Open Access Article
Open Access ArticleA palette of background-free tame fluorescent probes for intracellular multi-color labelling in live cells†
Samira Husen
Alamudi‡
a,
Dongdong
Su‡
a,
Kyung Jin
Lee
b,
Jung Yeol
Lee
c,
José Luis
Belmonte-Vázquez
d,
Hee-Sung
Park
b,
Eduardo
Peña-Cabrera
 d and
Young-Tae
Chang
d and
Young-Tae
Chang
 *ace
*ace
aLaboratory of Bioimaging Probe Development, Singapore Bioimaging Consortium, Agency for Science, Technology and Research (A*STAR), 11 Biopolis Way, Helios #02-02, Singapore 138667
bDepartment of Chemistry, Korea Advanced Institute of Science and Technology, Republic of Korea 305701
cDepartment of Chemistry, Pohang University of Science and Technology, Pohang, Republic of Korea 37673. E-mail: ytchang@postech.ac.kr
dDepartamento de Quimica DCNE, Campus Guanajuato, Universidad de Guanajuato, Guanajuato, Mexico 36050
eCenter for Self-assembly and Complexity, Institute for Basic Science (IBS), Pohang, Republic of Korea 37673
First published on 23rd January 2018
Abstract
A multi-color labelling technique for visualizing multiple intracellular apparatuses in their native environment using small fluorescent probes remains challenging. This approach requires both orthogonal and biocompatible coupling reactions in heterogeneous biological systems with minimum fluorescence background noise. Here, we present a palette of BODIPY probes containing azide and cyclooctyne moieties for copper-free click chemistry in living cells. The probes, referred to as ‘tame probes’, are highly permeable and specific in nature, leaving no background noise in cells. Such probes, which are rationally designed through optimized lipophilicity, water solubility and charged van der Waals surface area, allow us to demonstrate rapid and efficient concurrent multi-labelling of intracellular target components. We show that these probes are capable of not only labelling organelles and engineered proteins, but also showing the intracellular glycoconjugates’ dynamics, through the use of metabolic oligosaccharide engineering technology in various cell types. The results demonstrated in this study thus provide flexibility for multi-spectral labelling strategies in native systems in a high spatiotemporal manner.
Introduction
Visualizing intracellular components in native systems is essential to understand the complex biological roles and processes. Due to the limitations of genetically encoded tags such as GFP,1,2 small fluorescent probes have now emerged as powerful tools to chemically label intracellular apparatuses and to assist with the investigation of their structural and functional properties.To label intracellular components, an orthogonal reporter is firstly directed into a specific biomolecule or cellular site, and subsequently, a fluorescent probe bearing its complementary orthogonal group is introduced to label the target through chemical ligation. Cu(I)-catalyzed alkyne–azide cycloaddition (CuAAC) has revolutionized this technique in an efficient and reliable manner.3,4 However, the major drawback of CuAAC is the toxicity of Cu(I), which causes terrible cellular damage.5,6 To overcome this issue, CuAAC has been enhanced via ligand-assisted CuAAC (such as TBTA,7 BTTPS,8 THPTA,9 BTTAA,10) and chelation-assisted CuAAC (such as 2-picolylazide11,12 and AIO-1).13 These additional ligands and chelates, however, are not particularly convenient for intracellular labelling on live samples.14,15 On the other hand, along with inverse electron-demand Diels–Alder (IEDDA),16–18 copper-free strain-promoted alkyne–azide cycloaddition (SPAAC) has successfully bypassed the toxic Cu(I) and surfaced as a preferred strategy to label target biomolecules under physiological conditions.19,20 Through a SPAAC strategy, non-permanent strained cycloalkynes were previously reported for imaging extracellular glycans,21,22 followed by another attempt to target whole proteins in cells.23
Despite the emergence of multi-spectral labelling strategies, such as fluorophore-conjugated antibodies, polymer nanoparticles, and peptide-functionalized quantum dots,24–26 a robust multi-color intracellular labelling method in mammalian systems remains a challenging task. Traditionally, this multiple-reaction strategy involves protection-deprotection steps and suffers from low coupling efficiency.27 A preferred strategy requires: (i) a reasonably small size of biorthogonal reporters, (ii) efficient cellular incorporation of multiple reporters onto the target sites, (iii) rapid and specific labelling under biocompatible conditions, and (iv) highly permeable probes without background noise from the unreacted ones.
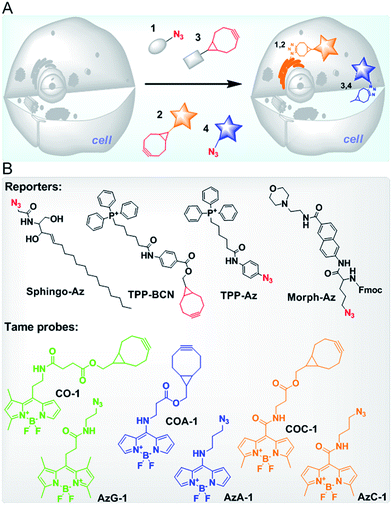
Ideally, biomolecules can be targeted anywhere in a cell, however, most probes reported so far suffer from either low permeability and poor water solubility or high background noise due to poor specificity.21–23,28–30 We previously demonstrated that the cellular behaviour of fluorescent probes is influenced by three key physicochemical properties (called molecular descriptors): lipophilicity, van der Waals charged surface area and water solubility.31 We have developed highly permeable probes with a low non-specific affinity towards intracellular organelles/biomolecules (also referred to as tame probes), namely AzG-1 and CO-1.31 Now, having advanced our understanding in fine-tuning the probe structural properties, we have sought to expand our work to provide a palette of tame probes consisting of azide and cyclooctyne functional groups (Fig. 1). Additionally, we now show that our wide range of probes could label not only intracellular organelles or engineered proteins, but also intracellular glycoconjugates in a native condition at the early stage of their biosynthetic pathway.
Results and discussion
Discovery of multi-color tame probes
To provide a wide range of spectral properties, we designed BODIPY-based fluorescent probes and modified the meso position of their core with different linkers to either shorten or lengthen the excitation and emission wavelengths.32,33 By introducing non-aromatic amines, a new type of stable meso-amino BODIPY can be constructed. The meso-amino BODIPYs exhibit a hypsochromic shift that results in a blue range emission and high laser efficiency.34,35 Likewise, by extending the π conjugation to the meso sites of the BODIPY core through generating a more push–pull structure, meso-amide BODIPYs can be constructed, and display a bathochromic shift in both the excitation and emission wavelengths.36We designed about 100 probes containing cyclooctyne and azide functional groups for SPAAC and then built their three-dimensional (3D) structures using Molecular Operating Environment (MOE CCG) software. After their 3D optimized conformations were set by a Hamiltonian AM1 energy minimization method, three key molecular descriptors, namely SlogP, logS and Q_VSA_FNEG, were calculated using the same software. By following our tame probes model guideline,31 we selected and synthesized four probes (cyclooctyne-containing probes COC-1 and COA-1, and azide-containing probes AzC-1 and AzA-1) whose descriptors are in a highly preferred range for being tamed (Table S1,†Fig. 1B). These probes with varying emission colors of blue and yellow were anticipated to behave as tame probes (Fig. S1, Table S2†). It was confirmed that the in vitro reactions between the probes and TPP-Az or TPP-BCN are rapid enough in 10 minutes of reaction (Fig. S2†). Interestingly, cellular retention and efflux studies showed that these probes are indeed highly permeable and readily washed out of the cells in the absence of reporters, in the two commonly used cell lines, U-2 OS and CHO, as predicted (Fig. S3 and S4†).
Single labelling of mitochondria in live cells
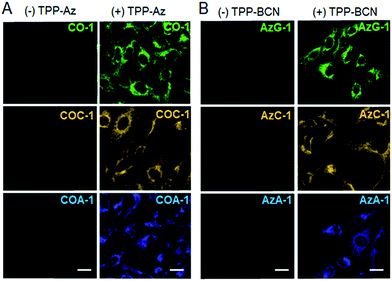
Next, we investigated the capability of the probes to label intracellular organelles in live U-2 OS. Azide- and cyclooctyne-containing triphenylphosphonium analogues which accumulate in mitochondria, TPP-Az and TPP-BCN, were synthesized as reported.31 These reporters can be ligated with tame probes through the SPAAC protocol. In vitro reactions between the reporters and the probes were clean and rapid. We treated the U-2 OS cells with either vehicle or 5 μM reporters for 20 min, followed by labelling with 2 μM respective probes for 30 min. CO-1 and AzG-1 were used as positive controls. As shown in Fig. 2A and B, the probes clearly labelled the tagged mitochondria out of a low background. However, in the absence of reporters, no probe signal was observed due to the washable property of the unreacted probes.Dual labelling using a pair of azide- and cyclooctyne-containing probes
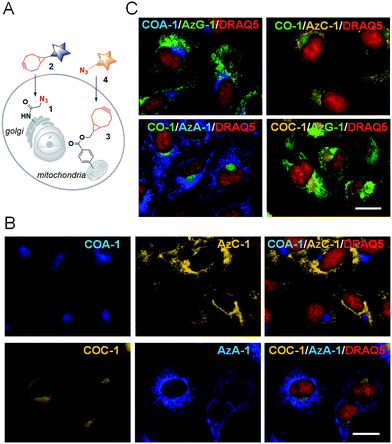
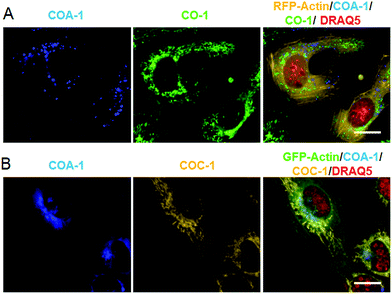
We then explored the probes’ potential for dual labelling with the examples of mitochondria and the Golgi apparatus in live U-2 OS. The Golgi apparatus is essential for the proper maturation of proteins and lipids. Most of the available Golgi targeting probes make use of this function using lipid-like molecules, such as fluorescent ceramides.37 We used the azide-bearing ceramide Sphingo-Az as a reporter for targeting the Golgi apparatus.31In vitro reactions between the Sphingo-Az and probe COC-1 or COA-1 were also found to be clean and rapid. For dual labelling, firstly U-2 OS cells pre-treated with 5 μM Sphingo-Az were labelled with either 2 μM COA-1 or 2 μM COC-1 in growth medium for 30 min at 37 °C. Afterwards, the medium was replaced with 5 μM TPP-BCN-containing growth medium for subsequent mitochondria labelling using either 2 μM AzC-1 or 2 μM AzA-1 for 30 min (Fig. 3A). Cells were then imaged after being washed with fresh medium.As can be seen in Fig. 3B, a strong signal showing the Golgi structure was observed from both the COA-1 (top panel) and COC-1 (bottom panel). As anticipated, a clear signal that shows the fine mitochondria structure was also observed from the AzC-1 (top panel) and AzA-1 (bottom panel). It is interesting to note that even though the two different classes of probe pairs (COA-1/COC-1 and AzC-1/AzA-1) can react with each other, cell images show distinguishable patterns from these two classes of probes. This indicates efficient labelling of the first organelle, which leaves minimal room to interfere with the second organelle.
Dual labelling using a pair of both cyclooctyne-containing probes
Following the results, we further examined whether dual labelling using the same class of probes is feasible. We opted to use only the cyclooctyne probes (COA-1 and COC-1) to label both organelles.We initially examined the probes’ labelling efficiency by sequentially incubating TPP-Az-treated cells with various concentrations of the first probe (COA-1) while keeping the same concentration of the second one (COC-1). We found that the labelling of mitochondria with TPP-Az (optimum cellular incorporation is at 5 μM, Fig. S5†) was efficiently accomplished at 2 μM for the first probe, and consequently, this leaves minimal room to interfere with the second probe (Fig. S6†).
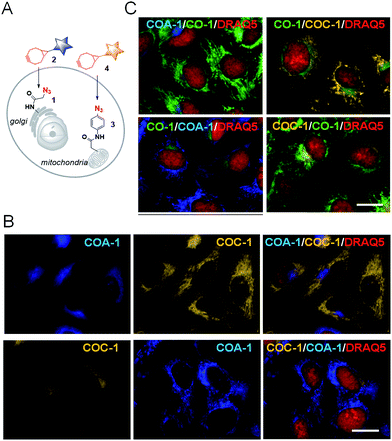
From this observation, we then performed dual labelling of Golgi apparatus and mitochondria with COA-1 and COC-1 (Fig. 4A). We firstly labelled Golgi apparatus by incubating the Sphingo-Az-treated cells with 2 μM COC-1 or COA-1 for 30 min. Then, the subsequent mitochondria labelling was done by replacing the medium with 5 μM TPP-Az-containing medium followed by 2 μM COA-1 or COC-1 for 30 min. The cells were washed to remove the unreacted probes and then imaged. Cell images in Fig. 4B show the unambiguous pattern of the Golgi apparatus from both COA-1 (top panel) and COC-1 (bottom panel). Excitingly, a fine structure of the mitochondria was also observed from subsequent labelling with COC-1 (top panel) and COA-1 (bottom panel) without background noise. Moreover, the labelling can be done in the native condition without permeabilization and/or fixation of the cells. We then successfully explored this specific intracellular dual labelling by means of different combinations of: (1) probes’ colors (blue, green and yellow) and (2) probes’ classes (azide/cyclooctyne pair and cyclooctyne/cyclooctyne pair) (Fig. 3C and 4C).
Dual labelling in FP-tagged overexpressing cell lines
We then investigated whether the dual labelling interferes with the overexpression of genetically coded recombinant proteins. Red and green fluorescent protein (RFP and GFP) were fused to actin. After the proteins were expressed in live U-2 OS, the cells were sequentially labelled with either lysosome/mitochondria or Golgi apparatus/mitochondria reporters. In RFP-tagged actin expressing cells (Fig. 5A), U-2 OS pre-treated with 5 μM Morph-Az31 (a morpholine-azide derivative which accumulates in lysosomes) for 1 h were firstly labelled with COA-1 (2 μM, 30 min). Afterwards, the medium was replaced with TPP-Az-containing medium (5 μM, 20 min) for subsequent mitochondria labelling using CO-1 (2 μM, 30 min).Similarly, in GFP-tagged actin expressing cells (Fig. 5B), Sphingo-Az (5 μM) pre-treated cells were firstly labelled with COA-1 (2 μM, 30 min) followed by mitochondria labelling using the TPP-Az (5 μM, 20 min) and COC-1 (2 μM, 30 min) pair. Clear organelle labelling in both RFP- and GFP-actin expressing cells was observed without interfering with the fused-protein signals. This signifies a distinct spatial labelling with a flexibility of color choices.
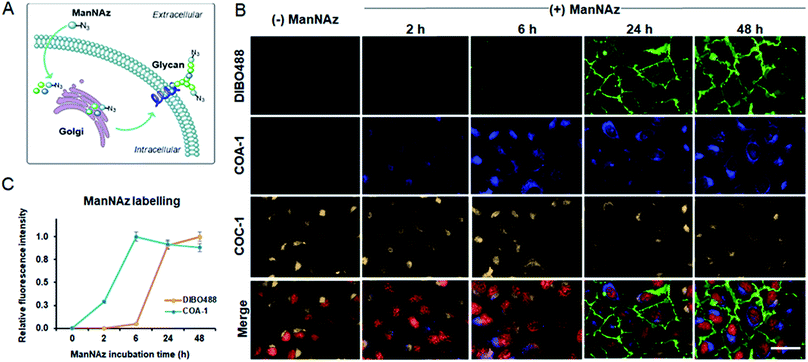
Imaging extra- and intracellular glycoconjugates in live cells
We previously showed that tame probes are capable of specifically labelling engineered intracellular proteins using in vivo unnatural amino acid technology.31 We now further challenge the probes to label intracellular azide-containing glycoconjugates through metabolic oligosaccharide engineering technology. Tetraacetylated N-azidoacetyl-D-mannosamine (ManNAz), an azide-functionalized sugar, can be metabolized by glycosylation machinery through sialic acid biosynthetic pathways for N-linked glycosylation to produce azide-containing sialic acid (Fig. S7A†).38,39 The pathways involve the transfer of newly synthesized tagged glycoconjugates from the lumen of Golgi compartments to finally incorporate into cell surface glycans (Fig. 6A). Typically, the imaging of these glycoconjugates has only been exploited after they are expressed on the cell surface (24–72 h post-incubation). Here, we show that tame probes could image intracellular tagged glycoconjugates at the early stage of their endogenous biosynthetic pathway.Live U-2 OS were treated with vehicle and ManNAz at the different time points of 2, 6, 24 and 48 h. Afterwards, the cells were labelled with 5 μM COA-1 and 15 μM dibenzocyclooctyne-modified DIBO488 (Alexa Fluor 488 DIBO Alkyne, Fig. S7B†) for 1 h. After cells were washed with fresh media and imaged, no fluorescent signals were observed in vehicle-treated cells from either probe as shown in Fig. 6B. This is due to the poor permeability of DIBO488 which hardly crosses cell membranes, and the background-free property of highly permeable COA-1 which can be readily washed out of cells in the absence of its reporters. A strong DIBO488 signal started to show from the extracellular membrane of cells treated with ManNAz after at least 24 h (first row). On the contrary, COA-1 labels intracellular glycoconjugates on cells incubated with ManNAz after as little as 2 h (second row). COA-1 signals were strongest at 6 h post-incubation with ManNAz, while DIBO488 signals were strongest at 48 h (Fig. 6C).
Additionally, to show that triple labelling is feasible with this technology, we further co-labelled the ManNAz-tagged U-2 OS with a pair of Golgi reporters Sphingo-Az and COC-1. We found that the COA-1 and COC-1 signals were co-localized, suggesting concurrent multi-color labelling of the intracellular ceramide and glycoconjugate derivatives in the Golgi apparatus (Fig. 6B, third and last row).
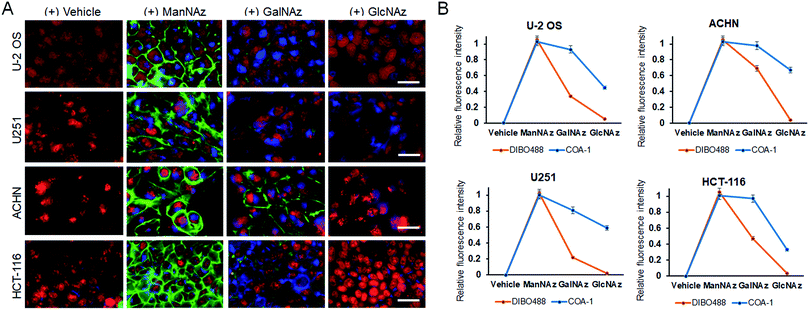
Delighted with the results, we then explored the system to image the dynamics of the metabolic incorporation of different sugar derivatives in various cell lines. We anticipated that our probes could image not only tagged sialic acid, but also tagged O-linked glycans. The strategy involves treating cells with tetraacetylated N-azidoacetylgalactosamine (GalNAz)40,41 and tetraacetylated N-azidoacetylglucosamine (GlcNAz)42,43 (Fig. S6C and D†) which can be metabolically incorporated into O-linked cellular glycoconjugates (O-GlcNAc attachment). Live U-2 OS, U251, ACHN, and HCT-116 cell lines were treated with vehicle and the three 50 μM azide sugars (ManNAz, GalNAz and GlcNAz) for 48 h. Afterwards, the cells were co-labelled with both 5 μM COA-1 and 15 μM DIBO488 for 1 h, washed to remove unreacted probes, and then imaged with the Operetta High-Content Imaging System.
As can be seen in Fig. 7A, first panel, negligible fluorescent background was observed from all cells treated with vehicle. On the other hand, cells incubated with ManNAz showed high levels of labelled sialic acid as characterized by strong fluorescent signals from both extracellular (green) and intracellular glycoconjugates (blue) (Fig. 7A, second panel).
O-Glycosylation can be found in the Golgi apparatus, cytoplasm and extracellular matrix to regulate fundamental cellular processes such as transcription and cell signalling. Interestingly, although both GalNAz and GlcNAz are incorporated into O-linked glycoconjugates, they show different labelling patterns. GalNAz is installed on a predominant form of O-linked glycoconjugate, which is the mucin-type, and its labelling efficiency seems to be greatly dependent on cell type. In Fig. 7A, the third panel shows that DIBO488 signals indicated that ACHN cells showed significantly higher levels of cell surface GalNAz-tagged glycoconjugates compared to other cell lines, while concurrently, COA-1 enables their detection specifically in the complex intracellular compartment (Fig. 7B). Individual images can be also seen in Fig. S8–S11.†
We observed that glycoconjugates from cells incubated with GlcNAz showed notably weaker signals, both extra- and intracellular, than cells treated with GalNAz (Fig. 7A, last panel). This observation is in agreement with previous study that suggested an inefficient step in the GlcNAc salvage pathway of UDP-GlcNAz biosynthesis, in addition to a more efficient conversion and incorporation of GalNAz to the O-GlcNAcylated conjugates,44 thus explaining weaker labelling by GlcNAz than GalNAz.
Conclusions
With growing interest in the development of improved techniques for visualizing multiple intracellular components in living systems, we demonstrate the wide-ranging utility of tame probes for concurrent multi-color labelling of various intracellular biomolecules. Our rational approach through optimized molecular properties yielded a palette of tame probes for SPAAC labelling in living cells; more importantly, we demonstrate that tame probes could label not only organelles and proteins but also intracellular sugar derivatives through metabolic oligosaccharide engineering.These sugar derivatives are tolerated by enzymes in their endogenous metabolic pathway;44 however, their metabolic labelling efficiency is thought to be highly cell type specific.45,46 We exploited azide-containing sugars as a functional group to allow specific labelling of different glycan populations by tame probes in various cell types, which is a valuable tool for future applications in studying the dynamics of posttranslational modifications.
In summary, our palette of tame probes offers multi-color labelling of intracellular apparatuses in living mammalian cells for copper-free click ligation. The ligation is rapid with minimal fluorescence background and is highly specific, as shown by high co-localization with respective organelle trackers (Fig. S12 and S13†). Our strategy has the advantage of providing a concurrent technique for robust and efficient labelling, allowing the flexibility to label virtually any intracellular apparatus in a spatiotemporal manner. Moreover, our probes did not interfere with cell proliferation (Fig. S14 to S19†) and had excellent photostability in aqueous solution (Fig. S20†). These properties together further demonstrate the high biocompatibility of this system for extensive applications in exploring biological dynamic processes in the native cellular environment.
Experimental section
Molecular modelling and molecular descriptor calculation
Three-dimensional structures of the probes were built using the Chemical Computing Group (CCG) Molecular Operating Environment (MOE) 2011 software. Their active conformation and energy minimization were set using the same software via a semi-empirical Hamiltonian AM1 method. Hydrogens and lone pair electrons were adjusted as required. Partial charges were set and the protonation state was corrected for each structure. Three descriptors (SlogP, logS and Q_VSA_FNEG) were calculated for each of the probes as reported.31Cell maintenance and preparation
The reagent DRAQ5 Fluorescent Probe (in DMSO) was purchased from Life Technologies (USA). 10 mM stock solutions of CO-1, COC-1, COA-1, AzG-1, AzC-1, AzA-1, TPP-Az, and TPP-BCN were first prepared in DMSO. The fluorescence excitation and emission spectra were measured using a Spectra Max M2 plate reader (Molecular Devices Corp, USA). U-2 OS cells were cultured at 37 °C with 5% CO2 in DMEM (Dulbecco’s modified eagle’s medium from Invitrogen, CA, USA) supplemented with fetal bovine serum (10%) and penicillin–streptomycin (1%). Cells were passaged twice a week into 10 cm cell culture dishes.Cellular retention test
Two cell lines were used to study the cellular retention of the probes: U-2 OS, a human osteosarcoma cell line, and CHO, a Chinese hamster ovary cell line, (from ATCC). The cells were cultured in DMEM (Dulbecco’s modified eagle’s medium from Invitrogen, CA, USA) supplemented with fetal bovine serum (10%) and penicillin–streptomycin (1%). Materials used in the cell culture were purchased from Invitrogen. These two cell lines were seeded onto a 96-well plate in growth medium at 37 °C and 5% CO2, then were allowed to grow to 70–80% confluence. During the experiment, the cells were incubated with 200 μL growth medium containing probes of 2 μM final concentration for 30 minutes at 37 °C and were imaged with the Operetta High-Content Imaging System (PerkinElmer) with a 10× objective lens (the before washing image). Afterwards, the cells were washed with fresh growth medium for 3 × 5 min at 37 °C and imaged again using the same imaging system (the after washing image). The images were acquired using appropriate filter sets according to the probes’ excitation and emission wavelength. Harmony High Content Imaging and Analysis Software was used to analyze the fluorescence intensity of the before washing and after washing images.Single labelling of mitochondria in live cells
U-2 OS cells were treated with TPP-Az or TPP-BCN (mitochondria reporters for COA-1/CO-1/COC-1 and AzA-1/AzG-1/AzC-1, respectively) at 5 μM concentration in DMEM growth medium for 20 min at 37 °C. After incubation, cells were washed one time with fresh medium, had 2 μM tame probes in DMEM growth medium added, and were incubated at 37 °C. Following incubation, the cells were counterstained with nuclear dye DRAQ5 Fluorescent Probe (1 μM final concentration). After 30 min, the cells were washed for 3 × 5 min with growth medium and imaged using the Operetta High-Content Imaging System (PerkinElmer) with a 40× objective lens.Single labelling of Golgi apparatus in live cells
U-2 OS cells were treated with Sphingo-Az (Golgi reporter) at 5 μM concentration in DMEM growth medium for 30 min at 4 °C. After incubation, the cells were washed once with fresh ice-cold growth medium and further incubated at 37 °C in the incubator chamber. After 30 min, the cells were washed again once and had 2 μM tame probes (COA-1, CO-1 or COC-1) in DMEM growth medium added, and were incubated at 37 °C. After 30 min, cells were washed for 3 × 5 min with growth medium and imaged using the Operetta High-Content Imaging System (PerkinElmer) with a 40× objective lens. Golgi labelling using commercial probe BODIPY TR Ceramide (Thermo Fisher Scientific) was done according to the manufacturer’s protocol.Dual labelling of Golgi apparatus and mitochondria in live cells
U-2 OS cells were treated with Sphingo-Az at 5 μM concentration in DMEM growth medium for 30 min at 4 °C. Afterwards, cells were washed once with fresh ice-cold growth medium and further incubated at 37 °C in the incubator chamber. After 30 min, the cells were washed again once and had 2 μM tame probes (COA-1, CO-1 or COC-1) in DMEM growth medium added, and were incubated at 37 °C. After 30 min, the cells were washed once and had TPP-Az or TPP-BCN (mitochondria reporters for tame probes COA-1/CO-1/COC-1 and AzA-1/AzG-1/AzC-1, respectively) at 5 μM concentration in DMEM growth medium added, for 20 min at 37 °C. Afterwards, the cells were washed once and had 2 μM tame probes (COA-1/CO-1/COC-1 or AzA-1/AzG-1/AzC-1) in DMEM growth medium added, and were incubated at 37 °C. Following incubation, the cells were counterstained with nuclear dye DRAQ5 Fluorescent Probe (1 μM final concentration). After 30 min, cells were washed for 3 × 5 min with fresh growth medium and imaged using the Operetta High-Content Imaging System (PerkinElmer) with a 40× objective lens.Dual labelling in FP-tagged actin expressing cell lines
pTAG-RFP-Actin was purchased from Evrogen and pAcGFP-Actin was purchased from Addgene. U-2 OS cells were transfected with pTAG-RFP-Actin and pAcGFP-Actin using Lipofectamine3000 transfection reagent (ThermoFisher) according to the manufacturer’s protocol. After RFP- or GFP-tagged actin was expressed in live U-2 OS (24 h), the cells were treated with 5 μM Morph-Az or Sphingo-Az for 1 h followed by 2 μM COA-1 for 30 min. After washing, cells were treated with 5 μM TPP-Az-containing growth medium for subsequent mitochondria labelling using 2 μM CO-1 or COC-1 for 30 min, followed by counterstaining with nuclear tracker, DRAQ5 (red). Cells were imaged using the Operetta High-Content Imaging System (PerkinElmer) with a 40× objective lens.Multi labelling of extracellular and intracellular tagged-glycoconjugates
Sugar ManNAz, GalNAz, GlcNAz and Alexa Fluor 488 DIBO Alkyne (DIBO488) were purchased from ThermoFisher. Live cells were treated with 50 μM sugars at either 2, 6, 24 and 48 hours. Afterwards, cells were washed and were labelled with 15 μM DIBO488 and 5 μM COA-1 for 1 h at 37 °C, followed by counterstaining with nuclear tracker, DRAQ5. Cells were imaged using the Operetta High-Content Imaging System (PerkinElmer) with a 40× objective lens.Conflicts of interest
Y.-T. C., S. H. A. and D. S. are the inventors of COC-1, COA-1, CO-1, AzC-1, AzA-1 and AzG-1 (PCT Int. Appl. WO 2017/078623 A1, 2017).Acknowledgements
The research was supported by intramural funding from the A*STAR Biomedical Research Council and the National Medical Research Council grant NMRC/TCR/016-NNI/2016. E. P.-C. wishes to thank Conacyt (grant No. 253623) for financial support.Notes and references
- D. Marguet, E. T. Spiliotis, T. Pentcheva, M. Lebowitz, J. Schneck and M. Edidin, Immunity, 1999, 11, 231 CrossRef CAS PubMed.
- C. S. Lisenbee, S. K. Karnik and R. N. Trelease, Traffic, 2003, 4, 491 CrossRef CAS PubMed.
- F. Himo, T. Lovell, R. Hilgraf, V. Rostovtsev, L. Noodleman, K. B. Sharpless and V. V. Fokin, J. Am. Chem. Soc., 2005, 127, 210 CrossRef CAS PubMed.
- V. O. Rodionov, S. I. Presolski, D. D. Diaz, V. V. Fokin and M. G. Finn, J. Am. Chem. Soc., 2007, 129, 12705 CrossRef CAS PubMed.
- D. C. Kennedy, C. S. McKay, M. C. Legault, D. C. Danielson, J. A. Blake, A. F. Pegoraro, A. Stolow, Z. Mester and J. P. Pezacki, J. Am. Chem. Soc., 2011, 133, 17993 CrossRef CAS PubMed.
- E. V. Soares, K. Hebbelinck and H. M. Soares, Can. J. Microbiol., 2003, 49, 336 CrossRef CAS PubMed.
- Q. Wang, T. R Chan, R. Hilgraf, V. V. Fokin, K. B. Sharpless and M. G. Finn, J. Am. Chem. Soc., 2003, 125, 3192 CrossRef CAS PubMed.
- W. Wang, S. Hong, A. Tran, H. Jiang, R. Triano, Y. Liu, X. Chen and P. Wu, Chem.–Asian J., 2011, 6, 2796 CrossRef CAS PubMed.
- V. Hong, S. I. Presolski, C. Ma and M. G. Finn, Angew. Chem., Int. Ed., 2009, 48, 9879 CrossRef CAS PubMed.
- C. Besanceney-Webler, H. Jiang, T. Zheng, L. Feng, D. Soriano del Amo, W. Wang, L. M. Klivansky, F. L. Marlow, Y. Liu and P. Wu, Angew. Chem., Int. Ed., 2011, 50, 8051 CrossRef CAS PubMed.
- W. S. Brotherton, H. A. Michaels, J. T. Simmons, R. J. Clark, N. S. Dalal and L. Zhu, Org. Lett., 2009, 11, 4954 CrossRef CAS PubMed.
- G.-C. Kuang, H. A. Michaels, J. T. Simmons, R. J. Clark and L. Zhu, J. Org. Chem., 2010, 75, 6540 CrossRef CAS PubMed.
- Y. Su, L. Li, H. Wang, X. Wang and Z. Zhang, Chem. Commun., 2016, 52, 2185 RSC.
- W. G. Lewis, F. G. Magallon, V. V. Fokin and M. G. Finn, J. Am. Chem. Soc., 2004, 126, 9152 CrossRef CAS PubMed.
- C. Besanceney-Webler, H. Jiang, T. Zheng, L. Feng, D. Soriano del Amo, W. Wang, L. M. Klivansky, F. L. Marlow, Y. Liu and P. Wu, Angew. Chem., Int. Ed., 2011, 50, 8051 CrossRef CAS PubMed.
- M. L. Blackman, M. Royzen and J. M. Fox, J. Am. Chem. Soc., 2008, 130, 13518 CrossRef CAS PubMed.
- N. K. Devaraj, R. Weissleder and S. A. Hilderbrand, Bioconjugate Chem., 2008, 19, 2297 CrossRef CAS PubMed.
- J. Yang, J. Seckute, C. M. Cole and N. K. Devaraj, Angew. Chem., Int. Ed., 2012, 51, 7476 CrossRef CAS PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046 CrossRef CAS PubMed.
- N. J. Agard, J. M. Baskin, J. A. Prescher, A. Lo and C. R. Bertozzi, ACS Chem. Biol., 2006, 1, 644 CrossRef CAS PubMed.
- J. C. Jewett, E. M. Sletten and C. R. Bertozzi, J. Am. Chem. Soc., 2010, 132, 3688 CrossRef CAS PubMed.
- S. T. Laughlin, J. M. Baskin, S. L. Amacher and C. R. Bertozzi, Science, 2008, 320, 664 CrossRef CAS PubMed.
- K. E. Beatty, J. Szychowski, J. D. Fisk and D. A. Tirrel, ChemBioChem, 2011, 12, 2137 CrossRef CAS PubMed.
- J. B. Delehanty, C. E. Bradburne, K. Susumu, K. Boeneman, B. C. Mei, D. Farrell, J. B. Blanco-Canosa, P. E. Dawson, H. Mattoussi and I. L. Medintz, J. Am. Chem. Soc., 2011, 133, 10482 CrossRef CAS PubMed.
- M. R. Karver, R. Weissleder and S. A. Hilderbrand, Angew. Chem., Int. Ed., 2012, 51, 920 CrossRef CAS PubMed.
- J. Pecher, J. Huber, M. Winterhalder, A. Zumbusch and S. Mecking, Biomacromolecules, 2010, 11, 2776 CrossRef CAS PubMed.
- H. Zong, S. N. Goonewardena, H.-N. Chang, J. B. Otis and J. R. Baker Jr, Bioorg. Med. Chem., 2014, 22, 6288 CrossRef CAS PubMed.
- M. King and A. Wagner, Bioconjugate Chem., 2014, 25, 825 CrossRef CAS PubMed.
- M. F. Debets, J. C. van Hest and F. P. Rutjes, Org. Biomol. Chem., 2013, 11, 6439 CAS.
- I. Nikic, T. Plass, O. Schraidt, J. Szymanski, J. A. G. Briggs, C. Schultz and E. A. Lemke, Angew. Chem., Int. Ed., 2014, 53, 2245 CrossRef CAS PubMed.
- S. H. Alamudi, R. Satapathy, J. Kim, D. Su, H. Ren, R. Das, L. Hu, A. Alvarado-Martınez, J. Y. Lee, C. Hoppmann, E. Pena-Cabrera, H.-H. Ha, H.-S. Park, L. Wang and Y.-T. Chang, Nat. Commun., 2016, 11, 11964 CrossRef PubMed.
- S. C. Dodani, D. W. Domaille, C. I. Nam, E. W. Miller, L. A. Finney, S. Vogt and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5980 CrossRef CAS PubMed.
- S. C. Dodani, S. C. Leary, P. A. Cobine, D. R. Winge and C. J. Chang, J. Am. Chem. Soc., 2011, 133, 8606 CrossRef CAS PubMed.
- R. I. Roacho, A. Metta-Magana, M. M. Portillo, E. Pena-Cabrera and K. H. Pannell, J. Org. Chem., 2013, 78, 4245–4250 CrossRef CAS PubMed.
- C. A. Osorio-Martinez, A. Urias-Benavides, C. F. Gomez-Duran, J. Banuelos, I. Esnal, I. Lopez Arbeloa and E. Pena-Cabrera, Org. Chem., 2012, 77, 5434 CrossRef CAS PubMed.
- Y. Ni, L. Zeng, N. Y. Kang, K. W. Huang, L. Wang, Z. Zeng, Y.-T. Chang and J. Wu, Chem.–Eur. J., 2014, 20, 2301 CrossRef CAS PubMed.
- U. Pütz and G. Schwarzmann, Eur. J. Cell Biol., 1995, 68, 113 Search PubMed.
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007 CrossRef CAS PubMed.
- E. Saxon, S. J. Luchansky, H. C. Hang, C. Yu, S. C. Lee and C. R. Bertozzi, J. Am. Chem. Soc., 2002, 124, 14893 CrossRef CAS PubMed.
- D. H. Dube, J. A. Prescher, C. N. Quang and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4819 CrossRef CAS PubMed.
- H. C. Hang, C. Yu, D. L. Kato and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14846 CrossRef CAS PubMed.
- D. J. Vocadlo, H. C. Hang, E. J. Kim, J. A. Hanover and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9116 CrossRef CAS PubMed.
- R. Sprung, A. Nandi, Y. Chen, S. C. Kim, D. Barma, J. R. Falck and Y. Zhao, J. Proteome Res., 2005, 4, 950 CrossRef CAS PubMed.
- M. Boyce, I. S. Carrico, A. S. Ganguli, S. H. Yu, M. J. Hangauer, S. C. Hubbard, J. J. Kohler and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3141 CrossRef CAS PubMed.
- S. T. Laughlin and C. R. Bertozzi, Nat. Protoc., 2007, 2, 2930 CrossRef CAS PubMed.
- S. J. Luchansky, S. Argade, B. K. Hayes and C. R. Bertozzi, Biochemistry, 2004, 43, 12358 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, synthetic procedures, characterization data of compounds and supporting imaging. See DOI: 10.1039/c7sc04716a |
| ‡ These authors have contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |