of enzyme activity19–22 and nucleotides23 by modulating the T2 relaxation by using the PRE effect. Yet another strategy by which the T2 relaxation can be modulated is through the restriction of molecular motions by self-assembling the probe. This was applied for the design of several assembly–disassembly-driven 19F ON/OFF probes for the detection and imaging of proteins.24–28 Though highly promising, no attempt has been made yet to develop a 19F-based biosensor for the detection of cancer biomarkers.
Herein, we report a universal supramolecular approach for the crafting of a 19F ON/OFF in vitro biosensor for the detection of ssDNA binding cancer biomarkers. Our strategy involves the initial electrostatic assembly between negatively charged cancer biomarker binding ssDNA (hydrophilic segment) and a cationic 19F probe (hydrophobic segment), leading to the formation of a supramolecular DNA amphiphile with the cationic 19F probe tethered along the anionic backbone of ssDNA.29–35 Subsequent self-assembly of the supramolecular amphiphile results in the formation of soft nanoparticles (NPs). The key to our design strategy is that in the self-assembled state with almost complete arrest of the molecular motions of the 19F probe moiety, the characteristic 19F signal of the probe has completely vanished (OFF state) due to the significant shortening of T2 relaxation. On the other hand, molecular recognition of ssDNA by the cancer biomarkers results in the disassembly of the NPs and the simultaneous release of the free probe. This disassembly causes the restoring of the characteristic 19F signal of the probe (ON state). The universal nature of our approach is demonstrated through the detection of various cancer biomarkers including miRNA (miRNA-21), ATP (small molecule-based biomarker), thrombin (enzyme-based biomarker), and telomerase (enzyme-based biomarker) (Scheme 1).
Synthesis of the cationic hydrophobic probe 1 is provided in the ESI.† Target cancer biomarkers selected in our study include thrombin,36 adenosine triphosphate (ATP),37 telomerase enzyme,38 and miRNA-21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 (Table 1). Accordingly, we have selected DNA1 (ATP binding aptamer), DNA2 (thrombin binding aptamer), DNA4 (complementary DNA to miRNA-21), and DNA5 (telomerase primer) as the target DNAs for the biomarkers. DNA3 is having two base mismatches with respect to the thrombin binding aptamer (DNA2), hence it acts as a control DNA to study the specificity of the aptamer binding to thrombin. Details of the synthesis and characterization of all DNAs are provided in the ESI.† Thrombin, ATP, and miRNA-21 were commercially purchased and used as such. Telomerase enzyme was extracted from HeLa cell lines by following a reported procedure.40
39 (Table 1). Accordingly, we have selected DNA1 (ATP binding aptamer), DNA2 (thrombin binding aptamer), DNA4 (complementary DNA to miRNA-21), and DNA5 (telomerase primer) as the target DNAs for the biomarkers. DNA3 is having two base mismatches with respect to the thrombin binding aptamer (DNA2), hence it acts as a control DNA to study the specificity of the aptamer binding to thrombin. Details of the synthesis and characterization of all DNAs are provided in the ESI.† Thrombin, ATP, and miRNA-21 were commercially purchased and used as such. Telomerase enzyme was extracted from HeLa cell lines by following a reported procedure.40
| DNA | Sequence (3′ → 5′) | Target |
|---|---|---|
| DNA1 | ACCTGGGGGAGTATTGCGGAGGAAGGT | ATP |
| DNA2 | GGTTGGTGTGGTTGG | Thrombin |
| DNA3 | GGTTGCTGTAGTTGG | Scrambled |
| DNA4 | TCAACATCAGTCTGATAAGCTA | miRNA-21 |
| DNA5 | AATCCGTCGAGCAGAGTT | Telomerase |
Noncovalent synthesis of supramolecular amphiphiles was achieved by annealing 1 (60 or 30 μM) and DNA (6 or 3 μM) at a molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
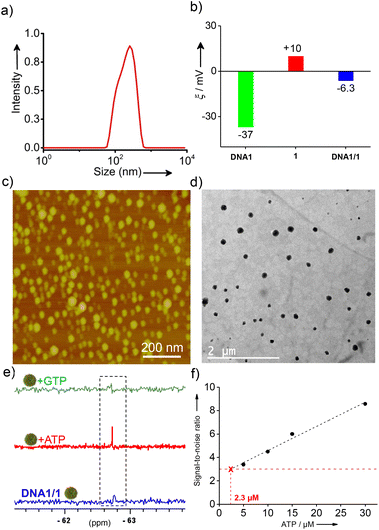
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in D2O at 90 °C for 10 minutes followed by slow cooling to room temperature at a cooling rate of 5 °C per minute over a period of 10 h. This particular procedure was adopted considering the uniform formation of NPs. Dynamic light scattering (DLS) analyses of DNA1/1 showed the formation of aggregated species in solution with a size distribution in the range of 60–615 nm (Fig. 1a). As expected, zeta potential measurements of DNA1 and 1 revealed values of −37.0 mV and +10.0 mV, respectively. Interestingly, a zeta potential value of −6.3 mV was observed for the aggregates of DNA1/1 (Fig. 1b). This almost neutral zeta potential value for DNA1/1 aggregates clearly indicates that the assembly between negatively charged DNA and positively charged 1 leads to the formation of almost neutral aggregated species in solution. Atomic force microscopy (AFM) (Fig. 1c) and transmission electron microscopy (TEM) (Fig. 1d) analyses of DNA1/1 showed the formation of spherical NPs. These observations conclude that the supramolecular assembly between DNA1 and 1 results in the formation of supramolecular amphiphiles (DNA1/1) by the non-covalent tethering of 1 along the anionic backbone of DNA through the strong electrostatic attraction. Subsequent self-assembly of the supramolecular amphiphile (DNA1/1) via hydrophobic and π-stacking interactions results in the formation of spherical NPs with the 19F containing hydrophobic moiety buried inside the NPs with no/minimum contact with the polar medium.
1 in D2O at 90 °C for 10 minutes followed by slow cooling to room temperature at a cooling rate of 5 °C per minute over a period of 10 h. This particular procedure was adopted considering the uniform formation of NPs. Dynamic light scattering (DLS) analyses of DNA1/1 showed the formation of aggregated species in solution with a size distribution in the range of 60–615 nm (Fig. 1a). As expected, zeta potential measurements of DNA1 and 1 revealed values of −37.0 mV and +10.0 mV, respectively. Interestingly, a zeta potential value of −6.3 mV was observed for the aggregates of DNA1/1 (Fig. 1b). This almost neutral zeta potential value for DNA1/1 aggregates clearly indicates that the assembly between negatively charged DNA and positively charged 1 leads to the formation of almost neutral aggregated species in solution. Atomic force microscopy (AFM) (Fig. 1c) and transmission electron microscopy (TEM) (Fig. 1d) analyses of DNA1/1 showed the formation of spherical NPs. These observations conclude that the supramolecular assembly between DNA1 and 1 results in the formation of supramolecular amphiphiles (DNA1/1) by the non-covalent tethering of 1 along the anionic backbone of DNA through the strong electrostatic attraction. Subsequent self-assembly of the supramolecular amphiphile (DNA1/1) via hydrophobic and π-stacking interactions results in the formation of spherical NPs with the 19F containing hydrophobic moiety buried inside the NPs with no/minimum contact with the polar medium.
The first cancer biomarker we targeted in our study was ATP. 19F NMR studies of the cationic probe 1 at a concentration of 60 μM in D2O revealed a sharp singlet at −67.72 ppm due to the presence of two magnetically equivalent –CF3 groups. Interestingly, a reduction in the intensity of the peak at −67.72 ppm was observed with the addition of DNA1 and the peak completely vanished at a concentration of 6 μM of DNA1 (Fig. S5a†). This suggests that the NPs synthesized at a molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 of DNA1 and 1 is 19F NMR silent due to the significant reduction of T2 relaxation caused by the complete arrest of the molecular motions of the probe in the aggregated state. Accordingly, the 1
10 of DNA1 and 1 is 19F NMR silent due to the significant reduction of T2 relaxation caused by the complete arrest of the molecular motions of the probe in the aggregated state. Accordingly, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 molar ratio of DNA1 and 1 was used for further experiments. Stability of the NPs is an extremely important requirement in our design strategy as the undesired disassembly of the NPs and the resulting emergence of the 19F signal may leads to wrong prediction. Stability of the NPs was then investigated under different conditions. Interestingly, no change was observed in the 19F NMR spectra of DNA1/1 NPs with time (0 → 48 h), temperature (25 °C → 85 °C) and pH (5.5 → 8.5), revealing that the NPs are stable with respect to time, temperature and pH (Fig. S5b−d†). Furthermore, no significant change was observed for the DLS size distribution of the NPs with respect to pH, time and temperature (Fig. S6a−c†). These results confirm that the NPs are stable and the possibility of their undesired disassembly is very unlikely.
10 molar ratio of DNA1 and 1 was used for further experiments. Stability of the NPs is an extremely important requirement in our design strategy as the undesired disassembly of the NPs and the resulting emergence of the 19F signal may leads to wrong prediction. Stability of the NPs was then investigated under different conditions. Interestingly, no change was observed in the 19F NMR spectra of DNA1/1 NPs with time (0 → 48 h), temperature (25 °C → 85 °C) and pH (5.5 → 8.5), revealing that the NPs are stable with respect to time, temperature and pH (Fig. S5b−d†). Furthermore, no significant change was observed for the DLS size distribution of the NPs with respect to pH, time and temperature (Fig. S6a−c†). These results confirm that the NPs are stable and the possibility of their undesired disassembly is very unlikely.
After establishing the stability of the NPs, we then enduced the interaction of DNA1/1 NPs with ATP by gradually adding ATP into the NP solution and the emergence of the 19F NMR signal was monitored. Interestingly, emergence of the characteristic 19F NMR peak of 1 at −67.72 ppm was observed with the addition of ATP into the DNA1/1 NP solution and an intense sharp singlet was observed with the addition 60 μM of ATP (Fig. 1e). These results clearly reveal that specific molecular recognition between ATP and DNA1 results in the disassembly of DNA1/1 NPs with the simultaneous formation of an ATP/DNA1 complex. This in turn disassembles the supramolecular amphiphile (DNA1/1) and releases the monomeric probe 1. This molecular recognition-driven disassembly leads to the ‘turn ON’ of the 19F NMR signal of the probe. Circular dichroism (CD) analyses revealed that DNA1 adopted a random coil conformation in the NP state as no specific CD signal was observed.41 However, after binding to ATP, an intense positive CD signal at 290 nm was clearly observed, indicating the formation of an antiparallel G-quadruplex conformation (Fig. S7†).41 This conformational switching of DNA1 from a random coil to an antiparallel G-quadruplex supports the binding of DNA1 with ATP. As expected, no ‘turn ON’ of the 19F NMR response was observed with the addition of guanosine-5′-triphosphate (GTP) (Fig. 1e), which clearly support our hypothesis that the specific molecular recognition between DNA1 and ATP is solely responsible for the disassembly of the NPs and the subsequent 19F NMR ‘turn ON’ response. Subsequently, we have determined the limit of detection (LoD) of the probe by titrating the DNA1/1 NPs with varying concentrations of ATP.42 For this, the 19F NMR silent DNA1/1 NPs (3 μM) were treated with increasing concentration of ATP (0 → 30 μM) and the signal-to-noise ratio of the 19F peak emerged at −67.72 ppm was plotted against the concentration of ATP. The LoD value for the detection of ATP was found to be 2.3 μM, which is well below the concentration range of ATP typically present inside the cancer cells.43
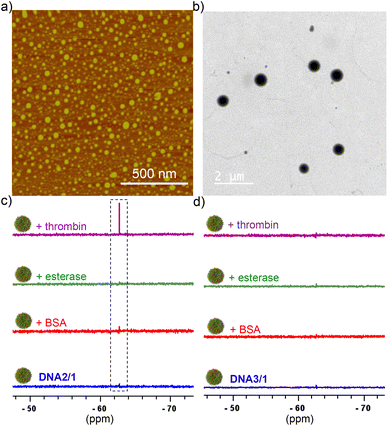
To establish the universal nature of the system for the detection of various cancer biomarkers, we have explored the detection of other potential cancer biomarkers including thrombin and miRNA-21. DNA2 was selected as the target for thrombin detection, which is a 15-mer thrombin binding DNA aptamer. Towards this, DNA2/1 NPs (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 μM) were first synthesized by following the general procedure. Nanoparticle formation was characterized using DLS (Fig. S3†), AFM (Fig. 2a) and TEM (Fig. 2b) analyses. DLS size distribution analyses revealed that the size of the NPs is in the range of 140–460 nm. As expected, the DNA2/1 NPs were 19F NMR silent (Fig. 2c). Interestingly, the addition of thrombin into the DNA2/1 NP solution showed the emergence of the characteristic 19F NMR peak of 1 at −67.72 ppm (Fig. 2c) and a sharp singlet at −67.72 ppm was observed with the addition of 5 μM of thrombin. This indicates that the molecular recognition between DNA2 and thrombin leads to the formation of a DNA2/thrombin complex, which in turn results in the disassembly of the NPs and releases the monomeric probe 1. As expected, no ‘turn ON’ of the 19F NMR response was observed with the addition of other enzymes such as BSA and esterase (Fig. 2c), which clearly support our hypothesis that the specific molecular recognition between DNA2 and thrombin is solely responsible for the disassembly of the NPs and the subsequent 19F NMR ‘turn ON’ response. Furthermore, no ‘turn ON’ response was observed for the DNA3/1 NPs with the addition of thrombin, BSA or esterase, disclosing that DNA2 (scrambled DNA sequence) has no interaction with any of the proteins and hence no molecular recognition-driven disassembly of the NPs (Fig. 2d).
60 μM) were first synthesized by following the general procedure. Nanoparticle formation was characterized using DLS (Fig. S3†), AFM (Fig. 2a) and TEM (Fig. 2b) analyses. DLS size distribution analyses revealed that the size of the NPs is in the range of 140–460 nm. As expected, the DNA2/1 NPs were 19F NMR silent (Fig. 2c). Interestingly, the addition of thrombin into the DNA2/1 NP solution showed the emergence of the characteristic 19F NMR peak of 1 at −67.72 ppm (Fig. 2c) and a sharp singlet at −67.72 ppm was observed with the addition of 5 μM of thrombin. This indicates that the molecular recognition between DNA2 and thrombin leads to the formation of a DNA2/thrombin complex, which in turn results in the disassembly of the NPs and releases the monomeric probe 1. As expected, no ‘turn ON’ of the 19F NMR response was observed with the addition of other enzymes such as BSA and esterase (Fig. 2c), which clearly support our hypothesis that the specific molecular recognition between DNA2 and thrombin is solely responsible for the disassembly of the NPs and the subsequent 19F NMR ‘turn ON’ response. Furthermore, no ‘turn ON’ response was observed for the DNA3/1 NPs with the addition of thrombin, BSA or esterase, disclosing that DNA2 (scrambled DNA sequence) has no interaction with any of the proteins and hence no molecular recognition-driven disassembly of the NPs (Fig. 2d).
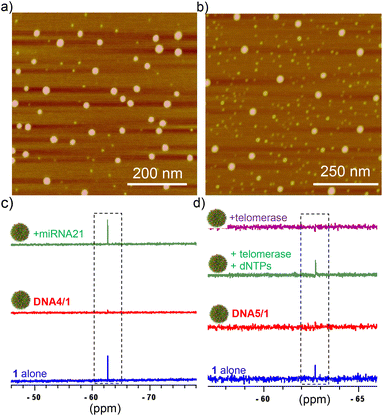
For the detection of miRNA-21, DNA4 was designed, which is fully complementary to miRNA-21. Subsequently, DNA4/1 (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 μM) NPs were synthesized and characterized by using various microscopic (Fig. 3b) and light scattering techniques (Fig. S3†). The average diameter of the particles from DLS analyses was found to be 125 nm. As expected, the NPs were 19F NMR silent due to the significant shortening of T2 relaxation (Fig. 3c). Interestingly, the characteristic singlet at −67.72 ppm of the probe emerged with the addition of miRNA-21 (Fig. 3c). This can be attributed to the duplex formation between DNA4 and miRNA-21, which in turn releases the probe 1 into the solution and thereby ‘turning ON’ the 19F NMR signal.
60 μM) NPs were synthesized and characterized by using various microscopic (Fig. 3b) and light scattering techniques (Fig. S3†). The average diameter of the particles from DLS analyses was found to be 125 nm. As expected, the NPs were 19F NMR silent due to the significant shortening of T2 relaxation (Fig. 3c). Interestingly, the characteristic singlet at −67.72 ppm of the probe emerged with the addition of miRNA-21 (Fig. 3c). This can be attributed to the duplex formation between DNA4 and miRNA-21, which in turn releases the probe 1 into the solution and thereby ‘turning ON’ the 19F NMR signal.
In order to demonstrate that our approach can be applied for practical applications, we have extended our study to the detection of telomerase activity (another potential cancer biomarker), that is directly extracted from cultured HeLa cells. For this purpose, DNA5 was designed, which is a short 22-mer telomerase primer and hence the primer ssDNA can be extended at its 3′-end by the telomerase enzyme to produce telomeric repeat sequences. Keeping this in mind, we initially synthesized DNA5/1 (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 μM) NPs and characterized using microscopic (Fig. 3b) and light scattering analyses (Fig. S3†). The average diameter of the particle is 60 nm from the DLS analysis. As anticipated, the DNA5/1 NPs were 19F NMR silent due to the shortening of T2 relaxation (Fig. 3d). The NPs were then incubated with deoxynucleotide triphosphates (dNTPs: dATP, dGTP, dCTP and dTTP; 100 μM each) and telomerase (10 μL from the extract of ∼2.5 × 106 cells) in phosphate-buffered saline (pH 7.5) for different time intervals. No appearance of the characteristic 19F NMR peak was observed in 10 minutes of incubation, indicating that the DNA5/1 NPs are still in the self-assembled state. However, the characteristic peak of 1 at −67.72 ppm emerged after 4 h of incubation, implying that the NPs are completely dissociated and caused the ‘turn ON’ of the 19F NMR signal (Fig. 3d). In the presence of telomerase and dNTPs, the telomerase primer (DNA5) is getting elongated from its 3′-end to produce the corresponding telomeric repeat sequences, which in turn transforms the supramolecular amphiphile (DNA5/1) into a longer hydrophilic ssDNA. This causes the disassembly of the NPs and leads to the ‘turn ON’ of the 19F NMR signal. No emergence of the 19F NMR signal was observed when the NPs were incubated just with the telomerase enzyme, indicating that primer elongation requires both telomerase and dNTPs (Fig. 3d).
60 μM) NPs and characterized using microscopic (Fig. 3b) and light scattering analyses (Fig. S3†). The average diameter of the particle is 60 nm from the DLS analysis. As anticipated, the DNA5/1 NPs were 19F NMR silent due to the shortening of T2 relaxation (Fig. 3d). The NPs were then incubated with deoxynucleotide triphosphates (dNTPs: dATP, dGTP, dCTP and dTTP; 100 μM each) and telomerase (10 μL from the extract of ∼2.5 × 106 cells) in phosphate-buffered saline (pH 7.5) for different time intervals. No appearance of the characteristic 19F NMR peak was observed in 10 minutes of incubation, indicating that the DNA5/1 NPs are still in the self-assembled state. However, the characteristic peak of 1 at −67.72 ppm emerged after 4 h of incubation, implying that the NPs are completely dissociated and caused the ‘turn ON’ of the 19F NMR signal (Fig. 3d). In the presence of telomerase and dNTPs, the telomerase primer (DNA5) is getting elongated from its 3′-end to produce the corresponding telomeric repeat sequences, which in turn transforms the supramolecular amphiphile (DNA5/1) into a longer hydrophilic ssDNA. This causes the disassembly of the NPs and leads to the ‘turn ON’ of the 19F NMR signal. No emergence of the 19F NMR signal was observed when the NPs were incubated just with the telomerase enzyme, indicating that primer elongation requires both telomerase and dNTPs (Fig. 3d).
In order to check the performance of the probe in a complex mixture of potential nontargets, we have carried out the detection of the analytes in the presence of other potential nontargets. For this purpose, DNA1/1 (probe for ATP), DNA2/1 (probe for thrombin) and DNA5/1 (probe for telomerase) NPs were prepared as described and their response was studied in the presence of a mixture of other targets. Initially, the response of DNA1/1 NPs towards ATP was studied. As expected, the DNA1/1 (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 μM) NPs were 19F NMR silent and no 19F NMR ‘turn ON’ response was observed with the addition of a mixture of other potential targets including thrombin (5 μM), telomerase (10 μL from the extract of ∼2.5 × 106 cells) and miRNA (10 μM). However, a strong ‘turn ON’ response was observed with the addition of ATP (100 μM, target cancer biomarker in this case) (Fig. 4a). This clearly reveals that our probe is highly specific to the target and is responsive even in the presence of a mixture of nontargets. Similarly, DNA2/1 (6
60 μM) NPs were 19F NMR silent and no 19F NMR ‘turn ON’ response was observed with the addition of a mixture of other potential targets including thrombin (5 μM), telomerase (10 μL from the extract of ∼2.5 × 106 cells) and miRNA (10 μM). However, a strong ‘turn ON’ response was observed with the addition of ATP (100 μM, target cancer biomarker in this case) (Fig. 4a). This clearly reveals that our probe is highly specific to the target and is responsive even in the presence of a mixture of nontargets. Similarly, DNA2/1 (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 μM) was 19F NMR silent in the presence of a mixture of ATP (100 μM), telomerase (10 μL from the extract of ∼2.5 × 106 cells) and miRNA (10 μM), but a strong 19F NMR signal emerged with the addition of target thrombin (5 μM) (Fig. 4b). The same observations were made for telomerase detection as well. The 19F NMR silent DNA5/1 (6
60 μM) was 19F NMR silent in the presence of a mixture of ATP (100 μM), telomerase (10 μL from the extract of ∼2.5 × 106 cells) and miRNA (10 μM), but a strong 19F NMR signal emerged with the addition of target thrombin (5 μM) (Fig. 4b). The same observations were made for telomerase detection as well. The 19F NMR silent DNA5/1 (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 μM) NPs in a mixture of ATP (100 μM), thrombin (5 μM), and miRNA (10 μM) showed the emergence of a strong 19F NMR signal with the addition of target telomerase (10 μL from the extract of ∼2.5 × 106 cells) along with dNTPs: dATP, dGTP, dCTP and dTTP (100 μM each) (Fig. 4c).
60 μM) NPs in a mixture of ATP (100 μM), thrombin (5 μM), and miRNA (10 μM) showed the emergence of a strong 19F NMR signal with the addition of target telomerase (10 μL from the extract of ∼2.5 × 106 cells) along with dNTPs: dATP, dGTP, dCTP and dTTP (100 μM each) (Fig. 4c).
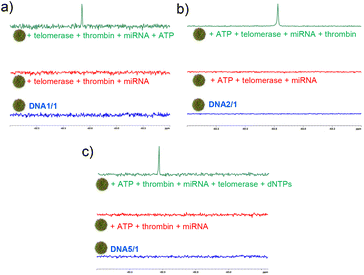 | ||
| Fig. 4 19F-NMR spectral responses of (a) DNA1/1 NPs towards ATP, (b) DNA2/1 NPs towards thrombin and (c) DNA5/1 NPs towards telomerase in the presence of a mixture of other nontargets. | ||
In summary, we have demonstrated a supramolecular approach for the design of assembly–disassembly-driven 19F ON/OFF NPs triggered by specific molecular recognition for the detection of ssDNA binding cancer biomarkers with excellent selectivity. To the best of our knowledge, this is the first report demonstrating the potential of 19F ON/OFF NPs for the specific detection of DNA binding cancer biomarkers in vitro. The universal nature of our approach is demonstrated through the detection of different types of targets including thrombin, ATP, miRNA, and telomerase. Because the molecular recognition between DNA and the analyte is highly specific in nature, this strategy offers excellent selectivity. Unlike the known 19F NMR probes, where the detection of different analytes involves the laborious synthetic modification of the probe, our approach is modular in nature and permits the detection of any DNA-based analyte with a single 19F NMR probe. Though unique in many ways, it is to be mentioned that the sensitivity of 19F NMR-based systems is considerably lower when compared to the traditional 1H-MRI probes. However, this can be addressed to a large extent by the appropriate molecular design of the 19F probe and improvement in the NMR instrumentation, and the 19F-MRI technique can emerge as a dominant tool for in vivo imaging applications. We hope that the universal nature and less laborious non-covalent strategy demonstrated here may encourage other researchers to explore this for the detection of other DNA-binding analytes for disease diagnosis and in vivo imaging applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial supports from DBT (BT/PR30172/NNT/28/1593/-2018) and CSIR (Research Fellowship) are acknowledged.References
- K. A. Cronin, A. J. Lake, S. Scott, R. L. Sherman, A. M. Noone, N. Howlader, S. J. Henley, R. N. Anderson, A. U. Firth, J. Ma, B. A. Kohler and A. Jemal, Cancer, 2018, 124, 2785–2800 CrossRef PubMed.
- L. Wuab and X. Qu, Chem. Soc. Rev., 2015, 44, 2963–2997 RSC.
- M. Perfezou, A. Turner and A. Merkoci, Chem. Soc. Rev., 2012, 41, 2606–2622 RSC.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530–10574 CrossRef CAS PubMed.
- B. V. Chikkaveeraiah, A. A. Bhirde, N. Y. Morgan, H. S. Eden and X. Chen, ACS Nano, 2012, 6, 6546–6561 CrossRef CAS PubMed.
- R. de la Rica and M. M. Stevens, Nat. Nanotechnol., 2012, 7, 821–824 CrossRef CAS PubMed.
- S. A. Kazane, D. Sok, E. H. Cho, M. L. Uson, P. Kuhn, P. G. Schultz and V. V. Smider, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 3731–3736 CrossRef CAS PubMed.
- S. R. Panikkanvalappil, M. A. Mackey and M. A. El-Sayed, J. Am. Chem. Soc., 2013, 135, 4815–4821 CrossRef CAS PubMed.
- S. Mizukami, R. Takikawa, F. Sugihara, Y. Hori, H. Tochio, M. W. Lchli, M. Shirakawa and K. Kikuchi, J. Am. Chem. Soc., 2008, 130, 794–795 CrossRef CAS PubMed.
- M. Srinivas, P. A. Morel, L. A. Ernst, D. H. Laidlaw and E. T. Ahrens, Magn. Reson. Med., 2007, 58, 725–734 CrossRef CAS PubMed.
- E. T. Ahrens, B. M. Helfer, C. F. O'Hanlon and C. Schirda, Magn. Reson. Med., 2014, 72, 1696–1701 CrossRef CAS PubMed.
- W. Cui, P. Otten, Y. Li, K. S. Koeneman, J. Yu and R. P. Mason, Magn. Reson. Med., 2004, 51, 616–620 CrossRef CAS PubMed.
- M. R. Baranowski, A. Nowicka, A. M. Rydzik, M. Warminski, R. Kasprzyk, B. A. Wojtczak, J. Wojcik, T. D. W. Claridge, J. Kowalska and J. Jemielity, J. Org. Chem., 2015, 80, 3982–3997 CrossRef CAS PubMed.
- A. R. Lippert, G. C. Van De Bittner and C. J. Chang, Acc. Chem. Res., 2011, 44, 793–804 CrossRef CAS PubMed.
- K. J. Bruemmer, S. Merrikhihaghi, C. T. Lollar, S. N. S. Morris, J. H. Bauer and A. R. F. Lippert, Chem. Commun., 2014, 50, 12311–12314 RSC.
- G. A. Smith, R. T. Hesketh, J. C. Metcalfe, J. Feeney and P. G. Morris, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 7178–7182 CrossRef CAS PubMed.
- Y. Zhao, G. Markopoulos and T. M. Swager, J. Am. Chem. Soc., 2014, 136, 10683–10690 CrossRef CAS PubMed.
- Y. Zhao, L. Chen and T. M. Swager, Angew. Chem., Int. Ed., 2016, 55, 917–921 CrossRef CAS PubMed.
- S. Mizukami, R. Takikawa, F. Sugihara, Y. Hori, H. Tochio, M. Wälchli, M. Shirakawa and K. Kikuchi, J. Am. Chem. Soc., 2008, 130, 794–795 CrossRef CAS PubMed.
- S. Mizukami, H. Matsushita, R. Takikawa, F. Sugihara, M. Shirakawa and K. F. Kikuchi, Chem. Sci., 2011, 2, 1151–1155 RSC.
- H. Matsushita, S. Mizukami, Y. Mori, F. Sugihara, M. Shirakawa, Y. Yoshioka and K. F. Kikuchi, ChemBioChem, 2012, 13, 1579–1583 CrossRef CAS PubMed.
- S. Mizukami, R. Takikawa, F. Sugihara, M. Shirakawa and K. Kikuchi, Angew. Chem., Int. Ed., 2009, 48, 3641–3643 CrossRef CAS PubMed.
- M. E. Dempsey, H. D. Marble, T.-L. Shen, N. L. Fawzi and E. M. Darling, Bioconjugate Chem., 2018, 29, 335–342 CrossRef CAS PubMed.
- Y. Takaoka, T. Sakamoto, S. Tsukiji, M. Narazaki, T. Matsuda, H. Tochio, M. Shirakawa and I. Hamachi, Nat. Chem., 2009, 1, 557–561 CrossRef CAS PubMed.
- Y. Takaoka, K. Kiminami, K. Mizusawa, K. Matsuo, M. Narazaki, T. Matsuda and I. Hamachi, J. Am. Chem. Soc., 2011, 133, 11725–11731 CrossRef CAS PubMed.
- K. Matsuo, R. Kamada, K. Mizusawa, H. Imai, Y. Takayama, M. Narazaki, T. Matsuda, Y. Takaoka and I. Hamachi, Chem. – Eur. J., 2013, 19, 12875–12883 CrossRef CAS PubMed.
- Y. Yuan, S. Ge, H. Sun, X. Dong, H. Zhao, L. An, J. Zhang, J. Wang, B. Hu and G. Liang, ACS Nano, 2015, 9, 5117–5124 CrossRef CAS PubMed.
- J. M. Perez, L. Josephson, T. O'Loughlin, D. Högemann and R. Weissleder, Nat. Biotechnol., 2002, 20, 816–820 CrossRef CAS PubMed.
- M. A. Azagarsamy, P. Sokkalingam and S. Thayumanavan, J. Am. Chem. Soc., 2009, 131, 14184–14185 CrossRef CAS PubMed.
- B. Narayan, S. P. Senanayak, A. Jain, K. S. Narayan and S. J. George, Adv. Funct. Mater., 2013, 23, 3053–3060 CrossRef CAS.
- C. Rest, M. J. Mayoral, K. Fucke, J. Schellheimer, V. Stepanenko and G. Fernández, Angew. Chem., Int. Ed., 2014, 53, 700–705 CrossRef CAS PubMed.
- S. Ghosh, D. S. Philips, A. Saeki and A. Ajayaghosh, Adv. Mater., 2017, 29, 1605408 CrossRef PubMed.
- A. Mukherjee, T. Sakurai, S. Seki and S. Ghosh, Langmuir, 2020, 36, 13096–13103 CrossRef CAS PubMed.
- S. K. Albert, M. Golla, N. Krishnan, D. Perumal and R. Varghese, Acc. Chem. Res., 2020, 53, 2668–2679 CrossRef CAS PubMed.
- S. K. Albert, I. Sivakumar, M. Golla, H. V. P. Thelu, N. Krishnan, K. L. J. Libin, Ashish and R. Varghese, J. Am. Chem. Soc., 2017, 139, 17799–17802 CrossRef CAS PubMed.
- M. Lundbech, A. E. Krag, T. D. Christensen and A.-M. Hvas, Thromb. Res., 2020, 186, 80–85 CrossRef CAS PubMed.
- S. Zhang, Y. Yan and S. Bi, Anal. Chem., 2009, 81, 8695–8701 CrossRef CAS PubMed.
- X. Lou, Y. Zhuang, X. Zuo, Y. Jia, Y. Hong, X. Min, Z. Zhang, X. Xu, N. Liu, F. Xia and B. Z. Tang, Anal. Chem., 2015, 87, 6822–6827 CrossRef CAS PubMed.
- Y. Wang, D. Zheng, Q. Tan, M. X. Wang and L.-Q. Gu, Nat. Nanotechnol., 2011, 6, 668–674 CrossRef CAS PubMed.
- R. Qian, L. Ding, L. Yan, M. Lin and H. Ju, J. Am. Chem. Soc., 2014, 136, 8205–8208 CrossRef CAS PubMed.
- S. Yan, R. Huang, Y. Zhou, M. Zhang, M. Deng, X. Wang, X. Weng and X. Zhou, Chem. Commun., 2011, 47, 1273–1275 RSC.
- M. E. Dempsey, H. D. Marble, T.-L. Shen, N. L. Fawzi and E. M. Darling, Bioconjugate Chem., 2018, 29, 335–342 CrossRef CAS PubMed.
- X.-R. Song, S.-H. Li, H. Guo, W. You, D. Tu, J. Li, C.-H. Lu, H.-H. Yang and X. Chen, Adv. Sci., 2018, 5, 1801201 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr01175e |
| This journal is © The Royal Society of Chemistry 2023 |