Nanomaterial-modified electrochemical aptasensors for tetracycline detection: a review
Jingjing
Huo
,
Rui
Guo
,
Juanjuan
Yin
,
Yanran
Liu
,
Ying
Zhang
,
Fangyi
Ruan
 ,
Yonghong
Shi
* and
Yang
Li
,
Yonghong
Shi
* and
Yang
Li
 *
*
Xi'an Rare Metal Materials Institute Co, Ltd., Northwest Institute for Non-ferrous Metal Research, Xi'an 710016, China. E-mail: shiyh_91@163.com; Yang_Li1990@hotmail.com
First published on 6th May 2025
Abstract
Excessive residues of tetracyclines in the livestock, food products and environment can lead to their accumulation in the human body through the food chain, unavoidably posing a threat to the human health. Therefore, it is essential to establish detection methods with high specificity, stability, and sensitivity. Among the numerous detecting techniques, electrochemical sensors with aptamers working as biorecognition elements have been increasingly applied to monitor tetracyclines. Notably, the synergy of a wide range of nanomaterials with aptamer-based sensors has improved the charge transfer efficiency and signal sensitivity. In this review, the advantages of aptamer-based recognition methods are discussed, and the measuring processes of electrochemical detection are introduced. Then, advances in electrochemical aptasensors used for detecting tetracyclines are summarized with an emphasis on the role of nanomaterials, such as carbon-based nanomaterials and gold-based nanomaterials, functioning as -transducing media and electrically conductive polymers. Finally, the current challenges and emerging trends in this field are also discussed, shedding light on the prospects for developing new aptasensors for tetracycline detection.
1. Introduction
Tetracyclines (TCs) are a group of broad-spectrum antibiotics that could fight infectious disease by penetrating bacterial cells and inhibiting protein synthesis.1,2 The extensive use of TCs as antibiotics or growth promoters include applications in human and veterinary medicines and aquaculture.3,4 However, the abuse and overuse of TCs can eventually lead to their accumulation in the human body through the food chain, resulting in negative effects, such as teeth discoloration, enamel hypoplasia, esophagus damage, and hearing loss.5 Hence, efficient analysis techniques for the detection of TCs in food products, pharmaceutical preparations, and water are needed. Currently, various methods have been developed to quantitatively detect TCs, such as chromatography, including liquid chromatography-mass spectrometry and high-performance liquid chromatography; capillary electrophoresis; enzyme-linked immunosorbent assays; and colorimetry.6 The combination of these methods could yield excellent analysis accuracy and sensitivity.7,8 However, various drawbacks of these methods, such as complex preparation procedures, time-consuming processes, high equipment costs, and the necessity for well-trained technicians, have restricted their widespread applications. Therefore, it is crucial to build a rapid, simple, accurate, and sensitive method for TC detection.Currently, biosensor-based analysis methods are gaining significant research attention.9–11 Biosensors typically consist of two basic components: a recognition element (biochemical receptors like nucleic acids, antibodies, peptides, or enzymes) and a signal transducer (like fluorescence, electrochemical signal, or chemiluminescence).12,13 Aptamers are short, single-stranded nucleic acid sequences isolated through in vitro SELEX processes (systematic evolution of ligands by exponential enrichment).14,15 The small RNA/DNA molecules are capable of specifically binding to diverse target molecules via their three-dimensional structure.16 Compared with traditional antibody recognition, aptamers feature the advantages of small physical sizes, flexible structures, quick chemical production, versatile chemical modification, and high stabilities.17–19 Biosensors that utilize aptamers as biorecognition elements are named as aptasensors. Among the different transduction types, electrochemical platforms integrating a biorecognition element could produce electrical signals, such as current and potential, through affinity recognition with the target analyte, leading to its quantitative detection.16 Besides desirable sensitivity, simplicity, low-cost, and robustness, electrochemical biosensors can be particularly miniaturized and integrated into portable testing devices.20–22 Thus, in recent years, aptasensor-based electrochemical detection has been increasingly applied in the detection of TC antibiotics.
Despite the above-mentioned advantages of aptasensors, major challenges still exist for their advanced utilization.23 First, the limited loading number of aptamers and potential inactivation might compromise the recognition efficiency of target molecules, reducing the specificity and accuracy of biosensors.24,25 Besides, physical adsorption and embedding are used for introducing recognition element owing to the lack of functional groups on the electrode surface. However, this unstable fixation faces unavoidable aptamer shedding from the surface. Meanwhile, ideal biosensors should have a low limit of detection (LOD) and excellent sensitivity, for which higher electrical conductivity of the electrodes are required. To address these issues and achieve increased electrochemical sensing performance, electrode surfaces modified with advanced nanomaterials are proposed.26,27 For one thing, the selected biocompatible nanomaterials usually feature a large surface-to-volume ratio, which could enhance the loading capacity of aptamers and help increase the recognition efficiency.28 For another, the aptamer could be covalently bridged onto the sensor's surface through the functional groups of nanomaterials such as amine groups and carboxyl groups, avoiding shedding and promoting sensing efficiency. Moreover, by coating electrically conductive nanomaterials on the electrode, the peak current changes could be amplified and the sensitivity could be significantly improved.29
Currently, various classes of nanomaterials have been functionalized onto the electrode surface, and serve as a multifunctional biosensing coating of the aptasensor.30,31 Among them, the most widely used and studied electrode materials are carbon and gold-based nanomaterials for TC detection.32,33 In addition, polymer-based modification layers have been reported, as shown in Scheme 1. These materials have the advantages of large surface areas, good biocompatibility, high surface reactivity, and strong adsorption capabilities.34 When immobilized on the biosensing surface, they help improve the stability, sensitivity, and signal-to-noise ratios of the constructed TC apatasensors. Therefore, the advances in nanomaterial-immobilized aptasensor electrodes in TC detection are summarized in this review. Herein, based on representative classification of carbon-based nanomaterials, gold-based nanomaterials and polymers, we systematically discuss the nanomaterial-immobilized electrode platform applied in aptasensors for TC detection. The focus is on how these platforms are properly designed to demonstrate preferable sensor sensitivity and efficiency. Moreover, we conclude with the future challenges faced by electrochemical aptasensors in TC detection and discuss the future prospects in this field. This review aims to provide a better understanding of immobilization strategies applied to the electrodes of TC aptasensors, to motivate further innovations and widen their applications in health diagnostics, food safety and environmental monitoring.
2. TC residues
TCs are a class of antibiotics that could work against a variety of bacterial infections, including Gram-positive and Gram-negative bacteria, atypical organisms such as chlamydiae, mycoplasma bacteria, rickettsiae, and protozoan parasites.35 By targeting the ribosomal machinery within the bacteria that assembles proteins from amino acids, TCs interfere with the ability to produce certain vital proteins required for bacterial growth. Due to this mode of action, TCs inhibit bacterial growth rather than killing them.36 In some countries, TCs are added at subtherapeutic levels into animal feeds and function as growth promoters. Since its discovery in the 1940s, the evolution of tetracycline antibiotics includes the development of first-generation compounds like chlortetracycline, followed by second-generation derivatives such as minocycline and doxycycline, which offer improved pharmacokinetics. Later, in response to emerging bacterial resistance challenges, ongoing optimization was made to the third-generation of tetracyclines, including tigecycline e and omadacycline.37Table 1 summarizes selected TCs and their molecular formula, molecular weight, chemical structure and other features.Nevertheless, the extensive employment of TCs in health care, livestock husbandry, aquaculture, etc. is becoming increasingly controversial out of safety concerns.37,38 Tetracycline residues in food products can be taken up by humans through the food chain and pose significant toxicological risks to health.39 It has been demonstrated that chronic exposure to residual TCs is associated with hepatotoxicity, nephrotoxicity, and gastrointestinal disturbances.40 Additionally, prolonged or excessive exposure may cause photosensitivity, dental discoloration, and skeletal abnormalities, particularly during the last half of pregnancy, infancy, and childhood.41 The accumulation of TC residues can also contribute to the development of antibiotic-resistant bacterial strains, complicating treatment of infections. These side effects highlight the importance of strict monitoring and regulation of tetracycline residue levels to ensure consumer safety and mitigate long-term health risks associated with exposure.
