Open Access Article
Open Access ArticleSurface charge influences protein corona, cell uptake and biological effects of carbon dots†
Yasmin
Arezki
a,
François
Delalande
bc,
Christine
Schaeffer-Reiss
bc,
Sarah
Cianférani
bc,
Mickaël
Rapp
a,
Luc
Lebeau
 a,
Françoise
Pons
a and
Carole
Ronzani
a,
Françoise
Pons
a and
Carole
Ronzani
 *a
*a
aLaboratoire de Conception et Application de Molécules Bioactives, UMR 7199 CNRS-Université de Strasbourg, 67400 Illkirch, France. E-mail: ronzani@unistra.fr
bLaboratoire de Spectrométrie de Masse BioOrganique (LSMBO), IPHC, UMR 7178, CNRS-Université de Strasbourg, 67087 Strasbourg, France
cInfrastructure Nationale de Protéomique ProFI – FR2048 CNRS, 67087 Strasbourg, France
First published on 28th September 2022
Abstract
Carbon dots are emerging nanoparticles (NPs) with tremendous applications, especially in the biomedical field. Herein is reported the first quantitative proteomic analysis of the protein corona formed on CDs with different surface charge properties. Four CDs were synthesized from citric acid and various amine group-containing passivation reagents, resulting in cationic NPs with increasing zeta (ζ)-potential and density of positive charges. After CD contact with serum, we show that protein corona identity is influenced by CD surface charge properties, which in turn impacts CD uptake and viability loss in macrophages. In particular, CDs with high ζ-potential (>+30 mV) and charge density (>2 μmol mg−1) are the most highly internalized, and their cell uptake is strongly correlated with a corona enriched in vitronectin, fibulin, fetuin, adiponectin and alpha-glycoprotein. On the contrary, CDs with a lower ζ-potential (+11 mV) and charge density (0.01 μmol mg−1) are poorly internalized, while having a corona with a very different protein signature characterized by a high abundance of apolipoproteins (APOA1, APOB and APOC), albumin and hemoglobin. These data illustrate how corona characterization may contribute to a better understanding of CD cellular fate and biological effects, and provide useful information for the development of CDs for biomedical applications.
1. Introduction
Nanoparticles (NPs) exhibit unique properties due to their small size (below 100 nm) and are therefore developed for a wide variety of applications in various fields, including bioimaging and drug delivery.1,2 In the last decade, carbon-based NPs called carbon dots (CDs) have emerged as especially attractive nanomaterials for biomedical applications.3–5 CDs are spherical NPs of very small size (less than 10 nm) that exhibit high water solubility, chemical stability, intrinsic fluorescence and resistance to photobleaching. They can be easily synthesized by top-down or bottom-up approaches.6,7 In the most popular bottom-up methods, CDs are synthesized through the carbonization of organic material (citric acid, glucose, food waste⋯) in the presence of catalysts and/or passivation reagents. Due to the high density of nitrogen- and oxygen-containing functional groups at their surface, the obtained CDs can be easily chemically functionalized, allowing some tuning of their physicochemical and optical properties or anchoring of various molecules of interest, e.g., for triggering specific recognition by receptors or for improving circulation half-life time in biological fluids.8–10 Most CDs are passivated with nitrogen-containing reagents, which may result in significant cationic charge density at their surface thus making these NPs potent gene delivery platforms.11,12 Obviously, CDs with both photoluminescent and drug delivery properties are promising candidates for theranostic applications, particularly in the field of cancer.13–15 Nevertheless, full and safe exploitation of the great potential of CDs in nanomedicine definitely goes first through a better understanding of their interactions with biological systems.In contact with biological fluids (blood, intestinal or respiratory tract fluid), NPs are rapidly covered with biomolecules, mainly proteins, forming the so-called protein corona.16,17 The protein corona alters NP surface and gives NPs a new biological identity, described as “what the cells see”.18 Indeed, identity and properties of proteins that compose the corona play a role in the recognition of NPs by the cells, their biodistribution and, therefore, their biological effects.19–23 Consequently, understanding formation of the protein corona on NPs and characterizing its composition are major challenges for successful development of nanomedicine.24–26 This task is especially challenging as many factors influence the protein corona formation, in particular the physicochemical properties of NPs,27,28 such as chemical composition,29 size,30,31 morphology,32 hydrophobicity,33 chirality,34 and surface charge. The latter has been extensively studied and there is broad consensus in the literature on the importance of electrostatic forces in protein corona formation, with positively charged NPs consistently attracting higher quantities of proteins than negatively charged ones, as reported for a range of nanomaterials such as polymeric,35 inorganic,36 gold,37 or lipid-based38 NPs. As well, particles with low surface charge, as determined by the ζ-potential value, tend to adsorb less proteins than strongly charged particles.28 However, NP surface chemistry also influences protein corona formation. Indeed, it has been reported that type (primary, secondary or tertiary), location, and density of amino groups on the surface of gold NPs are predominant factors in the protein corona formation, which has a subsequent impact on NP cellular uptake.39 Thus, understanding the synergy between the synthetic and the biological identities of NPs appears to be a necessary condition to achieve their rational design for biological applications.
To date, very few studies investigated the protein corona formed on the surface of CDs and its biological consequences. In the field of food toxicology, Song et al. reported that CDs from roast salmon can form a corona with human serum albumin.40 The resulting corona mitigated the cytotoxicity of CDs and decreased disturbances in energy, glucose and lipid metabolism in a rat kidney cell model.40 In another study, the same group found that the toxicity and mitochondrial damages induced by roast-beef derived CDs decreased upon the formation of corona with albumin.41 In nanomedicine, Peng et al. demonstrated by proteomic analysis that lipid-mimicking chlorophyll-based CDs can adsorb apolipoproteins when placed into mouse serum.42 This corona enhances CD uptake by breast cancer cells and thus could be used for tumor-targeting delivery. However, to the best of our knowledge, the relationship between physicochemical properties of CDs and protein corona formation, and how this influences the biological effects of CDs, has not been investigated in the literature yet. It is therefore now essential to understand the extent to which the protein corona formed on the surface of CDs with different charge properties may contribute to their biological responses, in order to fully exploit their therapeutic potential.
In this context, herein we provide a detailed characterization of the protein corona formed on cationic CDs and analyzed how this corona may influence CD biological effects. We focused on polyamine surface-passivated cationic CDs, which are one type of CDs currently developed for gene delivery and theranostic applications.43 Although these CDs are less toxic than the gold standard delivery reagent used for DNA transfection, namely bPEI25k,12 some studies pointed out possible safety concerns with cationic CDs.44,45 Indeed, our team found that the surface charge strongly impacts the CD toxicity, even if a cationic charge does not systematically confer toxicity to CDs.46 In an original way, we demonstrated that the surface charge density of a cationic CD is more predictive of its toxicity than the absolute value of its ζ-potential.47 Thus, in the present work, four cationic CDs with various amino-groups at their surface, resulting in different ζ-potential and surface charge density values were investigated. These NPs were produced from citric acid in the presence of various amine-containing passivation reagents, namely high molecular weight branched poly(ethylenimine) (MW = 25 kDa, bPEI25k), low molecular weight branched poly(ethylenimine) (MW = 600 Da, bPEI600), pentaethylene hexamine (PEHA), and N,N-dimethylethylene diamine (DMEDA). The protein corona formed around these NPs when in contact with fetal bovine serum (FBS) was characterized, proteins were identified and quantified by label-free MS-based quantitative proteomics, and biological functions of the identified proteins were analyzed with regards to cell uptake and viability in a human model of macrophages. This type of cells was selected because it is one of the main NP target cells in the body.48 Indeed, macrophages, which are a major class of phagocytic innate immune cells, can recognize and internalize foreign elements, including NPs, and potentially trigger an immunological response.48,49 As a consequence, macrophages play a key role in lifetime of NPs in the body and in their nanosafety. This work was performed on THP-1 monocyte cell line differentiated into macrophage-like cells by phorbol 12-myristate 13-acetate (PMA), which represents a suitable model for studying macrophage functions in vitro.50
2. Results and discussion
2.1. Design and physicochemical characterization of CDs
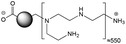
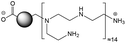
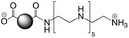
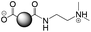
The CDs investigated herein were engineered so as to be cationic, their synthesis involving various oligo(ethyleneimine)-based passivation reagents. Their surface charge (ζ-potential) and charge density (electrokinetic charge, Qek) were tuned by using passivation reagents containing a decreasing number of amino groups per molecule (n): bPEI25k (n ≈ 550; CD1) > bPEI600 (n ≈ 14; CD2) > PEHA (n = 6; CD3) > DMEDA (n = 2; CD4) (Table 1). The ζ-potential of the CDs, at pH 7.4, was found as follows: +37.3 ± 5.2 mV (CD1), +31.8 ± 1.1 mV (CD2), +29.2 ± 2.2 mV (CD3) and +11.1 ± 2.2 mV (CD4). Though it decreased with the passivation reagent oligomerization state, it was not proportional to the latter. This may be a result of the differences in the activation process during the synthesis of the CDs (temperature, reaction time, concentration of the reagents⋯), that may lead 1-to the passivation reagent degradation to a variable extent, e.g., through cleavage or deoligomerization of the oligo(ethyleneimine) chains, and 2-to some difference in the incorporation ratio of the passivation reagent at the surface of the CDs. The surface charge density of the CDs was determined by polyelectrolyte titration as described previously.47 It ranged from 0.01 μmol mg−1 for CD4 to 4.70 μmol mg−1 for CD2. A higher oligomerization state of the passivation reagent also did not systematically translate into higher Qek, as was observed for CD1. Again, this may be related to differences in the experimental conditions leading to the formation of the various CDs. The size of the CDs was close to each other, with a mean hydrodynamic diameter, as determined by DLS, between 10.2 and 17.1 nm. As DLS method might overestimate the size of NPs due to the solvation molecules, our CDs were likely to be close to or slightly smaller than 10 nm in diameter, which is in line with the size range generally described for CDs in the literature.4 The NPs showed the classical UV-vis absorption pattern of CDs, i.e., a monotonous and decreasing absorption between 250 and 800 nm with a peak at 350 nm. Noteworthy, CD1 and CD4 only displayed the monotonous and decreasing absorption between 250 and 800 nm, with no other detectable peak. This was consistent with the full carbonization of the intermediate citric acid-derived fluorophores with blue emission that form at the beginning of the pyrolysis process, like imidazo[1,2-a]pyridine-7-carboxylic acid (IPCA) and derivatives.51,52 These compounds do not resist high temperature (>170 °C). The fluorescence excitation and emission spectra were similar for the four CDs, with a maximum excitation and emission wavelength at ca. 350 and 460 nm, respectively. Fluorescence quantum yields (Φ) were found rather similar for CD2, CD3, and CD4 (12.4, 9.8, and 16.5%, resp.), while CD1 displayed a lower value (1.4%). Many factors can influence the Φ of CDs, such as the reaction conditions used for their synthesis (i.e., starting material, solvent, concentration, activation mode, reaction temperature and time⋯).53 But this remains a difficult point to investigate as the exact origin of the CD fluorescence is not fully elucidated and is still the subject of intense debate.54 The spectroscopic properties of the CDs used in this work are provided as ESI (Fig. S1†). Thanks to their intrinsic photoluminescence properties, it was possible to follow the cellular uptake of these CDs without the need for conjugation to an additional fluorescent dye (vide infra).| CD1 | CD2 | CD3 | CD4 | |
|---|---|---|---|---|
| a Monotonous UV-vis absorption decreasing between 250 and 800 nm. | ||||
| Structure |
|
|
|
|
| Passivation reagent | bPEI25K | bPEI600 | PEHA | DMEDA |
| ζ-potential [mV] | +37.3 ± 5.2 | +31.8 ± 1.1 | +29.2 ± 2.2 | +11.1 ± 2.2 |
| Surface charge density Qek [μmol mg−1] | 2.30 | 4.70 | 3.25 | 0.01 |
| Hydrodynamic diameter [nm] | 17.1 ± 1.1 | 11.0 ± 3.4 | 10.2 ± 3.2 | 11.8 ± 3.3 |
| Photoluminescence λmax/λex/λem [nm] | /375/460 | 350/365/460 | 350/370/465 | /315/465 |
| Quantum yield Φ [%] | 1.4 | 12.4 | 9.8 | 16.5 |
2.2. Physicochemical properties of CD corona proteins
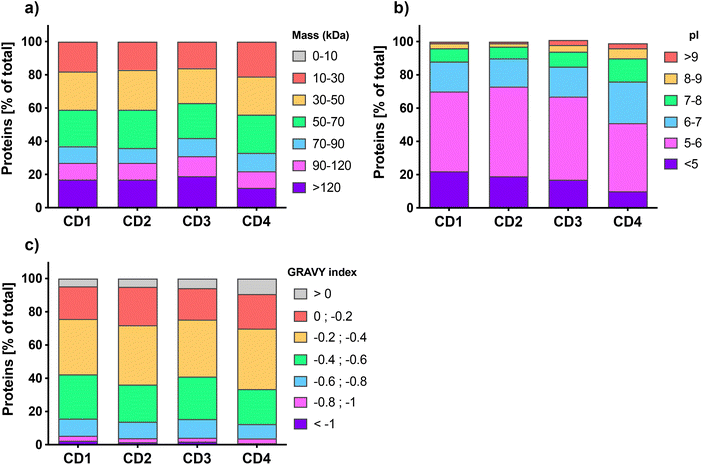
Quantitative label-free proteomic analysis was used to identify and quantify proteins adsorbed at the surface of the 4 engineered CDs. About five hundred proteins were identified on each type of CDs, with 539, 456, 575, and 465 proteins for CD1, CD2, CD3, and CD4, respectively. This number of proteins is in line with previous studies which reported that NP protein corona is generally composed of hundreds of proteins.55–57 We classified the proteins of CD corona according to their intrinsic physicochemical properties, namely molecular weight (MW), isoelectric point (pI) and Grand Average of Hydropathy (GRAVY) index, which is used to represent the hydrophobicity of the proteins. As shown in Fig. 1a, the distribution of protein MW was broadly homogeneous between the 4 CDs, with more than 80% of the proteins having a MW less than 120 kDa. This is in accordance with previous studies which also reported a high proportion of proteins <120 kDa in the corona of carbon,57,58 lipid,56,59 polystyrene,31 or gold NPs.60 Worth to note, the corona of CD4, which exhibits the lowest surface charge and charge density, contained slightly more small proteins (10–30 kDa) compared to the three other CDs. The pI analysis (Fig. 1b) showed that whatever the CDs, more than 75% of the corona proteins exhibited a negative net charge at pH 7.4 (i.e., pI < 7) and proteins with a pI between 5 and 6 were the most represented. Interestingly, CD4 corona contained about 25% more proteins with a positive net charge at pH 7.4 (i.e., pI > 7). These results do support the hypothesis that proteins bind to CDs thanks to electrostatic interactions, and are in agreement with the data obtained with other types of NPs.29,56,57,60 According to GRAVY index analysis (Fig. 1c), the hydrophilic or hydrophobic character of the proteins was fairly similar between the 4 CDs, with a mean GRAVY index of −0.37, −0.34, −0.36, and −0.31, for CD1, CD2, CD3, and CD4, respectively. In line with some previous studies, these negative GRAVY scores indicate that the CD corona proteins are rather hydrophilic.29,57,59 This is not surprising since serum proteins are relatively hydrophilic. But remarkably, almost 10% of the proteins bound to CD4 had a positive GRAVY index score, which evidences a preference of CD4 to bind hydrophobic proteins compared to other CDs. As recently reported for carbon nanotubes,61 this result indicates that hydrophobic interactions are also implicated in the formation of the protein corona on CDs, especially when CDs have a low surface charge. Thus, for the first time, thanks to proteomic analysis, we were able to characterize the physicochemical properties of proteins that compose CD corona and we showed that these physicochemical properties are comparable to the one described for other types of NPs, in particular other carbon NPs.57,58,61 Our data point to the role of electrostatic and, to a lesser extent, hydrophobic interactions in the formation of CD corona and suggest that the surface charge of CDs could have an impact on the corona protein composition.2.3. Biological functions of CD corona proteins
Before analyzing the biological functions of the proteins found in the corona, a Venn diagram was used to represent the common and specific proteins identified at the surface of the four CDs (Fig. 2). We found that 218 proteins (i.e., about 25% of the total proteins) do bind to all CDs. Furthermore, we established that 111 proteins were shared by CD1, CD2 and CD3, corresponding to 13% of the total proteins, whereas CD3 and CD4 had the highest number of specific proteins (115 and 110, respectively).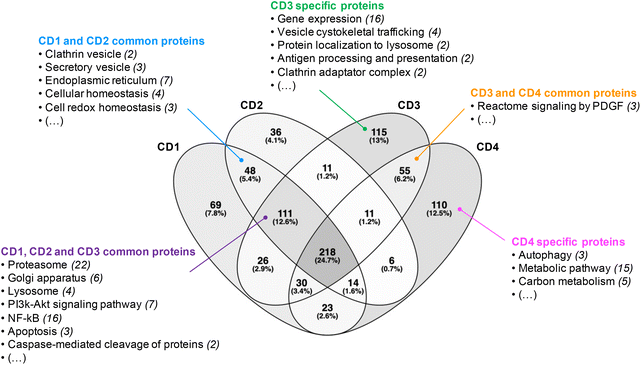 | ||
| Fig. 2 Venn diagram of the proteins identified at the surface of the four CDs investigated in this study. | ||
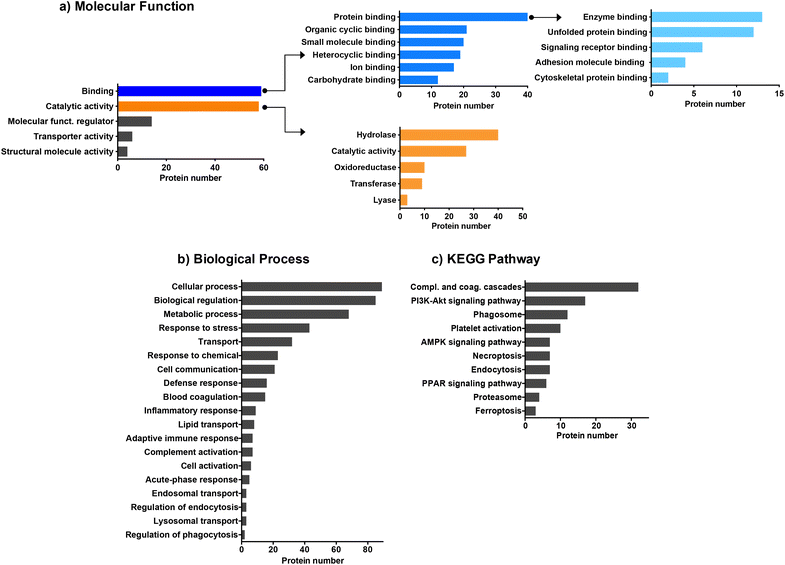
We then employed bioinformatic tools such as DAVID or Panther systems to classify the proteins according to their molecular function, as well as their involvement in biological processes and KEGG pathway. We first focused on the proteins common to the four CDs (Fig. 3). The protein classification based on their molecular function (Fig. 3a) revealed that most of the proteins common to the four CDs are involved in binding or catalytic activity functions. Proteins with binding function are the most represented with notably proteins involved in enzyme binding. This result may be of importance as it has been reported that interaction of NPs with enzymes can result in enzyme activation or inactivation in the cellular environment.62 Proteins with catalytic functions are mainly involved in the regulation of hydrolase, oxidoreductase, transferase or lyase activities, or carry out these activities directly. As shown in Fig. 3b, the proteins common to the four CDs are associated with a wide variety of biological processes, including cellular response such as response to stress, response to chemicals, defense response, and inflammatory or adaptative immune responses. A large number of proteins are also involved in the transport processes, including lipid transport, endosomal transport or lysosomal transport, as also described for carbon nanotubes.61 Besides, most proteins belonging to the KEGG pathway are involved in complement and coagulation cascade activation (Fig. 3c), in line with what has been previously observed for other types of NPs.29,31,60,61 While binding of complement factors at the surface of NPs has been reported to promote NP phagocytosis by immune cells,60 no study focused so far on CDs. This point will be discussed below.
Next, we focused on the biological function of the proteins specific to each CD or to some CD subsets (Fig. 2). While no particular category of biological functions was found for proteins specific to CD1 or CD2, proteins specific to CD3 or CD4 corona are involved in several biological functions, such as gene expression, vesicle cytoskeletal trafficking, or protein localization to lysosome for CD3, and autophagy, metabolic pathway, or carbon metabolism for CD4. CD1 and CD2 shared proteins involved in clathrin-coated vesicles or secretory vesicles, endoplasmic reticulum, cellular or cell redox homeostasis. Unlike CD4, CD1, CD2 and CD3 shared a high number of proteins which are linked to proteasome, Golgi apparatus, lysosome, PI3k-Akt signaling pathway, NF-kB or apoptosis. This is particularly interesting as these 3 CDs are the closest in terms of surface charge properties (ζ-potential between +29.2 and +37.3 mV and surface charge density between 2.30 and 4.70 μmol mg−1). Thus, we demonstrated that 25% of the corona proteins are common to the four CDs. These proteins are associated with a wide variety of biological functions, including binding function, catalytic activity, cellular response and complement pathway activation, suggesting that protein corona could contribute to the biological effects of CDs. Besides, we showed that the four CDs have specific proteins in their corona. This difference in corona composition is presumably due to the difference in surface charge properties among the four CDs, and it may play an important role in the NP cellular fate and safety. Therefore, we next assessed CD cellular uptake and toxicity in a human model of macrophages cultured in presence of FBS, and then analyzed the link between the responses evoked by the NPs, their surface charge properties and the composition of the protein corona formed around the NPs when incubated with FBS.
2.4. Cellular uptake and toxicity of the four CDs in human macrophages
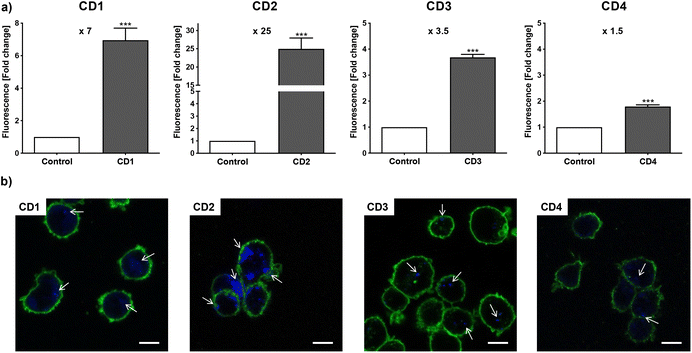
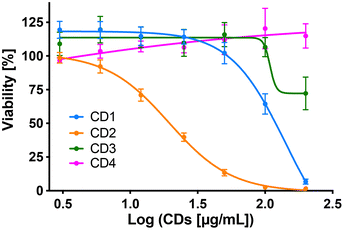
To assess internalization of the CDs by human macrophages (THP-1-derived macrophages cultured in medium containing 10% serum), the cells were exposed to non-cytotoxic concentration (25 μg mL−1) of the NPs for 4 h and NP uptake was determined by monitoring CD-associated fluorescence of cells by fluorescence activated cell sorting (FACS) and confocal laser scanning microscopy (CLSM). As shown in Fig. 4a, an increase in cell fluorescence was measured to various extents in the cells treated with the 4 CDs, as assessed by FACS. Indeed, among the 4 CDs, CD2 which exhibits ζ-potential higher than +30 mV and the highest charge density (Qek = 4.70 μmol mg−1) gave the greatest fluorescence increase, while CD4 which exhibits the lowest ζ-potential (+11.1 mV) and charge density (0.01 μmol mg−1) resulted in the lowest one. CD1 and CD3, with a ζ-potential around +30 mV and medium charge densities (2.30 and 3.25 μmol mg−1, respectively), translated into moderate fluorescence intensity. These results were confirmed by macrophage observation by CLSM (Fig. 4b). Indeed, a high intensity and punctuated fluorescence signal (blue spots) was observed in the cytoplasm of cells (membrane labeled in green) exposed to CD2, while a mild to moderate signal was observed in cells exposed to CD1, CD3 and CD4. As CD1 exhibited a lower fluorescence Φ than other CDs, it is possible that its cellular uptake was underestimated, which will be taken into account in subsequent analyses. Anyway, based on CD2, CD3 and CD4 data, and in line with our previous studies,46,47 we showed that not all cationic CDs are potently internalized by human macrophages, and that not only the ζ-potential but also the surface charge density influence cellular uptake of the NPs.To assess cytotoxicity of the CDs toward macrophages, the cells were exposed to increasing concentrations of CD1 to CD4 (3 to 200 μg mL−1, 10% serum) for 24 h, and their viability was determined by measuring cell mitochondrial activity using the MTT assay (Fig. 5). The 4 CDs exhibited various toxicity profiles. CD1 and CD2 induced a dose-dependent loss in cell viability that reached nearly 100% at the concentration of 200 μg mL−1, with a lower IC50 value for CD2 compared to CD1 (19 μg mL−1 and 141 μg mL−1, respectively). CD3 triggered some toxicity at 200 μg mL−1 with viability loss not exceeding 30%, whereas CD4 did not affect macrophage viability. Thus, in agreement with our previous data,47 we showed that CD2 which exhibited ζ-potential higher than +30 mV and the highest charge density was the most cytotoxic, while CD4 which exhibited the lowest ζ-potential and charge density was the least cytotoxic, and CD1 and CD3 exhibited a medium toxicity.
2.5. Potential role of protein corona in CD cell uptake
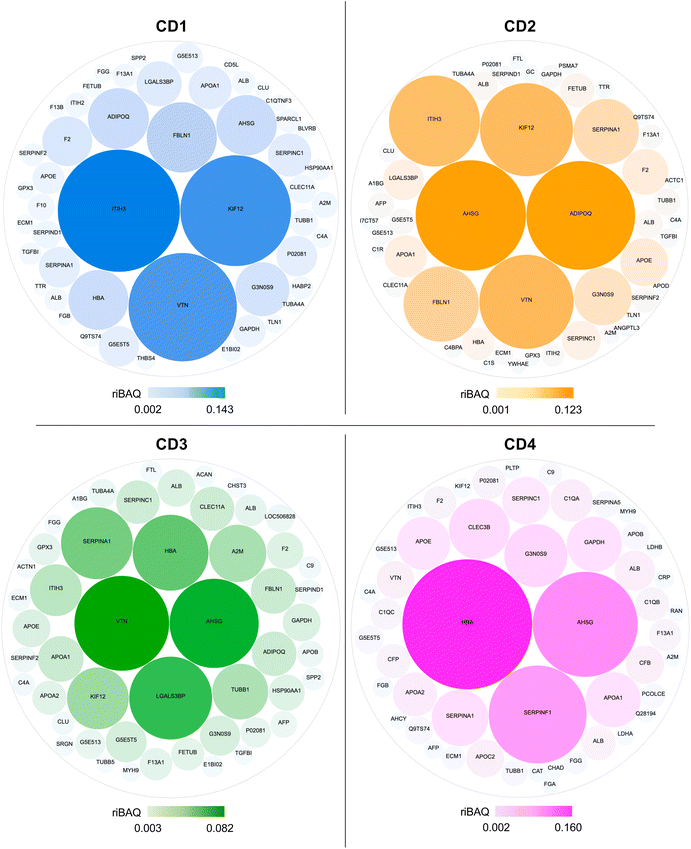
In order to understand to what extent the protein corona formed around CDs in the presence of serum could drive the cellular uptake of the NPs, we analyzed the top 50 most abundant proteins in the NP corona (Table S1, ESI†). Using the riBAQ approach, corona proteins were ranked from most to least abundant for each CD. In the literature, in-depth proteomic analyses often focus on the top 10 or 20 proteins.63–65 As this is here the first ever proteomic study carried out on CDs, we have chosen to conduct the analysis on the top 50 proteins. This is a more time-consuming and complex analysis to carry out, but it provides a more complete picture to analyze the protein corona in detail. The proteins ranked at rank 1 had a riBAQ of 0.08 to 0.14 according to CDs, which corresponds to a relative contribution of these proteins within the corona of 8 to 16%, whereas the proteins ranked at rank 50 had a riBAQ around 0.001, i.e., these proteins constituted about 0.1% of the protein corona.The composition and the contribution of the top 50 most abundant proteins of the corona of each CD are illustrated in Fig. 6. Eleven proteins were shared by CD1, CD2, and CD3, corresponding to about 20% of the top 50 proteins, whereas CD4 had the highest number of specific proteins (20 proteins). Notably, we identified proteins involved in the regulation of phagocytosis (AHSG, ADIPOQ) and of the phagosome (TUBB, TUBA4A, TUBB1, IGHM, THBS4, C1R) in the corona formed around CD1, CD2 and CD3, but not in that of CD4.
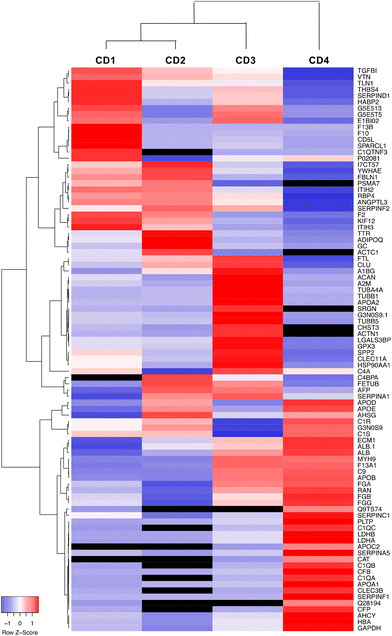
Data on the 50 most abundant proteins in the CD corona were also used to construct a heatmap (Fig. 7). This heatmap illustrates that the corona exhibited distinct proteomic profiles with different protein composition and abundance according to CDs. The column dendrogram shows that CD1 and CD2 are the closest CDs in terms of protein signature, followed by CD3, whereas CD4 is clearly distant from the three others CDs. Indeed, the proteins that were predominant (in red) in the CD4 corona were the least abundant or absent in the three others CDs. These proteins included apolipoproteins (APOD, APOE, APOB, APOC2, APOA1), albumin and hemoglobin which have been described to negatively correlate with the internalization of NPs.63,66,67
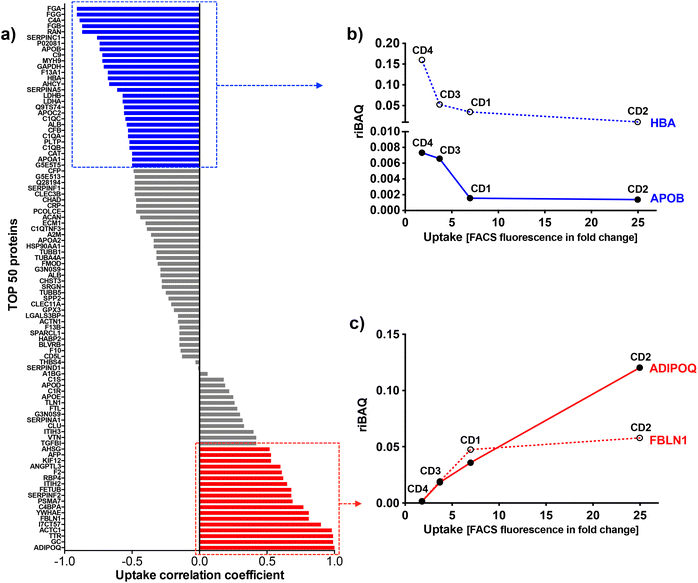
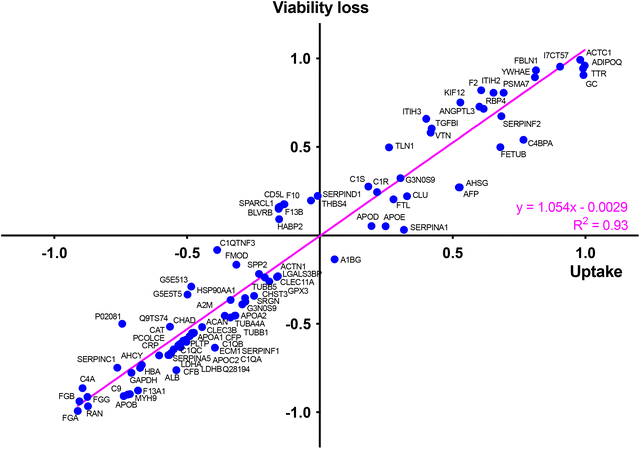
To identify the key corona proteins that could potentially contribute to CD cell uptake, we first determined the correlation between abundance of each top 50 protein in the corona and CD macrophage uptake (as assessed by FACS), using a Pearson correlation analysis (Fig. 8a). In agreement with our heatmap data, a negative correlation was found between albumin, hemoglobin (i.e., HBA) or some apolipoproteins (i.e., APOB) and the uptake of CDs by macrophages (Fig. 8b). Even if albumin is the most abundant protein in the blood and is therefore frequently identified in NP corona,68 it was not the highest abundant protein detected in the corona for any CDs investigated herein. Besides, in line with our data, a protein corona particularly enriched in dysopsonins, including albumin, could lead to a decrease in uptake by macrophages, as it has been shown for carbon nanotubes69 or silica NPs.67 Regarding apolipoproteins, it was suggested that their amount in the corona of silica NPs could limit phagocytosis of the latter through competitive surface binding with opsonins.63 However, this effect could depend on the type of apolipoprotein and NPs, since it has been reported that polystyrene NPs precoated with the apolipoproteins APOA4 or APOC3 exhibited a significant decreased cellular uptake, whereas NPs precoated with APOH displayed increased uptake.66 Recently, it was also demonstrated that APOA1, an abundant protein in graphene NP corona, could promote the internalization of these NPs via scavenger receptor B1,70 while pre-adsorption of gold and graphene NPs with APOE led to extended blood circulation of the NPs.71 In our case, the presence of APOB, APOC2 and APOA1 on the surface of CDs could lead to a decrease in CD recognition by macrophages and thus potentially increase their blood circulation time.
Among analyzed proteins, 18 proteins were positively correlated with CD uptake, with ADIPOQ (adiponectin) and FBLN1 (fibulin) as those exhibiting the highest correlation (Fig. 8c). Recently, it was observed that ADIPOQ is one of the most abundant proteins in the corona of positive carbon NPs (hydrogenated nanodiamonds, +60 mV).57 Although the link between ADIPOQ and NP cell internalization is not highly supported in the literature, it has been proposed that adiponectin-coated NPs exhibit enhanced macrophage targeting in atherosclerotic lesions.72 On its hand, FBLN1 was found in the corona of gold73 and silica67 NPs that were widely internalized by epithelial cells and macrophages. Qin et al. hypothesized that, as fibulin plays a role in cell adhesion and migration, its adsorption on the NP surface could indicate that the extracellular matrix-mediated interaction is one of the important triggers for NP uptake.73 We also found that C4BPA, a complement protein, correlated with CD cell uptake. In the literature, complement proteins were identified as key proteins constituting NP corona, and correlating with enhanced NP uptake by immune cells by promoting opsonization.63 In line with us, a study in particular has shown that complement protein C4BPA is abundant in the corona formed around cationic gold NPs leading to uptake of the NPs by macrophages.60 According to our data, AHSG (alpha 2-HS glycoprotein) and FETUB (fetuin) correlated also with CD cell uptake. In the literature, fetuin and alpha 2-HS glycoprotein were reported to enhance phagocytosis and macropinocytosis of labelled dextran by macrophages.74 In particular, fetuin was shown to mediate internalization of polystyrene NPs by Kupffer cells (liver macrophages) via scavenger receptor.75 We can also underline the positive correlation between CD uptake and ACTC1 (actin) found in the present study, because it has been shown that actin plays a crucial role in the phagocytosis of quartz particles in macrophages.76 In addition, the correlation with TTR (transthyretin) can be highlighted as TTR is a transport protein that interacts with receptor-associated proteins in the liver and has been found to associate with silica NPs.77 This correlation analysis was based on the uptake data of the 4 CDs. However, as the CD1 uptake may have been underestimated, the analysis was also performed using only CD2, CD3 and CD4 uptake data. The results of this analysis were consistent with those obtained when CD1 was included in the data set. Indeed, we found the same proteins positively or negatively correlated with CD uptake (Table S2, ESI†).
Taken together, our data establish for the first time a link between several proteins constituting the protein corona of CDs and internalization levels of the NPs by macrophages. In particular, we identified some proteins negatively (albumin, apolipoproteins, hemoglobin) or positively (complement, fibulin, fetuin, alpha-glycoprotein) associated with CD uptake. In order to determine the involvement of each of these proteins in the mechanisms of CD recognition and uptake, future studies could focus more precisely on the effect of each protein, for example by performing CD coating studies with one protein at a time.
2.6. Role of the CD surface charge on the corona composition and CD cell uptake
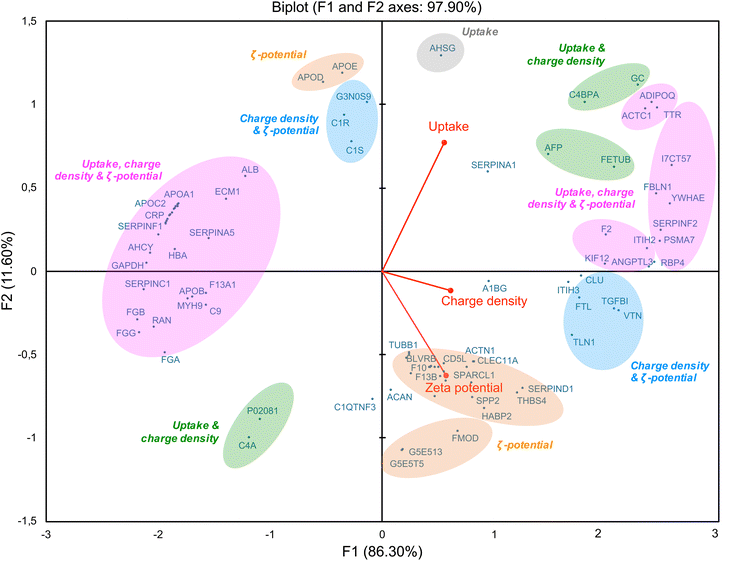
As we showed a correlation between some corona proteins and the cellular uptake of CDs, we further investigated the role of CD surface charge in this behavior. By using a multivariate approach based on principal component analysis (PCA),29,78 we studied association of CD protein corona composition (50 top proteins) with multiple CD variables, including ζ-potential, surface charge density and cellular uptake (Fig. 9). This analysis revealed a positive (right of the diagram, positive value on the F1-axis) or negative (left of the diagram, negative value on the F1-axis) correlation between the variables. Variables that are correlated with each other are grouped together. Thus, some proteins, including ITIH3, rather correlated with the ζ-potential of CDs (in orange), while others, including VTN, correlated with both ζ-potential and surface charge density (in blue). Indeed, ITIH3 was little abundant (rank 43) on the surface of CDs with the lowest ζ-potential (CD4, +11.1 mV), whereas it was the most abundant on the CDs with the highest ζ-potential (CD1, +37.3 mV). As ITIH3 acts as a hyaluronan transporter or as a binding protein with other matrix proteins, this protein can promote the interaction of NPs with the cell surface.79,80 Using quantitative structure–activity relationship (QSAR) based on gold NP protein corona dataset, it was also reported that ITIH3 was one of the 11 best descriptors of NP-cell association along with ζ-potential.81 In agreement with our data on VTN (vitronectin), it was shown that this protein was among the most abundant in the corona of cationic liposomes and PEI-coated gold NPs.22 Introducing positive charges into lipids NPs resulted in shifting of the NP corona from an apolipoprotein- to a vitronectin-enriched corona.82 Moreover, optimized cationic lipid NPs so that they preferentially recruit vitronectin in their corona led to enhanced gene delivery into tumor cells via the ανβ3 integrin receptor.83 In these studies, the parameter used to characterize the NP charge and establish a link between NP corona composition and cell internalization is ζ-potential. Remarkably, in our study, we also found a correlation between NP surface charge density and proteins involved in cell internalization. Some proteins are common with those in the ζ potential-based correlation (in pink), such as ACTC1, ADIPOQ, TTR, and FBLN1 which we discussed above. But other proteins specifically correlate with NP surface charge densities, such as C4PBA and FETUB (in green). Thus, the density of positive charges due to amino groups displayed on the surface of CDs may be an important factor in the formation of the protein corona and subsequent CD cell uptake. In line with us, in a study conducted on polymer NPs, it was proposed that amine density and type of amine (primary, secondary or tertiary) are both important parameters in the corona formation and NP cellular association.39 It has been also recently proposed that the interaction between some model proteins (albumin, lysozyme) and nanodiamond surface becomes stronger as the functional group density (amino- or carboxyl- groups) increases.84 Our multivariate approach also revealed a negative correlation between ζ-potential, surface charge density and cell uptake of CDs (Fig. 9, on the left part of the graph, in pink) and some proteins including apolipoproteins (APOA1, APOB, APOC2), albumin and hemoglobin. Thus, this confirms that modulating the ζ-potential but also the density of positive charges at the surface of CDs, has an impact on proteins constituting the CD corona and CD cellular uptake.2.7. Potential role of corona proteins in CD cytotoxicity
At last, to determine whether the protein corona formed around CDs in the presence of serum could drive the cytotoxicity of the NPs and identify key proteins in this phenomenon, we determined, in the same way as for the CD cell uptake, the correlation between abundance of each top 50 protein in the corona (Table S1, ESI†) and the cytotoxicity of CDs using a Pearson correlation analysis (Fig. S2, ESI†). To do so, a toxicity marker was chosen as the loss of viability induced by 125 μg mL−1 CDs, from viability curves described in Fig. 5. We identified some proteins negatively (in blue) and positively (in red) associated with CD cytotoxicity (Fig. S2, ESI†). Remarkably, most of these proteins were also associated with the CD uptake, such as APOA1, APOB, APOC2, ALB and HBA (negative correlation) or VTN, TTR, FBLN1, FETUB, ITIH3 and ADIPOQ (positive correlation) (Fig. S2, ESI†). This observation was confirmed by analyzing the relationship between CD uptake and macrophage viability loss depending on each top 50 protein present in the NP corona (Fig. 10). This analysis was based on the uptake data of the 4 CDs. Another analysis performed using only CD2, CD3 and CD4 data gave consistent results by showing also a correlation between CD uptake, viability loss and protein abundance (linear regression with R2 = 0.91, Fig. S3, ESI†). Thus, a direct correlation was observed between the uptake of CDs and their toxicity profile, as it is often described for NPs.85 We also showed that CD uptake and cytotoxicity were influenced by the CD protein corona, which in turn depends on the ζ-potential and surface charge density of the CDs.3. Conclusion
To the best of our knowledge, our study is the first proteomic analysis of the protein corona of CDs conducted in the literature. Our data showed that the physicochemical properties as well as the biological functions of the proteins constituting the corona formed around cationic CDs depend on the charge characteristics of the NPs. In particular, we demonstrated a role for the ζ-potential but also for the surface charge density. Moreover, we established a link between several proteins present in the CD protein corona and the uptake and cytotoxicity of the NPs. Thus, modulating the ζ-potential but also the density of positive charges at the surface of CDs has an impact on the composition of CD protein corona, which in turns drive the CD cellular uptake and toxicity. These data provide useful information for developing safe CDs for biomedical applications, and illustrate the potential contribution of corona study in the understanding of mechanisms of cellular fate and toxicity of NPs.4. Experimental
4.1. Synthesis of CDs
The NPs were prepared according to various procedures under solvent-free (CD1) or solvothermal conditions, at atmospheric (CD2 and CD3) or hyperbaric pressure (CD4) as detailed below.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da) against HCl (0.1 N, 12 h) and ultra-pure water (96 h). The dialysis bag content was filtered over a 0.22 μm polyethersulfone (PES) membrane (Millex) and the filtrate was freeze-dried to provide hygroscopic yellow-brown powder (0.68 g).
000 Da) against HCl (0.1 N, 12 h) and ultra-pure water (96 h). The dialysis bag content was filtered over a 0.22 μm polyethersulfone (PES) membrane (Millex) and the filtrate was freeze-dried to provide hygroscopic yellow-brown powder (0.68 g).
4.2. Characterization of CDs
All measurements were performed on fresh CD samples (1.0 mg mL−1 in 1.5 mM NaCl pH 7.4). The hydrodynamic diameter of CDs was measured by dynamic light scattering (DLS, Zetasizer Nano ZS, Malvern Instruments, France) and calculated from the number distribution graph. The ζ-potential was measured by DLS as well, and calculated with the Smoluchowski's equation. The surface charge density of CDs was determined by polyelectrolyte titration as previously described.47 In brief, ζ-potential variations in the sample were monitored along spiking with a solution of poly(acrylic acid) (PAA, MW ± 1800 Da, NaCl 1.5 mM pH 7.4). The amount of PAA required to reverse the sign of ζ-potential was approximated by linear interpolation of the titration curve at the isoelectric point. The density of surface charge of the CDs, Qek, was then calculated from the required amount of PAA using the equation:| Qek = V·c/w |
| ΦCD = Φq·(FCD/Fq)·(Aq/ACD) |
4.3. Sample preparation for proteomic analysis of CD protein corona
CD water suspensions (250 μL, 2.0 mg mL−1) were sonicated in a sonication bath (40 Hz, for 3 min) and mixed with non-diluted fetal bovine serum (250 μL, 40 mg mL−1 of protein, GIBCO, France) in Eppendorf® Protein Lobind tubes. The tubes were then incubated for 1 h at 37 °C under rocking agitation (70 oscillations per min). After that, the samples were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 20 °C for 45 min. Supernatant was discarded and the pellets containing the CD-protein complexes were resuspended in ultra-pure water and centrifuged again (14
000g and 20 °C for 45 min. Supernatant was discarded and the pellets containing the CD-protein complexes were resuspended in ultra-pure water and centrifuged again (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 45 min at 20 °C) to remove unbound proteins. The process was repeated twice. The final pellets were resuspended in 100 μL of ultrapure water. After determination of protein concentration by the Bicinchoninic acid assay (Sigma-Aldrich, MO, USA), 7.5 μg of samples were denaturated at 95 °C for 5 min in Laemmli buffer and concentrated in one stacking band using a 5% SDS-PAGE gel. The gel was fixed with 50% ethanol/3% phosphoric acid and stained with colloidal Coomassie Brilliant Blue. Each band was excised, cut into three pieces, and transferred into a 96-well microtiter plate. Gel slices were washed in 100 μL of 50
000g for 45 min at 20 °C) to remove unbound proteins. The process was repeated twice. The final pellets were resuspended in 100 μL of ultrapure water. After determination of protein concentration by the Bicinchoninic acid assay (Sigma-Aldrich, MO, USA), 7.5 μg of samples were denaturated at 95 °C for 5 min in Laemmli buffer and concentrated in one stacking band using a 5% SDS-PAGE gel. The gel was fixed with 50% ethanol/3% phosphoric acid and stained with colloidal Coomassie Brilliant Blue. Each band was excised, cut into three pieces, and transferred into a 96-well microtiter plate. Gel slices were washed in 100 μL of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (v/v) 25 mM NH4HCO3/acetonitrile (ACN) (3 washes of 10 min each). Gel bands were then dehydrated with 50 μL ACN (100%) and reduced with 50 μL DTT (10 mM) for 30 min at 60 °C, followed by 30 min at RT. Proteins were then alkylated with 50 μL iodoacetamide (55 mM) for 20 min in the dark at RT, before addition of 100 μL ACN for 5 min. Samples were washed with 50 μL NH4HCO3 (25 mM) for 10 min and 50 μL ACN for 5 min, before being dehydrated with two cycles of incubations in 50 μL ACN for 5 min. Then proteins were digested overnight at 37 °C with a modified porcine trypsin solution (Promega, WI, USA) at a 1
50 (v/v) 25 mM NH4HCO3/acetonitrile (ACN) (3 washes of 10 min each). Gel bands were then dehydrated with 50 μL ACN (100%) and reduced with 50 μL DTT (10 mM) for 30 min at 60 °C, followed by 30 min at RT. Proteins were then alkylated with 50 μL iodoacetamide (55 mM) for 20 min in the dark at RT, before addition of 100 μL ACN for 5 min. Samples were washed with 50 μL NH4HCO3 (25 mM) for 10 min and 50 μL ACN for 5 min, before being dehydrated with two cycles of incubations in 50 μL ACN for 5 min. Then proteins were digested overnight at 37 °C with a modified porcine trypsin solution (Promega, WI, USA) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (w/w) enzyme/protein ratio. Tryptic peptides were extracted under agitation at RT with 60 μL 60% ACN/0.1% formic acid (FA) for 45 min, and then 100% ACN for 10 min. The extraction supernatants were pooled and vacuum-dried before re-suspension in 20 μL 2% ACN/0.1% FA.
100 (w/w) enzyme/protein ratio. Tryptic peptides were extracted under agitation at RT with 60 μL 60% ACN/0.1% formic acid (FA) for 45 min, and then 100% ACN for 10 min. The extraction supernatants were pooled and vacuum-dried before re-suspension in 20 μL 2% ACN/0.1% FA.
4.4. Mass spectrometry analysis
NanoLC-MS/MS analyses were performed on a nanoAcquity UPLC device (Waters, MA, USA) coupled to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, France). The solvents consisted of 0.1% FA in H2O (solvent A) and 0.1% in ACN (solvent B). The digested samples (2 μL) were loaded onto a Symmetry C18 pre-column (20 mm × 180 μm, 5 μm diameter particles; Waters, Milford, MA) over 3 min at 5 μL min−1 with 1% solvent B. Peptides were eluted on an Acquity UPLC BEH130 C18 column (250 mm × 75 μm, 1.7 μm particles; Waters, Milford, MA) at 450 μL min−1 with the following gradient of solvent B: linear from 1% to 8% in 2 min, linear from 8% to 35% in 77 min, linear from 35% to 90% in 1 min, isocratic at 90% for 5 min, down to 1% in 2 min, isocratic at 1% for 2 min. The Q-Exactive Plus mass spectrometer was operated in data-dependent acquisition mode by automatically switching between full MS and consecutive MS/MS acquisitions. Full-scan MS spectra were collected from 300–1800 m/z at a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at 200 m/z with an automatic gain control target fixed at 3 × 106 ions and a maximum injection time of 50 ms. The top 10 precursor ions with an intensity exceeding 2 × 105 ions and charge states ≥2 were selected on each MS spectrum for fragmentation by higher-energy collisional dissociation. MS/MS spectra were collected at a resolution of 17
000 at 200 m/z with an automatic gain control target fixed at 3 × 106 ions and a maximum injection time of 50 ms. The top 10 precursor ions with an intensity exceeding 2 × 105 ions and charge states ≥2 were selected on each MS spectrum for fragmentation by higher-energy collisional dissociation. MS/MS spectra were collected at a resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 at 200 m/z with a fixed first mass at 100 m/z, an automatic gain control target fixed at 1 × 105 ions and a maximum injection time of 100 ms. A dynamic exclusion time was set to 60 s.
500 at 200 m/z with a fixed first mass at 100 m/z, an automatic gain control target fixed at 1 × 105 ions and a maximum injection time of 100 ms. A dynamic exclusion time was set to 60 s.
4.5. Proteomics data processing
A complete data set has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Raw mass spectrometry (MS) data processing was performed using the MaxQuant software v1.6.7.0.86 Peak lists were searched against a database including Bos Taurus protein sequences extracted from Uniprot (17-07-2020; 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 497 sequences, taxonomy ID = 9913) supplemented with 35 keratins and 1 trypsin, generated with the database toolbox from MSDA.87 MaxQuant parameters were set as follows: MS tolerance set to 20 ppm for the first search and 5 ppm for the main search, MS/MS tolerance set to 40 ppm, maximum number of missed cleavage set to 1, carbamidomethyl (C) set as fixed modification, oxidation (M) and protein N-term acetylation set as variables modifications. False-discovery rates (FDRs) were estimated based on the number of hits after searching a reverse database and were set to 1% for both peptide spectrum (with a minimum length of seven amino acids) and proteins matches. The LFQ (label-free quantification) and the “match between runs” options were not used. All other MaxQuant parameters were set as default. To be considered, proteins must be identified in all four replicates of at least one condition. To determine the relative abundance for each protein and classify them, we determined the relative intensity-based absolute quantification (riBAQ) using the MaxQuant software for each protein.88 Then, a cut-off was applied to select only proteins present in all four replicates.
497 sequences, taxonomy ID = 9913) supplemented with 35 keratins and 1 trypsin, generated with the database toolbox from MSDA.87 MaxQuant parameters were set as follows: MS tolerance set to 20 ppm for the first search and 5 ppm for the main search, MS/MS tolerance set to 40 ppm, maximum number of missed cleavage set to 1, carbamidomethyl (C) set as fixed modification, oxidation (M) and protein N-term acetylation set as variables modifications. False-discovery rates (FDRs) were estimated based on the number of hits after searching a reverse database and were set to 1% for both peptide spectrum (with a minimum length of seven amino acids) and proteins matches. The LFQ (label-free quantification) and the “match between runs” options were not used. All other MaxQuant parameters were set as default. To be considered, proteins must be identified in all four replicates of at least one condition. To determine the relative abundance for each protein and classify them, we determined the relative intensity-based absolute quantification (riBAQ) using the MaxQuant software for each protein.88 Then, a cut-off was applied to select only proteins present in all four replicates.
4.6. Bioinformatics analysis
The theoretical physicochemical properties of the proteins were calculated based on their sequences. The molecular weight (MW) and isoelectric point (pI) were computed viahttps://isoelectric.org/index.html and the GRAVY index viahttps://www.bioinformatics.org/sms2/protein_gravy.html. The Venn diagram used to decompose the common and specific proteins identified within corona of the different CDs was obtained using the Venny bioinformatic tool (v 2.1.0). The list of proteins was also organized according to the respective molecular function, biological process and involvement in KEGG pathway employing the classification generated from the DAVID (v 6.8), STRING (v 11.5) and Panther (v 16.0) systems. The graphs illustrating the top 50 most abundant proteins composing CD corona were constructed using the Rawgraph software (https://rawgraphs.io). The heatmap of the top 50 proteins was obtained from the heatmapper software (https://www.heatmapper.ca/expression/). The Pearson's correlation coefficients were calculated to determine the relationship between uptake or cytotoxicity of CDs and the top 50 corona proteins using the GraphPad Prism software (v 6.0). A multivariate approach based on principal component analysis (PCA) was used to study the influence of ζ-potential, surface charge density and cellular uptake on the top 50 corona proteins.4.7. Cell culture
THP-1 (TIB-202™, ATCC) cells were grown in culture flasks at 37 °C in a 5% CO2 humidified chamber using RPMI-1640 culture medium containing L-glutamine (2 mM), 2-mercaptoethanol (0.05 mM), penicillin (100 UI mL−1), streptomycin (100 μg mL−1), and 10% heat inactivated FBS (all reagents from GIBCO, France). For experiments, cells were seeded in appropriate culture devices and differentiated into macrophages overnight by adding 10 ng mL−1 phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, MO, USA) to the culture medium. The expression of surface markers such as CD11b (CR3), CD14 and CD36 was checked by FACS (data not shown), attesting to transition of the cells from a monocyte to a macrophage phenotype.504.8. Assessment of CD cell uptake
Thanks to the intrinsic fluorescence properties of CDs, it was possible to assess their cellular uptake by macrophages using CLSM and fluorescence activated cell sorting (FACS). CLSM experiments were carried out as follow. Cells were seeded into 8-well IbiTreat μ-slides (1.5 polymer coverslip, IBIDI®, Ibidi GmbH, Germany) at a density of 105 cells per well, differentiated into macrophages, and incubated with CDs at a concentration of 25 μg mL−1, for 4 h. At the end of the incubation time, the cells were washed twice with culture medium. In order to label the cell membrane, cells were exposed to the fluorescent probe DSQ12S (10 nM in PBS)89 shortly before their observation under the microscope. The cellular uptake of CDs was observed using a Leica SP2 microscope equipped with a 63X oil immersion objective (NA = 1.2). The CDs and the membrane probe DSQ12S were excited with 405 and 635 nm laser sources, respectively. For FACS experiments, cells were seeded into 24-well plates at a density of 5.105 cells per well, differentiated into macrophages, and incubated with CDs at a concentration of 25 μg mL−1 for 4 h. After CD exposure, the supernatant was discarded and the cells were rinsed with PBS, harvested by trypsin treatment, transferred into microtubes, and centrifuged for 5 min at 200g. The pelleted cells were rinsed with PBS and resuspended in culture medium without serum. Cell suspensions were then analyzed with a LSRFortessa X 20™ flow cytometer (BD Biosciences, France) and fluorescence of each sample (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events) was collected using a BV421 (violet laser) channel. CD uptake was quantified by determining changes in the mean fluorescence intensity (MFI) of CD-treated cells compared to untreated cells. Results were expressed as the ratio of the MFI of CD-treated cells to the MFI of untreated cells. They were means ± SEM of n = 3 experiments. Statistical differences were determined by Student's t-test, using the GraphPad Prism software (v 6.0).
000 events) was collected using a BV421 (violet laser) channel. CD uptake was quantified by determining changes in the mean fluorescence intensity (MFI) of CD-treated cells compared to untreated cells. Results were expressed as the ratio of the MFI of CD-treated cells to the MFI of untreated cells. They were means ± SEM of n = 3 experiments. Statistical differences were determined by Student's t-test, using the GraphPad Prism software (v 6.0).
4.9. Cell viability assay
Cell viability was assessed by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Cells were seeded into 96-well plates at a density of 105 cells per well, differentiated into macrophages and incubated for 24 h with increasing concentrations of CDs (3–200 μg mL−1). Then, cells were washed with PBS and incubated with 100 μL of MTT (1.0 mg mL−1 diluted in culture medium) for 1 h at 37 °C. At the end of the incubation period, the culture medium was removed, and the cells were lysed with DMSO. Absorbance of the resulting samples was read at 570 nm with a correction at 690 nm using a Varioskan Lux microplate reader (Thermo Fisher Scientific, France). Cell viability was expressed as the percentage of the absorbance of CD-treated cells relative to the absorbance of the non-exposed cells. The results were means ± SEM of n = 3–6 experiments. Concentration–response curves were obtained after logarithmic transformation of the data and fit with the Hill equation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Pascal Didier for its help with CLSM (Laboratoire de Bioimagerie et Pathologies, UMR 7021, Faculté de Pharmacie, CNRS-Université de Strasbourg, Illkirch, France), as well as Claudine Ebel and Muriel Koch for their help with FACS experiments (Cytometry platform, IGBMC, Strasbourg). This research was supported by the Agence Nationale de la Recherche (ANR, ANR-18-CE34-0005-01), by the Agence Nationale de Sécurité Sanitaire de l'Alimentation, de l'Environnement et du Travail (ANSES, EST-2015/1/005), by the Interdisciplinary Thematic Institute 2021–2028 program of the University of Strasbourg, CNRS and Inserm (ANR-10-IDEX-0002 and ANR-20-SFRI-0012) in the frame of the InnoVec Institute, and by the French Proteomic Infrastructure (ProFI, ANR-10-INBS-08-03).References
- X. Han, K. Xu, O. Taratula and K. Farsad, Nanoscale, 2019, 11, 799–819 RSC.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- A. L. Himaja, P. S. Karthik and S. P. Singh, Chem. Rec., 2015, 15, 595–615 CrossRef CAS PubMed.
- A. Truskewycz, H. Yin, N. Halberg, D. T. H. Lai, A. S. Ball, V. K. Truong, A. M. Rybicka and I. Cole, Small, 2022, 18, 2106342 CrossRef CAS PubMed.
- Y. He, C. Hu, Z. Li, C. Wu, Y. Zeng and C. Peng, Mater. Today Bio, 2022, 14, 100231 CrossRef CAS PubMed.
- A. Sciortino, A. Cannizzo and F. Messina, C-J. Carbon Res., 2018, 4, 67 CrossRef CAS.
- D. P. Huang, H. F. Zhou, Y. Q. Wu, T. Wang, L. L. Sun, P. Gao, Y. Z. Sun, H. N. Huang, G. J. Zhou and J. F. Hu, Carbon, 2019, 142, 673–684 CrossRef CAS.
- N. N. Shi, K. Y. Sun, Z. D. Zhang, J. Zhao, L. N. Geng and Y. H. Lei, J. Ind. Eng. Chem., 2021, 101, 372–378 CrossRef CAS.
- J. Liu, T. Jiang, C. Li, Y. Wu, M. He, J. Zhao, L. Zheng and X. Zhang, Stem Cells Transl. Med., 2019, 8, 724–736 CrossRef CAS PubMed.
- L. Yang, W. H. Jiang, L. P. Qiu, X. W. Jiang, D. Y. Zuo, D. K. Wang and L. Yang, Nanoscale, 2015, 7, 6104–6113 RSC.
- C. J. Liu, P. Zhang, X. Y. Zhai, F. Tian, W. C. Li, J. H. Yang, Y. Liu, H. B. Wang, W. Wang and W. G. Liu, Biomaterials, 2012, 33, 3604–3613 CrossRef CAS PubMed.
- P. Pierrat, R. R. Wang, D. Kereselidze, M. Lux, P. Didier, A. Kichler, F. Pons and L. Lebeau, Biomaterials, 2015, 51, 290–302 CrossRef CAS PubMed.
- S. Ross, R. S. Wu, S. C. Wie, G. M. Ross and H. T. Chang, J. Food Drug Anal., 2020, 28, 677–695 CAS.
- H. Wu, W. Su, H. M. Xu, Y. Zhang, Y. C. Li, X. H. Li and L. Z. Fan, View, 2021, 2, 20200061 CrossRef CAS.
- J. F. Wan, X. Y. Zhang, K. Fu, X. Zhang, L. Shang and Z. Q. Su, Nanoscale, 2021, 13, 17236–17253 RSC.
- M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B. Bombelli and S. Laurent, Chem. Rev., 2011, 111, 5610–5637 CrossRef CAS.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed.
- A. B. Chinen, C. X. M. Guan, C. H. Ko and C. A. Mirkin, Small, 2017, 13, 1603847 CrossRef.
- L. Ding, C. J. Yao, X. F. Yin, C. C. Li, Y. A. Huang, M. Wu, B. Wang, X. Y. Guo, Y. L. Wang and M. H. Wu, Small, 2018, 14, 1801451 CrossRef.
- Y. T. Ho, R. D. Kamm and J. C. Y. Kah, Nanoscale, 2018, 10, 12386–12397 RSC.
- N. Liu, M. Tang and J. D. Ding, Chemosphere, 2020, 245, 125624 CrossRef CAS.
- S. Juling, A. Niedzwiecka, L. Bohmert, D. Lichtenstein, S. Selve, A. Braeuning, A. F. Thunemann, E. Krause and A. Lampen, J. Proteome Res., 2017, 16, 4020–4034 CrossRef CAS PubMed.
- M. Lundqvist and T. Cedervall, Small, 2020, 16, 2000892 CrossRef CAS PubMed.
- D. Chakraborty, K. R. Ethiraj and A. Mukherjee, RSC Adv., 2020, 10, 27161–27172 RSC.
- K. Liu, A. Salvati and A. Sabirsh, Nanoscale, 2022, 14, 2136–2154 RSC.
- N. Singh, C. Marets, J. Boudon, N. Millot, L. Saviot and L. Maurizi, Nanoscale Adv., 2021, 3, 1209 RSC.
- R. Bilardo, F. Traldi, A. Vdovchenko and M. Resmini, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 1788 Search PubMed.
- W. J. Lai, Q. S. Wang, L. M. Li, Z. Y. Hu, J. K. Chen and Q. J. Fang, Colloids Surf., B, 2017, 152, 317–325 CrossRef CAS PubMed.
- J. Piella, N. G. Bastus and V. Puntes, Bioconjugate Chem., 2017, 28, 88–97 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed.
- W. Richtering, I. Alberg and R. Zentel, Small, 2020, 16, 2002162 CrossRef CAS PubMed.
- Q. H. Yu, L. X. Zhao, C. C. Guo, B. Yan and G. X. Su, Front. Bioeng. Biotechnol., 2020, 8, 210 CrossRef PubMed.
- H. J. Yan, M. Cacioppo, S. Megahed, F. Arcudi, L. Dordevic, D. C. Zhu, F. Schulz, M. Prato, W. J. Parak and N. Feliu, Nat. Commun., 2021, 12, 7208 CrossRef CAS PubMed.
- K. Abstiens, S. M. Figueroa, M. Gregoritza and A. M. Goepferich, Soft Matter, 2019, 15, 709–720 RSC.
- C. Grafe, M. von der Luhe, A. Weidner, P. Globig, J. H. Clement, S. Dutz and F. H. Schacher, Nanotechnology, 2019, 30, 265707 CrossRef PubMed.
- M. Y. Wen, Y. X. Li, W. C. Zhong, Q. F. Li, L. P. Cao, L. L. Tan and L. Shang, J. Colloid Interface Sci., 2022, 610, 116–125 CrossRef CAS PubMed.
- F. Giulimondi, L. Digiacomo, D. Pozzi, S. Palchetti, E. Vulpis, A. L. Capriotti, R. Z. Chiozzi, A. Lagana, H. Amenitsch, L. Masuelli, M. Mahmoudi, I. Screpanti, A. Zingoni and G. Caracciolo, Nat. Commun., 2019, 10, 3686 CrossRef CAS PubMed.
- D. Burnand, A. Milosevic, S. Balog, M. Spuch-Calvar, B. Rothen-Rutishauser, J. Dengjel, C. Kinnear, T. L. Moore and A. Petri-Fink, Small, 2018, 14, 1802088 CrossRef PubMed.
- Y. K. Song, H. T. Wang, L. J. Zhang, B. Lai, K. J. Liu and M. Q. Tan, Food Funct., 2020, 11, 2358–2367 RSC.
- K. J. Liu, Y. K. Song and M. Q. Tan, J. Agric. Food Chem., 2020, 68, 9789–9795 CrossRef CAS.
- Y. Y. Peng, Y. Y. Cong, Y. Z. Lei, F. W. Sun, M. H. Xu, J. Z. Zhang, L. Fang, H. Hong and T. Cai, Adv. Healthcare Mater., 2022, 11, 2102270 CrossRef CAS PubMed.
- J. J. Du, N. Xu, J. L. Fan, W. Sun and X. J. Peng, Small, 2019, 15, 1805087 CrossRef PubMed.
- M. Havrdova, K. Hola, J. Skopalik, K. Tomankova, P. A. Martin, K. Cepe, K. Polakova, J. Tucek, A. B. Bourlinos and R. Zboril, Carbon, 2016, 99, 238–248 CrossRef CAS.
- M. Weiss, J. Fan, M. Claudel, L. Lebeau, F. Pons and C. Ronzani, Nanomaterials, 2021, 11, 180 CrossRef CAS.
- J. H. Fan, M. Claudel, C. Ronzani, Y. Arezki, L. Lebeau and F. Pons, Int. J. Pharm., 2019, 569, 118521 CrossRef CAS PubMed.
- M. Weiss, J. H. Fan, M. Claudel, T. Sonntag, P. Didier, C. Ronzani, L. Lebeau and F. Pons, J. Nanobiotechnol., 2021, 19, 5 CrossRef CAS PubMed.
- M. Nakayama, Front. Immunol., 2018, 9, 103 CrossRef PubMed.
- D. L. Laskin, V. R. Sunil, C. R. Gardner and J. D. Laskin, Annu. Rev. Pharmacol. Toxicol., 2011, 51, 267–288 CrossRef CAS PubMed.
- W. Chanput, J. J. Mes and H. J. Wichers, Int. Immunopharmacol., 2014, 23, 37–45 CrossRef CAS.
- S. J. Zhu, X. H. Zhao, Y. B. Song, S. Y. Lu and B. Yang, Nano Today, 2016, 11, 128–132 CrossRef CAS.
- D. Qu and Z. C. Sun, Mater. Chem. Front., 2020, 4, 400–420 RSC.
- T. Garcia-Millan, T. A. Swift, D. J. Morgan, R. L. Harniman, B. Masheder, S. Hughes, S. A. Davis, T. A. A. Oliver and M. C. Galan, Nanoscale, 2022, 14, 6930–6940 RSC.
- F. Ehrat, S. Bhattacharyya, J. Schneider, A. Lof, R. Wyrwich, A. L. Rogach, J. K. Stolarczyk, A. S. Urban and J. Feldmann, Nano Lett., 2017, 17, 7710–7716 CrossRef CAS.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772 CrossRef CAS PubMed.
- A. Amici, G. Caracciolo, L. Digiacomo, V. Gambini, C. Marchini, M. Tilio, A. L. Capriotti, V. Colapicchioni, R. Matassa, G. Familiari, S. Palchetti, D. Pozzi, M. Mahmoudi and A. Lagana, RSC Adv., 2017, 7, 1137–1145 RSC.
- I. Machova, M. Hubalek, T. Belinova, A. Fucikova, S. Stehlik, B. Rezek and M. H. Kalbacova, Carbon, 2020, 162, 650–661 CrossRef CAS.
- X. N. Cai, R. Ramalingam, H. S. Wong, J. P. Cheng, P. Ajuh, S. H. Cheng and Y. W. Lam, Nanomed-Nanotechnol., 2013, 9, 583–593 CrossRef CAS.
- D. Y. Chen, S. Ganesh, W. M. Wang and M. Amiji, Nanoscale, 2019, 11, 8760–8775 RSC.
- K. Saha, M. Rahimi, M. Yazdani, S. T. Kim, D. F. Moyano, S. Hou, R. Das, R. Mout, F. Rezaee, M. Mahmoudi and V. M. Rotello, ACS Nano, 2016, 10, 4421–4430 CrossRef CAS PubMed.
- R. L. Pinals, D. Yang, D. J. Rosenberg, T. Chaudhary, A. R. Crothers, A. T. Iavarone, M. Hammel and M. P. Landry, Angew. Chem., Int. Ed., 2020, 59, 23668–23677 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. B. Guo, A. Emili and W. C. W. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS.
- Y. Ju, H. G. Kelly, L. F. Dagley, A. Reynaldi, T. E. Schlub, S. K. Spall, C. A. Bell, J. W. Cui, A. J. Mitchell, Z. X. Lin, A. K. Wheatley, K. J. Thurecht, M. P. Davenport, A. I. Webb, F. Caruso and S. J. Kent, ACS Nano, 2020, 14, 15723–15737 CrossRef PubMed.
- R. M. Visalakshan, L. E. G. Garcia, M. R. Benzigar, A. Ghazaryan, J. Simon, A. Mierczynska-Vasilev, T. D. Michl, A. Vinu, V. Mailander, S. Morsbach, K. Landfester and K. Vasilev, Small, 2020, 16, 2000285 CrossRef PubMed.
- K. N. Yang, B. Mesquita, P. Horvatovich and A. Salvati, Acta Biomater., 2020, 106, 314–327 CrossRef CAS PubMed.
- S. Ritz, S. Schottler, N. Kotman, G. Baier, A. Musyanovych, J. Kuharev, K. Landfester, H. Schild, O. Jahn, S. Tenzer and V. Mailander, Biomacromolecules, 2015, 16, 1311–1321 CrossRef CAS PubMed.
- H. Kim, D. Roth, Y. Isoe, K. Hayashi, C. Mochizuki, M. Kalkum and M. Nakamura, Colloids Surf., B, 2021, 199, 111527 CrossRef CAS PubMed.
- V. H. Nguyen, N. M. Meghani, H. H. Amin, T. T. D. Tran, P. H. L. Tran, C. Park and B. J. Lee, Colloids Surf., B, 2018, 170, 179–186 CrossRef CAS PubMed.
- T. Zhang, M. Tang, Y. Yao, Y. Ma and Y. P. Pu, Int. J. Nanomed., 2019, 14, 993–1009 CrossRef CAS PubMed.
- F. Alnasser, V. Castagnola, L. Boselli, M. Esquivel-Gaon, E. Efeoglu, J. McIntyre, H. J. Byrne and K. A. Dawson, Nano Lett., 2019, 19, 1260–1268 CrossRef CAS PubMed.
- X. Lu, P. P. Xu, H. M. Ding, Y. S. Yu, D. Huo and Y. Q. Ma, Nat. Commun., 2019, 10, 4520 CrossRef.
- G. Almer, K. Wernig, M. Saba-Lepek, S. Haj-Yahya, J. Rattenberger, J. Wagner, K. Gradauer, D. Frascione, G. Pabst, G. Leitinger, H. Mangge, A. Zimmer and R. Prassl, Int. J. Nanomed., 2011, 6, 1279–1290 CAS.
- M. M. Qin, J. Zhang, M. H. Li, D. Yang, D. C. Liu, S. Y. Song, J. J. Fu, H. Zhang, W. B. Dai, X. Q. Wang, Y. G. Wang, B. He and Q. Zhang, Theranostics, 2020, 10, 1213–1229 CrossRef CAS.
- H. P. Jersmann, I. Dransfield and S. P. Hart, Clin. Sci., 2003, 105, 273–278 CrossRef CAS PubMed.
- S. Nagayama, K. Ogawara, K. Minato, Y. Fukuoka, Y. Takakura, M. Hashida, K. Higaki and T. Kimura, Int. J. Pharm., 2007, 329, 192–198 CrossRef CAS PubMed.
- P. Haberzettl, R. Duffin, U. Kramer, D. Hohr, R. P. Schins, P. J. Borm and C. Albrecht, Arch. Toxicol., 2007, 81, 459–470 CrossRef CAS PubMed.
- Y. M. Kim, S. I. Chung and S. Y. Lee, Toxicol. Lett., 2005, 158, 1–9 CrossRef CAS PubMed.
- I. T. Jolliffe and J. Cadima, Philos. Trans. R. Soc., A, 2016, 374, 20150202 CrossRef PubMed.
- D. Pozzi, G. Caracciolo, L. Digiacomo, V. Colapicchioni, S. Palchetti, A. L. Capriotti, C. Cavaliere, R. Z. Chiozzi, A. Puglisi and A. Lagana, Nanoscale, 2015, 7, 13958–13966 RSC.
- G. Caracciolo, O. C. Farokhzad and M. Mahmoudi, Trends Biotechnol., 2017, 35, 257–264 CrossRef CAS PubMed.
- R. Liu, W. Jiang, C. D. Walkey, W. C. W. Chan and Y. Cohen, Nanoscale, 2015, 7, 9664–9675 RSC.
- D. Y. Chen, N. Parayath, S. Ganesh, W. M. Wang and M. Amiji, Nanoscale, 2019, 11, 18806–18824 RSC.
- D. Y. Chen, S. Ganesh, W. M. Wang, A. Lupieri and M. Amiji, Nanomedicine, 2021, 16, 535–551 CrossRef CAS.
- Y. J. Zou and N. Komatsu, Carbon, 2020, 163, 395–401 CrossRef CAS.
- F. Marano, S. Hussain, F. Rodrigues-Lima, A. Baeza-Squiban and S. Boland, Arch. Toxicol., 2011, 85, 733–741 CrossRef CAS PubMed.
- J. Cox, M. Y. Hein, C. A. Luber, I. Paron, N. Nagaraj and M. Mann, Mol. Cell. Proteomics, 2014, 13, 2513–2526 CrossRef CAS PubMed.
- C. Carapito, A. Burel, P. Guterl, A. Walter, F. Varrier, F. Bertile and A. Van Dorsselaer, Proteomics, 2014, 14, 1014–1019 CrossRef CAS PubMed.
- C. Lenz and H. Dihazi, Methods Mol. Biol., 2016, 1362, 3–27 CrossRef CAS PubMed.
- M. Collot, R. Kreder, A. L. Tatarets, L. D. Patsenker, Y. Mely and A. S. Klymchenko, Chem. Commun., 2015, 51, 17136–17139 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr03611h |
| This journal is © The Royal Society of Chemistry 2022 |