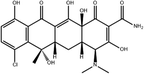
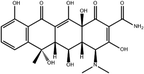
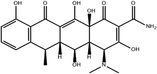
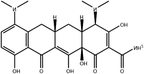
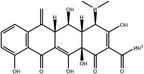
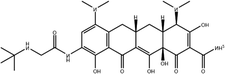
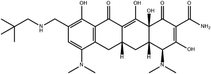
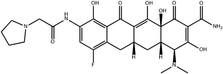
To safeguard human health, regulatory agencies such as the European Commission (EC), the U.S. Food and Drug Administration (FDA), and the Codex Alimentarius Commission (CAC) have established maximum residue limits (MRLs) for TCs in food products to ensure consumer safety. The EC has determined MRLs of each TC derivative at 100 μg kg−1, 300 μg kg−1 and 600 μg kg−1 for the muscle, liver and kidney, respectively for all food-producing species. The MRL of 100 μg L−1 for each TC in milk has been prescribed by the CAC. The FDA has established an acceptable daily intake for OTC, TC and CTC as 25 μg per kg bw per day.42,43 Undoubtedly, monitoring and detection of the antibiotic residues are essential for assessing human health risk and maintaining public safety.
3. Electrochemical aptasensor
In early 1990, two separate groups isolated a class of RNA sequences, which can bind specifically to target molecules by folding into three-dimensional structures.44,45 These RNA oligonucleotides were termed as aptamers. Later, DNA-based aptamers were also found. These functional oligonucleotides were then termed as aptamers. Since then, aptamer technology has been extended to a broad range of targets, such as proteins, amino acids, drugs, metal ions and even cells, attracting considerable interests from both the scientific and industrial sectors.46 Compared with protein-based biosensor recognition elements of peptides, antibodies or enzymes, aptamers feature higher chemical stability, greater design flexibility and low-cost. Moreover, once selected, they can be facilely produced in large quantities with desirable purity.47 Aptamers that have been developed for the detection of TCs and residues are listed in Table 2.| Target | Aptamer sequences | kD (nM) | Size (mer) | Type | Ref. |
|---|---|---|---|---|---|
| kD refers to the dissociation constant, the binding affinity of the aptamer against its receptor; — refers to not given in the references. | |||||
| TC | 5′-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCGTGGATCCGAGCTCCACGTG-3′ | 63.6 | 76 | DNA | 48 |
| OTC | 5′-CGTACGGAATTCGCTAGCCGAGTTGAGCCGGGCGCGGTACGGGTACTGGTATGTGTGGGGATCCGAGCTCCACGTGCCCCCCCCCCCC-3′ | 56.84 | 88 | DNA | 49 |
| OTC | 5′-CGTACGGAATTCGCTAGCGGGCGGGGGTGCTGGGGGAATGGAGTGCTGCGTGCTGCGGGGATCCGAGCTCCACGTG-3′ | 11.13 | 76 | DNA | 50 |
| TC | 5′-NH2-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCG TGGATCCGAGCTCCACGTG-3′ | 1.00 | 76 | DNA | 51 |
| TC | 5′-Biotin-TTTTTCGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCGTGGATCCGAGCTCCACGTG-3′ | 63.6 | 81 | DNA | 52 |
| TC | Biotin-5′-GAGCCUAAAACAUACCAGAGAAAUCUGGAGAGGUGAAGAAUACGACCACCUAGGCUC-3′ | 0.77 | 57 | RNA | 53 |
| TC | 5′-GGGCCUAAAACAUACCAGAUCGCCACCCGCGCUUUAAUCUGGAGAGGUGAAGAAUACGACCACCUAGGCUC-3 | 0.77 | 71 | RNA | 54 |
| TC | 5′-GTTTGTGTATTACAGTTATGTTACCCTCATTTTTCTGAAC-3′ | 2.94 | 40 | DNA | 55 |
| TC | 5′-CTCTCTCGGTGGTGTCTCTC-3′ | — | Truncated 8 | DNA | 56 |
Among various aptamer derived detection measurement, biosensing that employs electrochemical transducers holds great promise for the successful development of commercial devices.57,58 In the typical electrochemical measuring process (as shown in Fig. 1), there occurs oxidation, reduction or affinity interactions with the specific analyte through aptamer receptor at the electrode surface, or the conformational changes involving signaling probe separation from or closing to the electrode surface happen. This can be transduced into detectable electrochemical output signals, basically including cyclic voltammetry (CV), differential pulse voltammetry (DPV), or impedance spectroscopy.14,59,60 These signals can be collected and analyzed for calculating the concentration of the analyte by portable and low-cost instruments with low power consumption, enabling on-site testing and diagnosis in the daily routines.46,61,62
As compared in Table 3, most of the TC detection strategies possess excellent testing stability in about 7 days and have satisfactory relative standard deviation (RSD) ranging from 0.9–5.6%, showing the repeatability of the method. Notably, electrochemical-based analyzing methods feature wider range of detection and the LOD can reach as low as the fM level, outperforming the sensitivity of other methods. The sensors exhibit high sensitivity especially when a complementary aptamer was introduced as the recognition probe. Besides, unlike fluorescent/colorimetric sensors requiring bulky and costly optical instruments, electrochemical systems use simple and low-cost electrical measurements to realize quantitative detection; thus, they have excellent portability for a wearable or miniaturized design, making them ideal for point-of-care diagnostics.
| Detection strategy | Range | LOD | Time | RSD | Stability | Ref. |
|---|---|---|---|---|---|---|
| EC refers to electrochemical method; ECL refers to electrochemiluminescent method; PEC refers to photoelectrochemical method; CL refers to colorimetric method; FL refers to fluorescent method; — refers to not given in references. | ||||||
| EC aptasensor | 1 × 10−16–1 × 10−6 M | 3.7 × 10−17 M | 60 min | 5.6% | Repeated 5 times after 7 days | 63 |
| ECL | 1 × 10−6–1 × 10−4 M | 2.3 × 10−7 M | — | 4.4% | Continuous usage within 7 days | 64 |
| PEC | 1 × 10−12–1 × 10−7 M | 8 × 10−14 M | 0.23 s | 1.82% | 7 days | 65 |
| CL | 0–4 × 10−4 M | 1 × 10−6 M | 18 min | — | 7 times reusage | 66 |
| FL | 0–5 × 10−5 M | 1.9 × 10−8 M | 5 min | 0.90%–3.77% | — | 67 |
To get a better analytical performance of this determination method, chemical modification of the electrode surface could be employed without decreasing the signal recognition. For example, Gao's group reported a wearable aptamer nanobiosensor for non-invasive monitoring of hormone oestradiol through sweat analysis.9 First, they immobilized AuNPs and MXene on the electrode surface for increased electrode conductivity and electrochemical sensitivity. Then, a strand displacement aptamer switch was successfully designed. Through the log-linear relationship between peak current density height of the square wave voltammogram (SWV) and target concentrations, a clear sweat oestradiol quantification was realized with the detection limit as low as 0.14 pM, enabling convenient at-home hormone monitoring.
4. Nanomaterial-modified biosensing surface
For electrochemical detection of TCs by the aptasensor, the basic principle involves the introduction of aptamers and target analytes to a biochemical reaction on the biosensing surface of the electrode.68 To significantly improve biosensor sensitivity as well as enhance selectivity and stability while reducing analysis time, nanomaterials are gradually synergized with aptamers to construct aptamer-based nanoprobes.69–71 Therefore, the selection of appropriate nanomaterials and the immobilization strategy of the biosensing layer play a key role in the sensitivity and efficiency of the electrochemical aptasensor.72,734.1. Carbon-based biosensing layer
Carbon-based nanomaterials, including carbon nanotubes (CNTs), carbon nanowires, and graphene feature large surface areas, high electron transfer rates and good biocompatibility.69,72,74 When used in a biosensing layer, they can facilitate electron transfer between bioactive substances and the electrode.75 Additionally, carbon materials can form a wide variety of chemical bonds and have complex surface chemistry, allowing for an increased loading of bioactive species and amplified electrochemical signal response.72,76By introducing functional group modified CNTs into the biosensor electrode, TC-specific aptamers could be stably conjugated to the electrode surfaces via the formation of covalent bonds. This is an effective strategy to enhance the loading capability of TC aptamer on the electrode and realize sensitive detection of the target analyte. As shown in Fig. 2a, Zhou and coworkers designed the TC determination aptasensor by covalently attaching an amino-modified anti-TC aptamer to the carboxylized multi-walled carbon nanotubes (MWCNTs) coated glassy carbon electrode (GCE).77 The increased roughness after MWCNTs modification increased the loading amount of the TCs aptamer; hence, MWCNTs are employed as the carriers of the electrochemical capture probe to amplify the currents signal upon combining of TCs. The ferricyanide solution is used as a mediator to generate the electron flow between bulk solution and the working electrode. In the absence of TCs, the stable and free aptamer configuration presented a significantly strong current, while after TC treatment, the complexed formation hindered the electron diffusion of Fe(CN)63−/Fe(CN)64− towards the electrode surface and produced a detectable signal decrease. From Fig. 2b, it can be obviously seen that the peak currents of DPV decreased with increasing concentrations of TC. Correspondingly, the calibration plot was obtained and the LOD can be determined to be as low as 5 nM.
 | ||
| Fig. 2 (a) Mechanism of electrochemical aptamer-based sensors. (b) DPV of an aptasensor incubated with different concentrations of TC. Inset shows the linear relationship between the anodic peak current changes of DPV to the logarithm of TC concentrations (reprinted with permission from ref. 77. Copyright 2012 Elsevier). (c) Schematic of the fabrication of anti-TC/GO/GCE aptasensor structures of three states. (d) DPV of anti-TC/GO/GCE aptasensor before and after hybridization with different concentrations of TC. Inset A: dependence of ΔI with log (TC concentration /M) (reprinted with permission from ref. 78. Copyright 2016 Springer Nature). | ||
Similarly, Benvidi and colleagues reported an aptasensor for TC detection using a graphene oxide nanosheet-modified GCE. The amino-modified anti-TC aptamer was attached to carboxylated graphene nanosheets on the electrode to construct the aptasensor (Fig. 2c).78 This is because the highly specific surface area of graphene enables the sensor to afford an ultrahigh loading capacity for DNA aptamer, and thus amplify the peak current changes for the binding of TET. The formation of an anti-TC/TC complex increased the impedance signal of the aptasensor owing to more repulsion between the negative charge of the complex and Fe(CN)63−/Fe(CN)64− anions. Additionally, the sensitivity and linear range of the aptasensor were evaluated using DPV. From Fig. 2d, it can be observed that the DPV peak current decreases as the TC concentration increases within the range of 1 × 10−10 to 1 × 10−4 M. This is because the formation of anti-TC/TC blocked the diffusion of Fe(CN)63−/Fe(CN)64− towards the electrode surface. The detection limit was 29 fM based on the plot of peak current versus TC concentration.
Except for carboxylated carbon nanomaterials, some biocompatible and easily prepared amino acid molecules can also be introduced into carbon nanomaterials, and then covalently linked to the anti-TC aptamer. For example, Benvidi's group developed a TC aptasensor by electro-depositing polyglutamic acid (PGA) on MWCNTs/GCE (Fig. 3a).63 The amazing fibrous structure of CNTs and the free carboxylic groups in the PGA structure provide better linkage of the anti-TC aptamer. Besides, the high electrical and thermal conductivity of the modified CNTs help the electrode surface work better as electron mediators. As shown in Fig. 3b, since it was more difficult for probe ions to reach the electrode surface in the presence of TC, the DPV peak current of K3[Fe (CN)6] decreased as the TC concentration increased, with the LOD determined as 0.037 fM. In the above example, by virtue of the amazing fibrous structure and high electrical conductivity, MWCNTs functioned as both immobilization matrices and electron mediators.
 | ||
| Fig. 3 (a) Schematic of the modification process of electrochemical aptasensor fabrication (anti-TC/PGA/MWCNTs/GCE). (b) DPV recorded at anti-TC/PGA/MWCNTs/GCE in the presence of different concentrations of TC. Inset: dependence of ΔI on the concentration of tetracycline (reprinted with permission from ref. 63. Copyright 2018 Elsevier). (c) Schematic of the fabrication of ss-tet/GO/GCE electrode. (d) DPV recorded at the ss-tet/GO/GCE in PBS containing ss-tet′ of different concentrations; (e) calibration curves of the logarithm of concentrations (pM) of ss-tetX′ and peak currents (reprinted with permission from ref. 79. Copyright 2024 Elsevier). | ||
The aptamer can also be physically absorbed onto the modified electrode material via non-covalent bonds, simplifying the sensor fabrication process. For example, Qu's group successfully prepared a sensor by adsorbing the single-stranded (ss)-tet probe onto a GO/GCE surface through π–π stacking (Fig. 3c).79 The ss-tet includes five subtypes, among which ss-tet X was discussed while other subtypes have similar properties. Under optimal conditions, the prepared ss-tet X/GO/GCE was used for DPV detection of complementary ss-tet (ss-tet′), as shown in Fig. 3d. The affinity between ds (double-stranded)-tet and GO was weaker compared with ss-tet, breaking the π–π stacking between GO and nucleobase. Thus, in the presence of ss-tet′, the ss-tet probes on the GO/GCE hybridized and formed ds-tet, which would be released from GO, leading to an increased peak current of DPV. Under optimal conditions, the prepared ss-tet X/GO/GCE was used for DPV detection of complementary ss-tet (ss-tet′), as shown in Fig. 3d. ss-tet Probes on the GO surface could be released from GO in the presence of ss-tet′, leading to an increased peak current of DPV. Within the range of 50.0 pM to 1.0 nM, the peak current showed a linear relationship with the logarithm of ss-tet′ concentration, with a LOD of 50.0 pM (Fig. 3e).
4.2. Gold-based biosensing layer
Gold materials have gained significant attention in biosensor application due to their remarkable electrochemical properties and good biocompatibility.80,81 Gold can be easily modified with bioactive species through various functional groups, such as Au–N and Au–S covalent bonding.52 Additionally, the resistance to corrosion and oxidation makes gold stable over a wide range of conditions, ensuring repeated use and long-term reliability.82 This enables the design and creation of tailored gold-based aptamer sensors for a wide array of applications.It is well known that thiolated aptamer can be immobilized on the surface of a gold electrode through a direct Au–S bond. Yu's group bound sulfhydryl to the gold interface to construct the inverted Y-type DNA sensor.88 Also, ferrocene was immobilized on a DNA strand near the interface to serve as a reference signal to support the accurate measurement, while methylene blue (MB) was attached to a DNA strand away from the interface as an index signal, as exhibited in Fig. 4a. Upon target TC binding to the aptamer, the MB-modified strand was released to replace another DNA strand near the interface through a toe-hold strand displacement effect. This replacement transformed the self-structure of the Y-type DNA and significantly enhanced the MB signal, leading to a linear range of 1.0 × 10−1 to 1.0 × 103 nM for TC with a detection limit of 28.4 pM, indicating high potential for antibiotic monitoring. Similarly, Adaszynska and co-workers covalently anchored a thiolated aptamer on the gold electrode surface with 6-mercaptohexan-1-ol.89 The variations in the redox activity of ferrocene serve as the foundation for analytical signal generation detected by square-wave voltammetry. The “signal-on” mechanism was as follows: upon recognition, aptamer molecules changed their structure so that the ferrocene had a better chance of exchanging electrons with the electrode surface, leading to the increased peak current. Successful detection of TC has been achieved, with a LOD of 0.16 nM in buffer and 0.20 nM in spiked milk, which are over 1000 times lower than the maximum residue level of 225 nM.
 | ||
| Fig. 4 (a) Scheme of inverted Y-type electrochemical aptasensor for ultrasensitive determination of TC (reprinted with permission from ref. 88. Copyright 2022 Elsevier). (b) Sensing mechanism of the electrochemical aptasensor based on a triblock DNA probe with CV measurements of different concentrations of target DNA. (c) SWV curves and radar plot of the electrochemical sensor in response to target DNA with concentrations from 10 pM to 10 μM (reprinted with permission from ref. 90. Copyright 2023 Elsevier). | ||
Rather than conventionally utilizing the Au–S bond to anchor the aptamer probe, Li's group developed a simple and label-free electrochemical aptasensor through the covalent binding between the gold surface and adenine.90 As shown in Fig. 4b and c, a triblock DNA probe featuring two DNA probes at each end and a poly A fragment in the middle that can be quickly anchored on the gold electrode was constructed. When the target DNA hybridizes with both capture probes, the probe sequences changed from an upright conformation into extensively stepped forward due to the robust base stacking effect. Thus, the signal indicator [Ru (NH3)6]3+ electrostatically adsorbed not only on the polyA fragments and the DNA probes, but also on the hybridized target DNA, leading to an electrochemical signal increment. This approach exhibited a wide linear range (10 pM to 10 μM) with a detection limit of 2.9 pM, exhibiting excellent repeatability, stability, and specificity.
By integrating host–guest recognition of β-cyclodextrin (β-CD) with an electrochemical assay, enriched sensitive DPV signals were obtained, contributing to the quantified sensing of TC. Wang and co-workers fixed HS-β-CD onto the surface of the gold electrode through bonds between gold and sulfhydryl groups.55 Ferrocene-modified aptamer was applied as the guest and captured by β-cyclodextrin (β-CD) to attach on the gold surface through the subject-object recognition. In the presence of TC, the configuration of the aptamer changed from upright linear to the reunited state, moving the electron donors (ferrocene moieties) away from the electrode surface and weakening the signal. Thereafter, the decreased signal indicates the presence of the target. The sensing principle is schematically illustrated in Fig. 5a. Results showed that the TC concentration had a linear relationship with the signal within the range of 0.01 nM to 100 nM (Fig. 5b and c). In another example, Liu's group reported a novel strategy incorporating elements of triple-helix aptamer probes (TAP) and catalyzed hairpin assembly (CHA) signal amplification.91 Specifically, when there are TC targets, the TAP conformation was altered and the trigger was liberated owing to the robust affinity between the aptamer and the target. Then, the CHA reaction was initiated, and hybridized Fc-DNA hairpins transformed into double helices. Through the cleavage of Exo III, signaling Fc was liberated and diffused onto the electrode surface via the host–guest recognition, yielding enriched DPV signals proportional to the TC content. Under optimal conditions, the signal intensity showed a linear relationship with the logarithm of target concentrations from 0.2 nM to 100 nM.
 | ||
| Fig. 5 (a) Scheme of the electrochemical sensor for TC detection based on Ferrocene (Fc)-modified aptamer. (b) Electrochemical impedance of different electrodes. (a: bare gold electrode, b: HS-β-CD modified gold electrode, c: HS-β-CD and Fc-aptamer, d: HS-β-CD and Fc-aptamer after 10 nM TCs). (c) DPV response of the electrochemical assay with different concentrations of TET. a–f (0, 0.01, 0.1, 1, 10, and 100 nM) (reprinted with permission from ref. 63. Copyright 2020 Springer Nature). | ||
In 2022, Song and co-workers used a simple hydrothermal reaction to wrap WO3 around CNTs, creating a highly conductive electrode material called C-WO3.95 Then, a layer of AuNPs was electro-deposited onto the surface of the electrode material (C-WO3@AuNPs), which could not only amplify the electrical signal, but provide more attachment sites to fix the aptamer via Au–S bonds, significantly improving the sensor's ability to capture TCs (Fig. 6a). Under optimal conditions, the C-WO3@AuNPs aptasensor was used to assess the sensitivity for detecting TC. The hydrolysis of the phosphodiester bond on the nucleotide caused the aptamer to be negatively charged, resulting in a change of the surface charge. The current increased alongside rising TC concentrations (0.1–100 nM) with a LOD of 48 pM, showing excellent sensitivity (Fig. 6b). In 2024, Xu's group developed octahedral Cu2O@Au@Apta nanomaterial for electrochemical aptasensors to detect TCs. In this platform, less conductive complex was formed on the electrode surface when the antibiotic TC is captured by the aptamer, impeding electron transfer and further reducing the electrochemical response signal. Notably, the nanocomposite synergistically enhanced electrochemical performance and increased binding sites for thiol-modified TCs aptamers without adding oxidants or reducing agents, which improved the detection sensitivity.96
 | ||
| Fig. 6 (a) Schematic of the stepwise fabrication of an aptasensor. (b) Linear relationship between the logarithmic values of TC concentrations (0.1, 0.5, 1, 5, 10, 50, and 100 nM) and the inset shows the DPV responses of different concentrations of TC based on the aptasensor (reprinted with permission from ref. 95. Copyright 2022 Elsevier). (c) Schematic of the preparation procedure of NH2-MIL-101(Fe)/CNF aptasensor. (d) EIS responses of different concentrations of TC based on the aptasensor (reprinted with permission from ref. 96. Copyright 2022 Elsevier). | ||
Metal–organic frameworks (MOFs) are porous crystalline materials composed of metal ions and organic linkers.97,98 MOFs not only have the advantages of large specific surface area and pore size, but are also non-toxic and biocompatible, making them highly promising carriers for aptamers.99 For example, Huang and co-workers developed a novel electrochemical aptasensor based on a MOF of NH2-MIL-101(Fe).100 To improve the sensitivity for antibiotic detection, they synthesized NH2-MIL-101 (Fe)/carbon nanofiber (CNF)@AuNPs using a combination of hydrothermal, electrospinning, pyrolysis, and electrodeposition methods (Fig. 6c). Then, the aptamer was attached to NH2-MIL-101(Fe)/CNF@AuNPs through Au–S bonds. The deposited AuNPs accelerated the electron transfer and promoted subsequent electron injection to the carbon fibers on NH2-MIL-101(Fe)/CNF. Accordingly, signal regulation and amplification were realized. When TC molecules were present, they are adsorbed onto the electrode surface and form an aptamer-TC complex, which hinders the electron transfer of the redox pair Fe(CN)63−/Fe(CN)64− and results in electrical signals changes. Under optimized conditions, the TC concentration detected by the sensor has a linear relationship with the impedance in the range of 0.1–105 nM. The lowest detection limit was 10 pM, exhibiting good selectivity and stability (Fig. 6d).
To amplify electrochemical signals and improve detection sensitivity, aptamers can also be labeled with active molecules or compounds, such as ferrocene, MB, horseradish peroxidase (HRP), and AuNPs. For example, in 2024, Liang's group reported the assembly of a novel MB-encapsulated COF(MB@COF) to create a TC-responsive dual-mode sensor for both electrochemical and fluorescence detection (Fig. 7a).101 The electrochemically active MB was effectively embedded into the fluorescent COF, forming the dual-mode probe MB@COF, where the results from the two detection modes corroborated each other. AuNPs and molybdenum disulfide nanosheets enhanced the conductivity of the GCE and MB@COF. The aptamer and its complementary strand (single-stranded DNA, ssDNA) were introduced into the system for specific TC recognition. When TC was present, it was recognized by the aptamer to trigger the release of ssDNA-MB@AuNPs@COF from the GCE, resulting in enhanced fluorescence and reduced electrochemical signals. The TC detection concentration ranges from 10 nM to 10 mM and the detection limit is 0.03 nM (Fig. 7b). Similarly, Zhao and co-workers developed a highly sensitive aptasensor for TC detection using a triple signal amplification strategy with GO-MWCNTs, Exo I, and AuNPs probes (Fig. 7c).102 The desirable conductivity and large surface area of GO-MWCNTs had been testified through current signal amplification on the electrode surface. Besides, the double-stranded DNA cleaving ability of Exo I could help detach the TC aptamer from the electrode, and significantly enhance the current signal. Moreover, in the presence of hydrogen peroxide, HRP adsorbs on the AuNPs probes and catalyzes the hydroquinone, further amplifying the electrochemical signal. The fabricated sensor has a detection range of 1 × 10−3 to 1 × 103 ng mL−1 and a detection limit of 3.3 × 10−4 ng mL−1 (Fig. 7d).
 | ||
| Fig. 7 (a) Principle of electrochemical/fluorescence dual-mode sensor based on COF for TC detection. (b) Linear relationship between current and logarithm of TC concentration (reprinted with permission from ref. 101. Copyright 2024 Elsevier). (c) Stepwise fabrication process of the electrochemical aptasensor. (d) DPV curves of the fabricated electrochemical sensor for detecting tetracyclines with different concentrations ranging from 103 to 10−3 ng mL−1 (reprinted with permission from ref. 102. Copyright 2024 Elsevier). | ||
4.3. Polymer-based biosensing layer
Electrode surfaces can be efficiently modified with electrically conductive polymers such as polydopamine (PDA), polyaniline (PAN), poly(3,4-ethylenedioxythiophene) (PEDOT), and form one-dimensional nanowires, two dimensional monomers, or three-dimensional matrices.103–105 These polymer layers can not only provide active sites for further immobilization of aptamer molecules with good stability, but also improve the conductive performance of electrodes, which could significantly enhance the electrochemical response and target recognition efficiency of aptasensors.106,107 Undoubtedly, electrochemical polymerization offers a facile and versatile modification strategy for aptamer-based TC sensor application.108,109In 2024, Gu's group developed an anti-fouling electrochemical aptasensor for sensitive and selective analysis of TCs in milk using a zwitterionic a poly(dopamine)-poly(sulfobetaine methacrylate) (PDA-PSBMA), as shown in Fig. 8a.3 The anchor of the TC aptamer relies on a Michael addition reaction between –SH and PDA-PSBMA. Once the TC aptamers recognize TC in the sample, the current signal of the apt/PDA-PSBMA aptasensor decreases. Notably, the antifouling aptasensor not only obviated the need for additional pretreatments, but facilitated the sensitivity and selectivity of TC analysis by means of its commendable resistance to nonspecific adhesion. The sensor has a linear response range of 0.1 to 1000.0 ng mL−1, with a detection limit of 68.0 pg mL−1 (Fig. 8b). Only TC (100.0 ng mL−1) causes significant signal suppression (Fig. 8c).
 | ||
| Fig. 8 (a) Schematic for the fabrication of an antifouling electrochemical aptasensor based on PDA-PSBMA copolymer. (b) DPV curves of the developed aptasensor after incubation with various concentrations of TC (reprinted with permission from ref. 3. Copyright 2024 Elsevier). (c) Signal suppression for TC, GLU, LAC, AMX, KAN, DOX, OXY and their mixture, respectively. (d) Construction and working principle of the electrochemical aptasensor for TC based on the bifunctional matrix of PAN@Cu-BTC. (e) SWV of TC-Apt/PAN@Cu-BTC/SPE after interaction with TC concentrations ranging from 0 to 1 μM (reprinted with permission from ref. 7. Copyright 2024 Elsevier). | ||
Zhang and co-workers synthesized a polyaniline@copper-1,3,5-benzenetricarboxylate (PAN@Cu-BTC) nanocomposite with excellent electrochemical activity on a screen-printed electrode surface using a two-step electrochemical synthesis method.7 Then, a TC aptamer (TC-Apt) was assembled onto the electrode surface through coordination between the 5′-PO43− group on the aptamer and Cu2+ in the MOF, creating a novel electrochemical aptasensor for TC detection (Fig. 8d). Notably, Cu-BTC provides excellent electrochemical activity from the Cu2+ metal center, while the highly conductive doped PAN significantly enhances the electrochemical response. Upon specific binding with the TC target, TC-Apt strands form a complex with the TC, hindering the electron transfer of Cu-BTC and decreasing the electrochemical signal of the aptasensor. Based on this mechanism, TC could be analyzed over a wide range of 10 pM to 1 μM, with a detection limit as low as 0.32 pM (Fig. 8e).
As summarized in Table 4, by comparing all these electrochemical aptasensor-derived TCs detection measurements, it can be learned that the DNA aptamer is the most widely utilized probe type in TC aptasensors by virtue of the inherent stability and the relatively lower manufacturing costs.110 In addition, among different immobilization methods for the aptamers, the formation of an amide bond between activated –COOH of nanomaterials and the –NH2 terminal end of aptamer is the most widely used immobilization approach on the carbon-based biosensing surface. Meanwhile, immobilization through Au–S covalent binding is the most common technique for the gold-based biosensing surface. The LOD can reach as low as fM-level when carbon-based nanomaterials are introduced into the sensing surface.63
| Nanomaterial-modified biosensing surface | Aptamer sequences | Type | Immobilization method | Range | LOD | Sample | Ref. | |
|---|---|---|---|---|---|---|---|---|
| — refers to not given in references. | ||||||||
| Carbon | MWCNTs | 5′-NH2-(CH2)-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCG TGGATCCGAGCTCCACGTG-3′ | DNA | Amidation reaction | 1 × 10−8–5 × 10−5 M | 5 × 10−9 M | Milk | 77 |
| Graphene | 5′-NH2-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGC GCGTGGATCCGAGCTCCACGTG-3′ | DNA | Amidation reaction | 1 × 10−10–1 × 10−4 M | 2.9 × 10−14 M | Tablet and serum | 78 | |
| Poly (L-glutamic acid)/MWCNTs | 5′-NH2-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGC GCGTGGATCCGAGCTCCACGTG-3′ | DNA | Amidation reaction | 1 × 10−16–1 × 10−6 M | 3.7 × 10−17 M | Drug and honey | 63 | |
| GO | 5′-TACCGGAGTGGTTTTATTAT-3′ | DNA | Physical adsorption | 5 × 10−11–1 × 10−9 M | 5 × 10−11 M | Water | 79 | |
| Gold | Gold film | 5′-SH-C6-TCGCAGGTCACGGATGTGTCTTCGTGGTGTAAT-3′ | DNA | Au–S covalent binding | 1 × 10−10–1 × 10−6 M | 2.84 × 10−11 M | Fish | 88 |
| Gold film | — | DNA | Au–S covalent binding | 1 × 10−10–1 × 10−9 M | 1.6 × 10−10 M in buffer, 2 × 10−10 M in milk | Buffer and milk | 89 | |
| Gold film | 5′-GCGCTTTCCCACCAACACACACACAAAAAAAAAAAAAAAAAAAAACACACACACATTGCC CGGCTTTCTT-3′ | DNA | Au-adenine covalent binding | 1 × 10−11–1 × 10−5 M | 2.9 × 10−12 M | Serum | 90 | |
| Gold film | 5′-GTTTGTGTATTACAGTTATGTTACCCTCATTTTTCTGAAC-3′ | DNA | Subject-object recognition | 1 × 10−11–1 × 10−7 M | 8 × 10−12 M | Water, milk, and bacteria culture medium | 55 | |
| Gold film | 5′-TCTCTCCTCGGTGGTGTCCTCTCT-3′ | DNA | — | 2 × 10−10–1 × 10−7 M | 1.3 × 10−10 M | Milk | 91 | |
| CWO3@AuNPs | 5′-Thiol-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGC GCG TGGATCCGAGCTCCACGTG-3′ | DNA | Au–S covalent binding | 1 × 10−10–1 × 10−7 M | 4.8 × 10−11 M | Water | 95 | |
| Cu2O@AuNPs | 5′-SH-(CH2)6-GAGAGACGGTGGTG-3′ | DNA | Au–S covalent binding | 1 × 10−9–1 × 10−3 M | 1.6 × 10−10 M | Milk | 96 | |
| NH2-MIL-101 (Fe)/CNF@AuNPs | 5′-Thiol-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGC GCGTGGATCCGAGCTCCACGTG-3′ | DNA | Au–S covalent binding | 1 × 10−10–1 × 10−4 M | 1 × 10−11 M | Water | 100 | |
| AuNPs@MoS2 NS | 5′-SH-(CH2)6-GTTTGTGTATTACAGTTATGTTACCCTCATTTTTCTGAAC-3′ | DNA | Au–S covalent binding | 1 × 10−8–1 × 10−2 M | 3 × 10−11 M | Milk and water | 101 | |
| GO-MWCNTs/AuNPs | 5′-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCGTGGATCCGAGCTCCACGTG-3′ | DNA | — | 1 × 10−3–1 × 103 ng/mL | 3.3 × 10−4 ng/mL | Milk | 102 | |
| Polymer | PDA-PSBMA | 5′-SH-(CH2)6-CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCGTGGATCCGAGCTCCACGTG-3′ | DNA | Michael addition reaction | 0.1–1000 ng/mL | 68 pg/mL | Protein solutions and milk | 3 |
| PAN@Cu-BTC | 5′-PO43-GTTTGTGTATTACAGTTATGTTACCCTCATTTTTCTGAAC-3′ | DNA | Coordination bonding | 1 × 10−11–1 × 10−6 M | 3.2 × 10−13 M | Fish | 7 | |
5. Conclusions and future perspectives
In this review, recently reported nanomaterials that are modified on the aptasensor electrode for TC detection were summarized and systematically discussed. According to chemical components, they can be classified into three categories, including carbon-based nanomaterials (like CNTs, carbon nanosheets, carbon nanofibers, and graphene), gold-based nanomaterial (like gold film and AuNPs) and polymer (like PPY, PAN, and PEDOT). Compared to traditional TC detecting methods, surface functionalized aptasensors exhibit notable advantages due to their enhanced sensitivity, rapidity and portability. Among those successfully fabricated sensing platforms mentioned above, a voltammetric method including DPV and an impedimetric method including the EIS technique are the most widely employed in the testing process.Despite the advantages of electrochemical aptasensors over traditional methods, it should be noted that several challenges remain to be addressed for their wider application. First, the configuration and binding affinities of aptamers can be changed upon temperature, ionic strength, or pH variations, resulting in a significant reduction in their sensitivity and selectivity. Thus, a significant obstruction for TC sensors lies in their practical implementation, particularly for on-site analysis in complex real-world scenarios. Furthermore, the process of selecting receptor elements and signal transduction components such as nanomaterials and substrates is both complicated and time-consuming. Also, the synthesis complexity and the associated high-cost have limited their broad use. Indeed, after all these years effort in proof-of-concept, only a handful of aptasensors are approved and used commercially in food industry, pharmaceutical or environmental monitoring.111
To overcome these challenges, there is a pressing need for technology that can efficiently produce large quantities of high-quality antibiotic sensors at a relatively low cost. Future efforts should focus on designing a multifunctional platform to make antibiotic sensors more robust. For real-world applications, the receptor elements, including biological and chemical recognition elements, and signal transducers should be improved to withstand more complex and demanding environmental conditions. Additionally, low-cost, biodegradable, and reusable nanomaterials need to be developed to reduce the costs of large-scale fabrication. Ultimately, we hope that electrochemical aptasensors will become more integrated, portable and widely used routine tools in the future for antibiotic and drug detection.
Author contributions
Jingjing Huo and Rui Guo: writing the original draft, editing, and reviewing. Juanjuan Yin, Yanran Liu, Ying Zhang and Fangyi Ruan: reviewing, and editing. Yonghong Shi and Yang Li: reviewing, editing, and supervision.Data availability
No primary research results, software or code have been included, and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (62301446) and the Advance Research Program of Xi'an Rare Metal Materials Research Institute Co. Ltd (Y2208S).References
- D. Wu, H. Karimi-Maleh, X. Liu and L. Fu, J. Anal. Methods Chem., 2023, 2023, 6443610 CrossRef PubMed.
- Y. Liu, Y. Jia, K. Yang, R. Li, X. Xiao, K. Zhu and Z. Wang, Adv. Sci., 2020, 7, 1902227 CrossRef CAS PubMed.
- Q. Wu, M. Niu, D. Ren, L. Yi, K. Ge and Y. Gu, Talanta, 2024, 271, 125623 CrossRef CAS PubMed.
- T. H. Grossman, Cold Spring Harbor Perspect. Med., 2016, 6, a025387 CrossRef PubMed.
- X. Liu, D. Huang, C. Lai, G. Zeng, L. Qin, C. Zhang, H. Yi, B. Li, R. Deng, S. Liu and Y. Zhang, TrAC, Trends Anal. Chem., 2018, 109, 260–274 CrossRef CAS.
- L. Zhang, M. Yin, X. Wei, Y. Sun, Y. Luo, H. Lin, R. Shu and D. Xu, Bioelectrochemistry, 2024, 157, 108668 CrossRef CAS PubMed.
- F. Zhan, Y. Zhao, X. Dai, J. Zeng and Q. Wang, Microchem. J., 2024, 196, 109512 CrossRef CAS.
- Y. Xing, H. Zheng, C. Wang, Z. Zhang, Y. Qian, J. Qu and X. Zhu, J. Environ. Chem. Eng., 2024, 12, 113642 CrossRef CAS.
- C. Ye, M. Wang, J. Min, R. Y. Tay, H. Lukas, J. R. Sempionatto, J. Li, C. Xu and W. Gao, Nat. Nanotechnol., 2024, 19, 330–337 CrossRef CAS PubMed.
- C. Xu, Y. Song, J. R. Sempionatto, S. A. Solomon, Y. Yu, H. Y. Y. Nyein, R. Y. Tay, J. Li, W. Heng, J. Min, A. Lao, T. K. Hsiai, J. A. Sumner and W. Gao, Nat. Electron., 2024, 7, 168–179 CrossRef CAS PubMed.
- S. Sharma, H. Byrne and R. J. O'Kennedy, Essays Biochem., 2016, 60, 9–18 CrossRef PubMed.
- D. Das, C. W. Lin and H. S. Chuang, Biosensors, 2022, 12, 1068 CrossRef CAS PubMed.
- M. Zhang, X. Guo and J. Wang, Biosens. Bioelectron., 2023, 224, 115077 CrossRef CAS PubMed.
- V. Leguillier, B. Heddi and J. Vidic, Biosensors, 2024, 14, 210 CrossRef CAS PubMed.
- S. Wang, Y. Dong and X. Liang, Biosens. Bioelectron., 2018, 109, 1–7 CrossRef CAS PubMed.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- Y. Xia, Y. Liu, X. Hu, F. Zhao and B. Zeng, ACS Sens., 2022, 7, 3077–3084 CrossRef CAS PubMed.
- J. Jiang, H. Wu, Y. Su, Y. Liang, B. Shu and C. Zhang, Anal. Chem., 2020, 92, 7708–7716 CrossRef CAS PubMed.
- M. Bai, F. Yue, S. Zhai, M. Hu, G. Qi, S. Chen, B. Li, X. Sun, Y. Guo and G. Marrazza, Sens. Actuators, B, 2024, 398, 134765 CrossRef CAS.
- W. R. Shin, G. Ahn, J. P. Lee, I. H. Oh, J. Y. Ahn, H. Kim and S. Chae, Chem. Eng. J., 2023, 472, 144742 CrossRef CAS.
- S. Liu, Y. Xu, X. Jiang, H. Tan and B. Ying, Biosens. Bioelectron., 2022, 208, 114168 CrossRef CAS PubMed.
- L. Huang, S. Tian, W. Zhao, K. Liu, X. Ma and J. Guo, Biosens. Bioelectron., 2021, 186, 113279 CrossRef CAS PubMed.
- K. Muzyka, M. Saqib, Z. Liu, W. Zhang and G. Xu, Biosens. Bioelectron., 2017, 92, 241–258 CrossRef CAS PubMed.
- A. Vasilescu and J. L. Marty, TrAC, Trends Anal. Chem., 2016, 79, 60–70 CrossRef CAS.
- M. Liu, A. Khan, Z. Wang, Y. Liu, G. Yang, Y. Deng and N. He, Biosens. Bioelectron., 2019, 130, 174–184 CrossRef CAS PubMed.
- Q. u. A. Zahra, S. Ullah, F. Shahzad, B. Qiu, X. Fang, A. Ammar, Z. Luo and S. A. Zaidi, Prog. Mater. Sci., 2022, 129, 100967 CrossRef CAS.
- R. Sharma, K. V. Ragavan, M. S. Thakur and K. S. Raghavarao, Biosens. Bioelectron., 2015, 74, 612–627 CrossRef CAS PubMed.
- J. Huang, F. Yang, L. Geng, X. Chen, G. Wang, J. Han, Y. Guo, X. Sun and G. Marrazza, Food Chem., 2023, 429, 136857 CrossRef CAS PubMed.
- F. Charbgoo, F. Soltani, S. M. Taghdisi, K. Abnous and M. Ramezani, TrAC, Trends Anal. Chem., 2016, 85, 85–97 CrossRef CAS.
- X. Meng, D. O'Hare and S. Ladame, Biosens. Bioelectron., 2023, 237, 115440 CrossRef CAS PubMed.
- T. Dong, N. M. M. Pires, Z. Yang and Z. Jiang, Adv. Sci., 2023, 10, e2205429 CrossRef PubMed.
- K. Mao, H. Zhang, Z. Wang, H. Cao, K. Zhang, X. Li and Z. Yang, Biosens. Bioelectron., 2020, 148, 111785 CrossRef CAS PubMed.
- M. Mahmoudpour, S. Ding, Z. Lyu, G. Ebrahimi, D. Du, J. E. N. Dolatabadi, M. Torbati and Y. Lin, Nano Today, 2021, 39, 101177 CrossRef CAS.
- Z. Hua, T. Yu, D. Liu and Y. Xianyu, Biosens. Bioelectron., 2021, 179, 113076 CrossRef CAS PubMed.
- R. Daghrir and P. Drogui, Environ. Chem. Lett., 2013, 11, 209–227 CrossRef CAS.
- I. Chopra and M. Roberts, Microbiol. Mol. Biol. Rev., 2001, 65, 232–260 CrossRef CAS PubMed.
- X. Cui, M. Jin, P. Du, G. Chen, C. Zhang, Y. Zhang, Y. Shao and J. Wang, Food Agric. Immunol., 2018, 29, 638–652 CrossRef CAS.
- Y. Ahn, J. Y. Jung, B. T. Veach, S. Khare, K. Gokulan, S. A. Pineiro and C. E. Cerniglia, Regul. Toxicol. Pharmacol., 2018, 99, 105–115 CrossRef CAS PubMed.
- K. Ziani, M. Pérez-López, A. Mansouri, M. B. Khaled, A. S. Rodriguez and M. Slimani, Food Anal. Methods, 2018, 11, 2528–2537 CrossRef.
- M. Guarddon, J. M. Miranda, B. I. Vazquez, A. Cepeda and C. M. Franco, Foods, 2015, 4, 306–317 CrossRef CAS PubMed.
- Y. Li, J. Fang, X. Yuan, Y. Chen, H. Yang and X. Fei, Int. J. Environ. Res. Public Health, 2018, 15, 1803 CrossRef CAS PubMed.
- K. Bahmani, Y. Shahbazi and Z. Nikousefat, Food Sci. Biotechnol., 2019, 29, 441–448 CrossRef PubMed.
- F. Granados-Chinchilla and C. Rodriguez, J. Anal. Methods Chem., 2017, 2017, 1315497 Search PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- I. Willner and M. Zayats, Angew. Chem., Int. Ed., 2007, 46, 6408–6418 CrossRef CAS PubMed.
- T. Hermann and D. J. Patel, Science, 2000, 287, 820–825 CrossRef CAS PubMed.
- A. O. Rad and A. Azadbakht, Mikrochim. Acta, 2019, 186, 56 CrossRef PubMed.
- M. Hosseini, F. Mehrabi, M. R. Ganjali and P. Norouzi, Luminescence, 2016, 31, 1339–1343 CrossRef CAS PubMed.
- Y. S. Kim, J. H. Kim, I. A. Kim, S. J. Lee, J. Jurng and M. B. Gu, Biosens. Bioelectron., 2010, 26, 1644–1649 CrossRef CAS PubMed.
- S. Jahanbani and A. Benvidi, Biosens. Bioelectron., 2016, 85, 553–562 CrossRef CAS PubMed.
- Y. J. Kim, Y. S. Kim, J. H. Niazi and M. B. Gu, Bioprocess Biosyst. Eng., 2010, 33, 31–37 CrossRef CAS PubMed.
- S. Jeong and I. R. Paeng, Sci. World J., 2012, 2012, 159456 Search PubMed.
- M. Mueller, J. E. Weigand, O. Weichenrieder and B. Suess, Nucleic Acids Res., 2006, 34, 2607–2617 CrossRef CAS PubMed.
- Y. H. Wu, H. Bi, G. Ning, Z. G. Xu, G. Q. Liu, Y. H. Wang and Y. L. Zhao, J. Solid State Electrochem., 2020, 24, 2365–2372 CrossRef CAS.
- M. Ramezani, N. M. Danesh, P. Lavaee, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2015, 70, 181–187 CrossRef CAS PubMed.
- G. K. Mishra, V. Sharma and R. K. Mishra, Biosensors, 2018, 8, 28 CrossRef PubMed.
- S. Song, L. Wang, J. Li, C. Fan and J. Zhao, TrAC, Trends Anal. Chem., 2008, 27, 108–117 CrossRef CAS.
- J. Aleman, T. Kilic, L. S. Mille, S. R. Shin and Y. S. Zhang, Nat. Protoc., 2021, 16, 2564–2593 CrossRef CAS PubMed.
- C. Loncaric, Y. Tang, C. Ho, M. A. Parameswaran and H. Z. Yu, Sens. Actuators, B, 2012, 161, 908–913 CrossRef CAS.
- S. L. Clark and V. T. Remcho, Electrophoresis, 2002, 23, 1335–1340 CrossRef CAS PubMed.
- K. Liu, K. S. Chen, D. Sen and H. Z. Yu, Electrochim. Acta, 2021, 379, 138125 CrossRef CAS.
- A. Benvidi, S. Yazdanparast, M. Rezaeinasab, M. D. Tezerjani and S. Abbasi, J. Electroanal. Chem., 2018, 808, 311–320 CrossRef CAS.
- X. Chen, L. Zhao, X. Tian, S. Lian, Z. Huang and X. Chen, Talanta, 2014, 129, 26–31 CrossRef CAS PubMed.
- H. Cao, K. Geng, X. Yan, H. Qiu, S. Lin, C. Dai, W. Liu and Y. Wang, Mater. Des., 2023, 233, 112219 CrossRef CAS.
- Y. Wei, Y. Xu, X. Zhang and X. Luo, Appl. Surf. Sci., 2025, 687, 162277 CrossRef CAS.
- X. Wang, L. Li, H. Jiang, H. Zhangsun, Q. Wang, X. Sun and L. Wang, Food Chem., 2022, 374, 131774 CrossRef CAS PubMed.
- V. Gupta and J. E. Dick, ACS Sens., 2023, 8, 1143–1150 CrossRef CAS PubMed.
- J. S. Del Rio, O. Y. F. Henry, P. Jolly and D. E. Ingber, Nat. Nanotechnol., 2019, 14, 1143–1149 CrossRef PubMed.
- Y. Xiong, W. Li, Q. Wen, D. Xu, J. Ren and Q. Lin, Food Control, 2022, 135, 108661 CrossRef CAS.
- J. Raina, G. Kaur and I. Singh, Talanta, 2024, 277, 126372 CrossRef CAS PubMed.
- S. S. Timilsina, N. Durr, M. Yafia, H. Sallum, P. Jolly and D. E. Ingber, Adv. Healthcare Mater., 2022, 11, e2102244 CrossRef PubMed.
- J. Li, S. Zhang, L. Zhang, Y. Zhang, H. Zhang, C. Zhang, X. Xuan, M. Wang, J. Zhang and Y. Yuan, Front. Chem., 2021, 9, 680593 CrossRef CAS PubMed.
- M. A. Sadique, S. Yadav, P. Ranjan, R. Khan, F. Khan, A. Kumar and D. Biswas, ACS Appl. Bio Mater., 2022, 5, 2421–2430 CrossRef CAS PubMed.
- F. Guan, H. Dong, Y. Xiang, M. Zhang, J. Huang, G. Wang, Z. Shen, D. Xu, X. Sun, Y. Guo and S. Zhao, J. Electrochem. Soc., 2022, 169, 127501 CrossRef CAS.
- C. Zhao, X. Li, S. An, D. Zheng, S. Pei, X. Zheng, Y. Liu, Q. Yao, M. Yang and L. Dai, Sci. Bull., 2019, 64, 1272–1279 CrossRef CAS PubMed.
- L. Zhou, D. J. Li, L. Gai, J. P. Wang and Y. B. Li, Sens. Actuators, B, 2012, 162, 201–208 CrossRef CAS.
- A. Benvidi, M. D. Tezerjani, S. M. Moshtaghiun and M. Mazloum-Ardakani, Microchim. Acta, 2016, 183, 1797–1804 CrossRef CAS.
- Y. Xing, Y. Zhang, X. Zhu, C. Wang, T. Zhang, F. Cheng, J. Qu and W. Peijnenburg, Sci. Total Environ., 2024, 906, 167615 CrossRef CAS PubMed.
- Q. Xu, Z. Liu, J. Fu, W. Zhao, Y. Guo, X. Sun and H. Zhang, Sci. Rep., 2017, 7, 14729 CrossRef PubMed.
- O. Hosu, G. Melinte, G. Ştefan, M. Casian and C. Cristea, Electrochim. Acta, 2023, 460, 142556 CrossRef CAS.
- Y. Huang, X. Yan, L. Zhao, X. Qi, S. Wang and X. Liang, Microchem. J., 2019, 150, 104179 CrossRef CAS.
- V. Mazzaracchio, M. R. Maciel, T. P. Santos, K. Toda-Peters and A. Q. Shen, Small, 2023, 19, e2207731 CrossRef PubMed.
- Y. Xiao, R. Y. Lai and K. W. Plaxco, Nat. Protoc., 2007, 2, 2875–2880 CrossRef CAS PubMed.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi, X. Lou, J. Shi and C. Fan, Nat. Protoc., 2016, 11, 1244–1263 CrossRef CAS PubMed.
- A. Li, J. Zhu, S. Chen, Y. Mei and H. Xu, Electroanalysis, 2021, 37, 625–631 CAS.
- X. Hu, Y. Xu, X. Cui, W. Li, X. Huang, Z. Li, J. Shi and X. Zou, Microchim. Acta, 2020, 187, 83 CrossRef CAS PubMed.
- L. Wang, X. Zeng, L. Zhang, Y. Yu, B. Lin, Y. Wang, M. Guo and Y. Cao, Microchem. J., 2022, 181, 107724 CrossRef CAS.
- K. Malecka-Baturo, A. Zaganiaris, I. Grabowska and K. Kurzatkowska-Adaszynska, Int. J. Mol. Sci., 2022, 23, 13785 CrossRef CAS PubMed.
- L. Li and Z. Chen, Bioelectrochemistry, 2023, 153, 108494 CrossRef CAS PubMed.
- Y. Wang, L. Yao, G. Ning, Y. Wu, S. Wu, S. Mao and G. Q. Liu, Biosens. Bioelectron., 2019, 143, 111613 CrossRef CAS PubMed.
- Z. Li, F. Chen, N. Zhu, L. Zhang and Z. Xie, ACS Nano, 2023, 17, 21935–21946 CrossRef PubMed.
- J. Li, H. Zheng, Y. He, B. Chen, L. Liu, Y. Ouyang, C. Zhu, Y. Zhou, J. Sun, Z. Hu and B. Wang, ACS Sens., 2019, 4, 3203–3209 CrossRef CAS PubMed.
- H. Abdulkarim and M. Siaj, Sci. Rep., 2022, 12, 9334 CrossRef CAS PubMed.
- J. Song, X. Lin, N. Jiang and M. Huang, Food Chem., 2022, 367, 130564 CrossRef CAS PubMed.
- X. Wei, M. Yin, L. Zhang, Y. Sun, Y. Luo and D. Xu, Spectrochim. Acta A: Mol. Biomol. Spectrosc., 2024, 304, 123361 CrossRef CAS PubMed.
- F. Gao, Y. Zhao, X. Dai, W. Xu, F. Zhan, Y. Liu and Q. Wang, Food Chem., 2024, 430, 137041 CrossRef CAS PubMed.
- Y. Meng, Y. Huang, G. Huang and Y. Song, RSC Adv., 2023, 13, 28148–28157 RSC.
- M. Lv, W. Zhou, H. Tavakoli, C. Bautista, J. Xia, Z. Wang and X. Li, Biosens. Bioelectron., 2021, 176, 112947 CrossRef CAS PubMed.
- J. Song, M. Huang, X. Lin, S. F. Y. Li, N. Jiang, Y. Liu, H. Guo and Y. Li, Chem. Eng. J., 2022, 427, 130913 CrossRef CAS.
- N. Liang, X. Hu, W. Li, Z. Guo, X. Huang, Z. Li, X. Zhang, X. Zou and J. Shi, Sens. Actuators, B, 2024, 405, 135261 CrossRef CAS.
- Z. Zhao, Q. Sheng, F. Guan, L. Wang, Y. Dong and D. Cai, Microchem. J., 2024, 204, 110952 CrossRef CAS.
- S. Kim, L. K. Jang, H. S. Park and J. Y. Lee, Sci. Rep., 2016, 6, 30475 CrossRef PubMed.
- M. Zhang, Y. Wang, K. Liu, Y. Liu, T. Xu, H. Du and C. Si, Carbohydr. Polym., 2023, 305, 120567 CrossRef CAS PubMed.
- J. Song, H. Teng, Z. Xu, N. Liu, L. Xu, L. Liu, F. Gao and X. Luo, Microchim. Acta, 2021, 188(6) DOI:10.1007/s00604-021-04859-1.
- M. Zhang, W. Yang, Z. Wang, H. Liu, R. Yin, C. Liu and C. Shen, Appl. Phys. Lett., 2023, 122, 043507 CrossRef CAS.
- C. Yang, D. Zhang, D. Wang, H. Luan, X. Chen and W. Yan, ACS Appl. Mater. Interfaces, 2023, 15, 5811–5821 CrossRef CAS PubMed.
- P. Salehan, A. A. Ensafi, K. Z. Mousaabadi, J. B. Ghasemi, E. Aghaee and B. Rezaei, Chemosphere, 2022, 292, 133460 CrossRef CAS PubMed.
- J. Liang, B. Wan, X. Tan, J. Li, Y. Zhan, Y. Huang, Z. Zhou and G. Li, Microchem. J., 2024, 205, 111182 CrossRef CAS.
- X. Xiao, H. Li, L. Zhao, Y. Zhang and Z. Liu, Biomed. Pharmacother., 2021, 143, 112232 CrossRef CAS PubMed.
- S. Wang, X. Yan, Y. Yang, X. Qi, Y. Zhao, L. Li, R. Ma, L. Wang, Y. Dong, J. Sun and X. Mao, Food Chem., 2021, 364, 130361 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |