Synthesis, characterization, DNA interaction, molecular docking, and α-amylase and α-glucosidase inhibition studies of a water soluble Zn(II) phthalocyanine†
Nagihan
Saglam Ertunga
 *a,
Ece Tugba
Saka
*a,
Ece Tugba
Saka
 a,
Tugba
Taskin-Tok
a,
Tugba
Taskin-Tok
 bc,
Kadriye
Inan Bektas
bc,
Kadriye
Inan Bektas
 d and
Melike
Yildirim Akatin
d and
Melike
Yildirim Akatin
 e
e
aKaradeniz Technical University, Faculty of Science, Department of Chemistry, Trabzon, Türkiye. E-mail: nagihan@ktu.edu.tr; Fax: +90 462 325 31 96; Tel: +90 462 377 42 74
bGaziantep University, Faculty of Arts and Sciences, Department of Chemistry, 27310-Gaziantep, Türkiye
cGaziantep University, Institute of Health Sciences, Department of Bioinformatics and Computational Biology, Gaziantep, Türkiye
dKaradeniz Technical University, Faculty of Science, Department of Molecular Biology and Genetics, Trabzon, Türkiye
eKaradeniz Technical University, Macka Vocational School, Trabzon, Türkiye
First published on 7th June 2024
Abstract
In this study, 2(3),9(10),16(17),23(24)-tetrakis-[(N-methyl-(1-benzylpiperidin-4-yl)oxy)phthalocyaninato]zinc(II) iodide (ZnPc-2) was synthesized and characterized using spectral methods (FT-IR, 1H-NMR, UV-Vis and mass spectroscopy). The interaction of ZnPc-2 with DNA was investigated by using the UV/Vis titrimetric method, thermal denaturation profile, agarose gel electrophoresis and molecular docking studies. Additionally, the antidiabetic activity of ZnPc-2 was revealed spectroscopically by studying α-amylase and α-glucosidase inhibition activities. The spectroscopic results indicated that ZnPc-2 effectively binds to calf thymus-DNA (CT-DNA) with a Kb value of 7.5 × 104 M−1 and interacts with CT-DNA via noncovalent binding mode. Gel electrophoresis results also show that ZnPc-2 binds strongly to DNA molecules and exhibits effective nuclease activity even at low concentrations. Furthermore, docking studies suggest that ZnPc-2 exhibits a stronger binding tendency with DNA than the control compounds ethidium bromide and cisplatin. Consequently, due to its strong DNA binding and nuclease activity, ZnPc-2 may be suitable for antimicrobial and anticancer applications after further toxicological tests. Additionally, antidiabetic studies showed that ZnPc-2 had both α-amylase and α-glucosidase inhibition activity. Moreover, the α-glucosidase inhibitory effect of ZnPc-2 was approximately 3500 times higher than that of the standard inhibitor, acarbose. Considering these results, it can be said that ZnPc-2 is a moderate α-amylase and a highly effective α-glucosidase inhibitor. This suggests that ZnPc-2 may have the potential to be used as a therapeutic agent for the treatment of type 2 diabetes.
1. Introduction
Phthalocyanine compounds are unique macrocyclic molecules with an 18 π electron system that are used in many areas such as catalysts, liquid crystals and organic semiconductors as well as therapeutic agents for anticancer, antibacterial and antiviral treatments.1–13 The piperidine ring is a valuable saturated heterocyclic compound with various biological activities found in the structures of many natural alkaloids.14–18The DNA molecule is one of the primary targets for many drug molecules used in anticancer, antibacterial, antiviral and antimalarial treatments, due to its main cellular functions such as replication and transcription.19,20 Small molecules interacting with DNA cause DNA damage preventing cell division and subsequently cause cell death. Therefore, studies on the interactions of small molecules with DNA are important in the discovery of new drug molecules.21,22 The DNA molecule tends to display binding activity to some metal compounds, including metallophthalocyanines, via intercalative or non-intercalative binding mechanisms such as groove binding and electrostatic interactions. It has been reported that especially cationic metallophthalocyanines display a high affinity for anionic nucleic acids. These complexes may also exhibit nuclease activity in a hydrolytic or oxidative manner.21,23
Phthalocyanines are considered to be good DNA binders, which makes them promising anti-tumor drug leads. In spite of a number of experimental studies that aim to enlighten the binding mechanism of phthalocyanines to DNA,24–27 there is a gap in the literature regarding the molecular docking studies on this subject. So we aimed to investigate the interactions between DNA and the ZnPc derivative in silico in order to contribute to filling this gap and to support our experimental studies.
α-Amylase and α-glucosidase are digestive enzymes that break down starch into glucose. Glucose is then absorbed through the small intestine, leading to an increase in the blood glucose level.28 Inhibiting the activity of α-amylase and α-glucosidase can slow down the digestion of starch and prevent postprandial hyperglycemia. This is one of the methods for treating type 2 diabetes. Currently available drugs, such as acarbose, voglibose, and miglitol, can effectively inhibit α-amylase and α-glucosidase, but they can also cause side effects. This limits their clinical use. Therefore, researchers are looking for new α-amylase and α-glucosidase inhibitors that are effective and have low toxicity.29 Due to the macrocyclic and bulky structure of metallophthalocyanines, enzyme inhibition studies of these complexes have not been widely reported. It seems that there are a limited number of studies revealing the antidiabetic effectiveness of metallophthalocyanines. In this respect, we think that this study is one of the limited number of studies, especially in terms of α-amylase inhibition activity.
In this work, we have chosen 3-[(1-benzylpiperidin-4-yl)oxy]phthalonitrile30 as the precursor molecule to obtain water soluble 2(3),9(10),16(17),23(24)-tetrakis-[(N-methyl-(1-benzylpiperidin-4-yl)oxy)phthalocyaninato]zinc(II) iodide (ZnPc-2). To reveal the interaction of the ZnPc-2 complex with DNA, UV/Vis absorption titration, agarose gel electrophoresis and thermal denaturation studies were performed. Furthermore, in the study, molecular docking calculations were performed on the basis of a computational perspective to predict the binding affinity of ZnPc-2 towards DNA. Lastly, the α-amylase and α-glucosidase inhibition activities of ZnPc-2 were also examined using spectroscopic techniques.
2. Experimental
2.1. Materials
3-[(1-Benzylpiperidin-4-yl)oxy]phthalonitrile and 2(3),9(10),16(17),23(24)-tetrakis-[(1-benzylpiperidin-4-yl)oxy]phthalocyaninato zinc(II) (ZnPc-1) were designed and prepared as given in the literature.30,31 All solvents were dried and purified as described in the reported procedure.32 3-Nitrophthalonitrile was purchased from commercial suppliers. Information on all materials, methods and devices is given in detail in the ESI.†2.2. Synthesis of 2(3),9(10),16(17),23(24)-tetrakis-[(N-methyl-(1-benzylpiperidin-4-yl)oxy)phthalocyaninato]zinc(II) iodide (ZnPc-2)
2(3),9(10),16(17),23(24)-Tetrakis-[(1-benzylpiperidin-4-yl)oxy]phthalocyaninato zinc(II) (ZnPc-1) (30 mg, 0.020 mmol)30 was dissolved in CHCl3 (3 ml), iodomethane (3 ml) was added, and stirred at room temperature for 3 days. The green solid product obtained by filtration was washed with CHCl3, diethyl ether, and acetone and dried over phosphorus pentoxide. Yield: 74 mg (93%); FT-IR: νmax/cm−1 3058, 3025 (Ar–CH), 2858–2810 (Aliph. C–H), 1603–1468, 1314, 1273, 1115, 1087, 1042, 964, 697, 653, 585; 1H-NMR. (DMSO), (δ: ppm): 7.92–7.79 (m, 12H, ArH), 7.68–7.45 (m, 20H, ArH), 4.65–4.48 (m, 4H, O-CH(1)) 4.34 (s, 8H, N+-CH2(5)), 3.80 (t, J: 6.8, 16H, N+-CH2(3)), 3.12 (s, 12H, N+-CH3(4)), 1.90–168 (m, 16H, Aliph. CH2(2)). UV/Vis (DMSO): λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 350 (4.43), 616 (4.25), 685 (4.98); MALDI-TOF-MS: m/z: calcd 1902.682; found 348.766 [M − 4I]4+.
ε) 350 (4.43), 616 (4.25), 685 (4.98); MALDI-TOF-MS: m/z: calcd 1902.682; found 348.766 [M − 4I]4+.
2.3. UV-Visible absorption spectroscopy assay
CT-DNA solution was prepared in 5 mM Tris-HCl/50 mM NaCl buffer (pH 7.2) and the concentration was measured by using NanoDrop™. A 2 mM stock solution of Zn-2 was prepared in deionized water with 4.8% DMSO (v/v) and used in all biological activity studies. Absorption spectrum experiments were carried out in the range of 300–800 nm with constant ZnPc-2 concentration (15 μM) and increasing CT-DNA concentrations (0–10 μM) in 5 mM Tris-HCl/50 mM NaCl buffer (pH 7.2). The mixtures were incubated for 10 min at room temperature and absorption changes were monitored. To eliminate dilution effects, control titrations were assayed with reaction buffer instead of CT-DNA. The binding constant (Kb) was used to quantitatively compare the DNA binding affinity of compounds and is calculated using the Wolfe–Shimer eqn (1).33| [DNA]/(εa − εf) = [DNA]/(εb − εf) + 1/[Kb(εb − εf)] | (1) |
In this equation, [DNA] is the concentration of DNA in base pairs, εa corresponds to Aobserved/[the complex] (Aobserved is the absorbance value of the complex titrated with DNA at the maximum of the spectra), and εf and εb are the extinction coefficients of the free complex (Afree/[free complex]) and that of the complex in the fully bound form (Afully bounded/[fully bound complex]), respectively. Free complex and fully bound complex are the complex in the absence and saturated concentration of DNA respectively. In the plots of [DNA]/(εa − εf) versus [DNA], Kb is given by the ratio of the slope to intercept.33
2.4. Determination of the change in the thermal denaturation profile of DNA
The DNA thermal denaturation studies were performed by measuring the absorbance of CT-DNA (50 μM) and CT-DNA-ZnPc-2 (50 μM) in 5 mM Tris-HCl/50 mM NaCl buffer (pH 7.2) at 260 nm with increasing temperature. The temperature was gradually increased from 50 °C to 95 °C, and absorbance readings were taken at 0.5 °C intervals on a Bio UV/VIS-spectrophotometer. The melting temperature of CT-DNA and CT-DNA-ZnPc-2 solutions were automatically calculated using a thermal melting program.342.5. DNA cleavage activity
The interaction of the water-soluble ZnPc-2 complex with CT-DNA and supercoiled pBR322 plasmid DNA was examined by agarose gel electrophoresis. For CT-DNA, reaction mixtures (10 μl) consisted of CT-DNA (65 ng), ZnPc-2 complex with varying concentrations (50 to 500 μM) and Tris-HCl buffer (50 mM, pH 7.2) without any external additives. These mixtures were incubated for 1 h at 37 °C in the dark, followed by the addition of 6 × DNA loading dye (1 μL) and loaded on 0.8% agarose gel containing ethidium bromide (0.5 μg ml−1). For plasmid DNA, reaction mixtures (10 μl) consisted of pBR322 plasmid DNA (240 ng), ZnPc-2 complex with varying concentrations (1 to 500 μM) and Tris-HCl buffer (50 mM, pH 7.2) in the absence and presence of reducing/oxidizing and radical scavenging agents. These mixtures were incubated for 1 h at 37 °C in the dark, followed by the addition of 6 × DNA loading dye (1 μL) and loaded on 1% agarose gel containing ethidium bromide (0.5 μg ml−1). After the gel was electrophoresed for 1 h at 100 V in 1 × TAE (Tris-acetate-EDTA) buffer, DNA was viewed on the Kodak gel logic gel imaging system and the electrophorograms were recorded on the computer.352.6. Molecular docking studies
Molecular docking allows researchers to visualize and model the best binding patterns and interactions between the ligand and the target.36 Before the docking process, ligand structures and target structure are prepared. The ZnPc-2 complex synthesized on this basis was drawn as a ligand with the help of Gaussian09 (G09) software,37 and its energy optimized structures were determined in the DFT/B3LYP/LAND2DZ basis set. Then, DNA (PDB: 1BNA, https://www.rcsb.org/structure/1bna, 1.90 Å) was prepared for docking with AutoDock Tools 1.5.7 software. The interaction studies of the ligand and target biomolecule were examined with AutoDock 4,38 a molecular docking application, as one of the molecular modeling methods. The positive compounds, ethidium bromide and cisplatin for DNA were compared with Zn(II) phthalocyanine.2.7. Effect of ZnPc-2 on α-amylase activity
Dinitrosalicylic acid (DNS) method was used to determine α-amylase inhibitory activity of ZnPc-2. A certain volume of complex solution (2 mM, dissolved in deionized water containing 4.8% DMSO, v/v) was added to the reaction mixture containing phosphate buffer (20 mM, pH 7.0, containing 6 mM NaCl) and soluble starch solution (50 μL, 1%, w/v). After the addition of 20 μL of α-amylase (1 U, α-amylase from porcine pancreas, Sigma), the reaction was carried out at 37 °C for 10 minutes. At the end of the reaction period, 500 μL of DNS reagent was added to the reaction mixture and kept in a boiling water bath for 10 minutes. Absorbance was measured at 540 nm using a UV-Vis spectrophotometer. Blind reaction solution was also prepared by the same method but without enzyme. ZnPc-2 concentration that inhibited 50% of α-amylase was given as the IC50 value. Acarbose, the standard inhibitor of the α-amylase, was used as a positive control. One unit of enzyme activity was defined as the amount of enzyme producing 1 μmol reducing sugar as maltose in 1 min under reaction conditions.39,40The enzyme reaction rates were determined with two different concentrations of ZnPc-2 when the concentrations of soluble starch were 0.10, 0.15, 0.20 and 0.30%. The maximum velocity (Vmax) of α-amylase and the Michaelis–Menten constant (Km) values were calculated from the Lineweaver–Burk plots.41
2.8. Effect of ZnPc-2 on α-glucosidase activity
The inhibition of α-glucosidase was determined according to the previously reported method with some modifications.29 In brief, 15 μL of α-glucosidase (1 U, α-glucosidase from Saccharomyces cerevisiae, Sigma) was added to the reaction mixture containing a certain volume of ZnPc-2 solution (2 mM, dissolved in deionized water containing 4.8% DMSO, v/v), phosphate buffer (100 mM, pH 6.8) and substrate solution (45 μL, 5 mM p-nitrophenyl glucopyranoside, p-NPG). The reaction was carried out at 37 °C for 10 minutes and was stopped by adding 0.1 M Na2CO3. The absorbance was recorded at 405 nm using a UV-Vis spectrophotometer. Blind reaction solution was also prepared by the same method but without enzyme. The sample concentration that inhibited 50% of α-glucosidase was given as the IC50 value. Acarbose, the standard inhibitor of the α-glucosidase, was used as a positive control. One unit of enzyme activity was defined as the amount of the enzyme which released 1 μmol of p-nitrophenol from p-NPG under reaction conditions.The enzyme reaction rates were determined with two different concentrations of ZnPc-2 when the concentrations of pNPG were 0.07, 0.20, 0.33, 0.67 and 1.00 μM. The maximum velocity (Vmax) and Michaelis–Menten constant (Km) values are calculated from the Lineweaver–Burk plots.42
3. Results and discussion
3.1. Synthesis and characterization
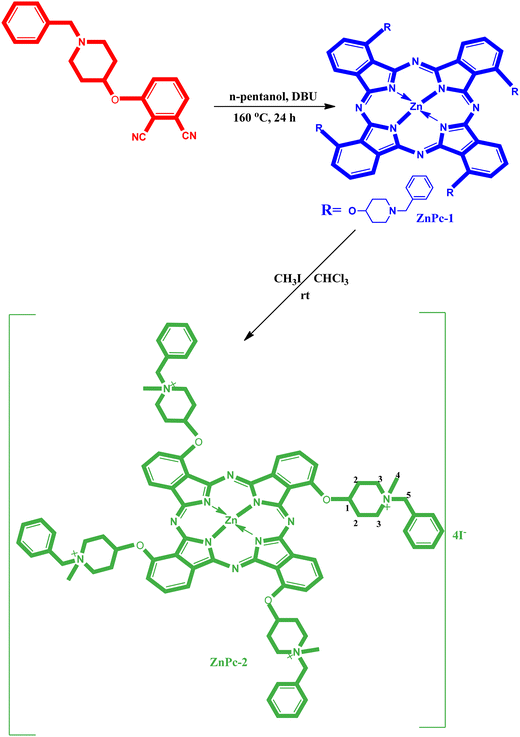
3-[(1-Benzylpiperidin-4-yl)oxy]phthalonitrile and 2(3),9(10),16(17),23(24)-tetrakis-[(1-benzylpiperidin-4-yl)oxy]phthalocyaninato zinc(II) (ZnPc-1) were synthesized according to literature methods, as mentioned in the Material and method section.30,31Fig. 1 shows the general synthesis of 2(3),9(10),16(17),23(24)-tetrakis-[(N-methyl-(1-benzylpiperidin-4-yl)oxy)phthalocyaninato]zinc(II) iodide (ZnPc-2). ZnPc-2 was synthesized by the reaction of ZnPc-1 with CH3-I in CHCl3. Quaternization of ZnPc-1 with an excess of methyl iodide in chloroform led to water soluble tetra cationic Zn phthalocyanine. After the reaction contents were stirred at room temperature for 72 hours, the dark green crude product was seen precipitated at the bottom of the reaction vessel. The crude product was filtered and washed three times with chloroform, ethanol, and diethyl ether. When looking at the infrared spectra of 2(3),9(10),16(17),23(24)-tetrakis-[(1-benzylpiperidin-4-yl)oxy]phthalocyaninato zinc(II) (ZnPc-1) and its water-soluble derivative (ZnPc-2), it can be seen that both are similar. Both of them have aromatic and aliphatic carbon resonance peaks and have no C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N sharp resonance peak around 2230 cm−1 that is identical to the peak for 3-[(1-benzylpiperidin-4-yl)oxy]phthalonitrile as the starting molecule. The FT-IR spectra of 2(3),9(10),16(17),23(24)-tetrakis-[(N-methyl-(1-benzylpiperidin-4-yl)oxy)phthalocyaninato]zinc(II) iodide (ZnPc-2) has aromatic carbon resonance peak at 3058 cm−1 and aliphatic carbon resonance peaks at 2858–2810. The 1H-NMR data of ZnPc-2 in DMSO showed signals due to the appearance of aromatic protons at 7.92–7.79 and 7.68–7.45. The positions of the aliphatic protons in the structure of the complex are numbered on one substituent (Fig. 1). CH3 groups bonded to N+ atom of ZnPc-2 are indicated as a singlet at 3.12 ppm. Other protons that have 4 different chemical environments and their chemical shift values are respectively 4.65–4.48 (m, 4H, O-CH(1)), 4.34 (s, 8H, +N-CH2(5)), 3.80 (t, J: 6.8, 16H, +N-CH2(3)), 1.90–168 (m, 16H, Aliph. CH2(2)). The molecular ion peaks of ZnPc-2 were seen at m/z = 348.766 [M − 4I]4+ (ESI Fig. 1†). The UV-Vis spectrum of ZnPc-2 was measured in DMSO and UV-vis spectrum of metallophthalocyanine is quite characteristic due to the strong π → π* transition in the phototherapeutic window (600–800 nm). In the UV-Vis spectrum of ZnPc-2, the intense single band absorption of π → π* transition was observed at 685 nm (ESI Fig. 2†).
N sharp resonance peak around 2230 cm−1 that is identical to the peak for 3-[(1-benzylpiperidin-4-yl)oxy]phthalonitrile as the starting molecule. The FT-IR spectra of 2(3),9(10),16(17),23(24)-tetrakis-[(N-methyl-(1-benzylpiperidin-4-yl)oxy)phthalocyaninato]zinc(II) iodide (ZnPc-2) has aromatic carbon resonance peak at 3058 cm−1 and aliphatic carbon resonance peaks at 2858–2810. The 1H-NMR data of ZnPc-2 in DMSO showed signals due to the appearance of aromatic protons at 7.92–7.79 and 7.68–7.45. The positions of the aliphatic protons in the structure of the complex are numbered on one substituent (Fig. 1). CH3 groups bonded to N+ atom of ZnPc-2 are indicated as a singlet at 3.12 ppm. Other protons that have 4 different chemical environments and their chemical shift values are respectively 4.65–4.48 (m, 4H, O-CH(1)), 4.34 (s, 8H, +N-CH2(5)), 3.80 (t, J: 6.8, 16H, +N-CH2(3)), 1.90–168 (m, 16H, Aliph. CH2(2)). The molecular ion peaks of ZnPc-2 were seen at m/z = 348.766 [M − 4I]4+ (ESI Fig. 1†). The UV-Vis spectrum of ZnPc-2 was measured in DMSO and UV-vis spectrum of metallophthalocyanine is quite characteristic due to the strong π → π* transition in the phototherapeutic window (600–800 nm). In the UV-Vis spectrum of ZnPc-2, the intense single band absorption of π → π* transition was observed at 685 nm (ESI Fig. 2†).
3.2. UV–Vis absorption spectroscopy
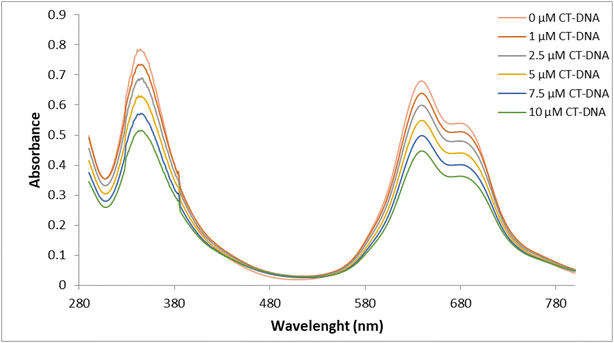
UV-Vis absorption spectroscopy is one of the most widely used methods to evaluate the interaction of DNA with metal complexes. In this technique, changes in the UV-Vis spectrum, increase or decrease in absorbance and/or red or blue shifts in wavelength, are monitored during the interaction of a complex with DNA.43 Hypochromicity, characteristic of intercalation, is generally attributed to the interaction between the electronic states of the chromophores and the electronic states of the DNA bases, while the red shift has been associated with the decrease in the energy gap between the highest and the lowest molecular orbitals after binding of the complex to DNA.44,45 The absorption spectra of the ZnPc-2 complex in the absence and presence of CT-DNA were recorded from 300 to 800 nm and observed in Fig. 2. It is seen in the figure that the characteristic absorption bands of ZnPc-2, at 344 nm (B band) and 639 nm (Q band), respectively, decreased with increasing concentrations of CT-DNA while the position of the peaks did not change significantly. The absorption spectrum of ZnPc-2 exhibited hypochromism of about 34% with a slight red shift of 2 nm in the B band and no shift in the Q band. Since it is generally accepted that hypochromism indicates a strong interaction between the chromophore and DNA bases,46 the absorption spectrum of ZnPc-2 with CT-DNA indicates a close proximity of ZnPc-2 to the CT-DNA bases. This proximity can also be attributed to the electrostatic interaction between the positively charged ZnPc-2 and the negatively charged DNA strand. However, the lack of red shift in the Q band can suggest that the binding mode is partial intercalation. The macrocyclic and bulky structure of ZnPc-2 can also support partial intercalation.44The absorbance values were used to construct the plot of [DNA]/(εa − εf) versus [DNA] (Fig. 3). The plot was linear and the Kb value was obtained as 7.5 × 104 M−1. While a large binding constant may indicate that the molecule has a strong interaction with CT-DNA, a smaller binding constant may indicate that the interaction of the molecule with DNA is weaker.47 However, in the literature, it has been observed that metal complexes that intercalate with DNA have different Kb values.
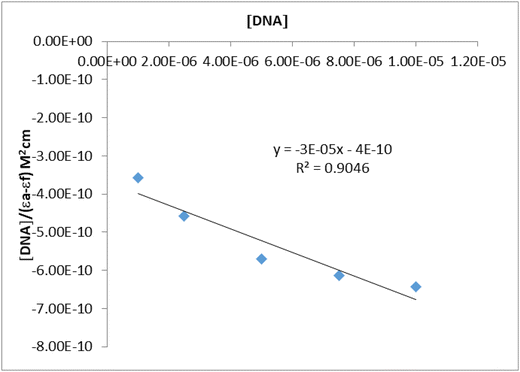 | ||
| Fig. 3 Plot of [DNA]/(εa − εf) vs. [DNA] for the absorption titration of CT-DNA with ZnPc-2 complex. | ||
Two water soluble quaternized Zn(II)Pc molecules showed a red shift and hypochromism as a result of DNA titration with Kb values of 1.13 × 104 and 5.2 × 103 M−1, so the authors suggested the compounds bind to DNA with the intercalation binding mode.48 The peripherally 2,10,16,24-tetrakis dimethyl 5-(phenoxy)-isophthalate-substituted Ni(II) phthalocyanine compound binds to CT-DNA via partial intercalation binding mode because of the hypochromism and the blue shift in UV/Vis absorption spectra shows a Kb value of 2.17 × 106 M−1.49 The silicon(IV) phthalocyanine complexes, showing hypochromism and slight red shift, interacted with DNA via the intercalative mode with Kb values of 1.25 × 104 M−1 and 1.13 × 104.50 Homodinuclear macrocyclic complexes of copper(II) and zinc(II) with CT-DNA showed hypochromism attributed to a partial intercalation mode. The intrinsic binding constant Kb of complexes were determined and found to be 4.25 × 104 M−1and 3.0 × 104 M−1, respectively.45 The Eu(III) complex strongly binds with CT-DNA with a large hypochromism of the absorption spectrum, presumably via an intercalation mechanism (Kb = 2.49 × 104 M−1).51 The Cu(II) complex with a Schiff base, showing a decrease in absorbance intensity without a shift, binds to DNA in an intercalative binding mode.52 The Zn(II) phthalocyanine derivative showed a hypochromic effect (25.98%) without any shift upon the addition of increasing concentrations of CT-DNA. Also, the intrinsic binding constant (Kb) of the compound was determined to be 2.45 × 104 M−1. UV–Vis spectroscopy studies showed that the compound bound to CT-DNA via non-covalent interaction.53
3.3. Thermal denaturation profile of DNA
To investigate the binding pattern of the ligand and CT-DNA, thermal denaturation experiments were conducted. The process of separating the DNA strands and completely disrupting the double-helical structure is referred to as denaturation. The temperature at which half of the total base pairs have separated is defined as the melting temperature (Tm) of DNA.54Tm is associated with the stability of the double helix and varies depending on the binding strength of the compound to DNA. Therefore, thermal denaturation studies serve as a technique used to demonstrate the stability of the structure formed as a result of binding. DNA melting studies provide advanced evidence of the intercalation of complexes into the DNA helix. These experiments are employed to distinguish between different binding modes, such as intercalation or non-intercalation binding. It is known that, as a result of the intercalation of small molecules into the double helix of DNA, the melting temperature increases due to the DNA double-stranded structure's stability. However, in non-intercalation modes such as groove and electrostatic binding, there is no significant alteration in the DNA's melting temperature.Under our experimental conditions, the Tm value for CT-DNA in the absence of ZnPc-2 complex was 76.50 °C, while the Tm value of the ZnPc-2-CT-DNA solution was 86.12 °C (Fig. 4). The addition of ZnPc-2 to CT-DNA resulted in a significant increase in the melting temperature (9.62 °C). A DNA intercalator, such as ethidium bromide, typically enhances the stability of the double helix, making thermal denaturation more challenging. The ΔTm value of CT-DNA in the presence of classical intercalators such as ethidium bromide was reported to be 11.2 °C.55 The significant increase observed in the Tm value of the ZnPc-2-CT-DNA solution supports the notion that ZnPc-2 can bind to CT-DNA in the intercalative binding mode.
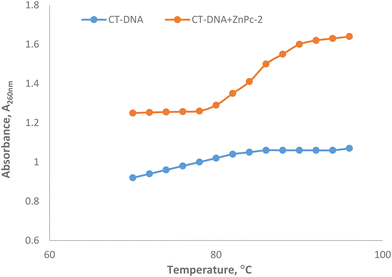 | ||
| Fig. 4 Thermal denatutation curves of CT-DNA in the absence and presence of ZnPc-2 in 5 mM Tris-HCl/50 mM NaCl buffer (pH 7.2). | ||
3.4. DNA cleavage activity
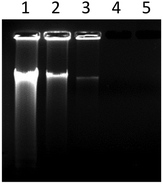
Nucleic acid chains can be cleaved either enzymatically or non-enzymatically. Non-enzymatic cleavage can occur by either the hydrolysis of phosphodiester linkages, called hydrolytic cleavage, or by the oxidative cleavage of deoxyribose residues. Hydrolytic cleavage is mostly produced by strong Lewis acidic metal complexes containing transition metals such as Zn(II), Cu(II) and Co(III). This process occurs through a metal–phosphate intermediate that facilitates the hydrolysis of the nucleic acid phosphate backbone. So, the design and synthesis of metal complexes that can hydrolytically cleave the phosphodiester bond are of great importance in elucidating the catalytic mechanisms of natural nucleases and in the development of nucleic acid-targeted drugs.56–58The ability of the complex to perform CT-DNA cleavage was examined by agarose gel electrophoresis. Fig. 5 showed the electrophoretic band profile of CT-DNA incubated at different complex concentrations. It was seen from the figure that complex concentrations of 50 and 100 μM caused more than 70 and 90% degradation of CT-DNA (65 ng), respectively, while a complex concentration of 250 μM completely degraded the DNA. The results showed that ZnPc-2 exhibited a concentration-dependent DNA degradation effect and the nuclease activity potential of the complex was effective even at low concentrations.
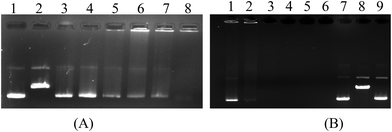
Metal compounds are known to cause random breaks in one of the double-stranded DNA molecules. However, a break on the DNA converts the supercoiled form of plasmid DNA (form I) to the open circular form (form II), and after cleavage of the second strand, to the linear form (form III). The pBR322 plasmid DNA cleavage activity of ZnPc-2 was also studied by agarose gel electrophoresis.59 The supercoiled pBR322 plasmid DNA was incubated with varying concentrations of ZnPc-2. The restriction endonuclease EcoRI was used to identify the linear form of pBR322 plasmid DNA.60 The agarose gel image showing the degradation efficiency of the complex on pBR322 DNA is displayed in Fig. 6. It can be seen from Fig. 6B that DNA form conversion could not be detected; instead, it was determined that 20–25 μM concentration of the complex significantly degraded plasmid DNA, while 50 μM concentration completely degraded it. Additionally, it was determined that the complex showed high nuclease activity even at low concentrations (Fig. 6A, lanes 3–5). Ultimately, pBR322 DNA was degraded into small fragments of various sizes that could not be determined in our study. According to this result, it can be said that ZnPc-2 demonstrated significant cleavage effect because DNA forms were fully cut into little parts and lost. In addition to these studies, DNA degradation efficiencies of the complex in the presence of some reducing/oxidizing agents and radical scavengers were also performed. The results from these experiments showed no change in the DNA cleavage activity of the complex (ESI Fig. 3 and 4†) compared to the results obtained without external additives. Since ZnPc-2 has DNA cleaving activity in the absence of any reducing/oxidizing agents and in the presence of radical scavengers, it can be concluded that the DNA cleavage reaction proceeds by hydrolysis. Due to the high nuclease activity of ZnPc-2, it can be a suitable agent for the hydrolytic cleavage of DNA.
3.5. Molecular docking
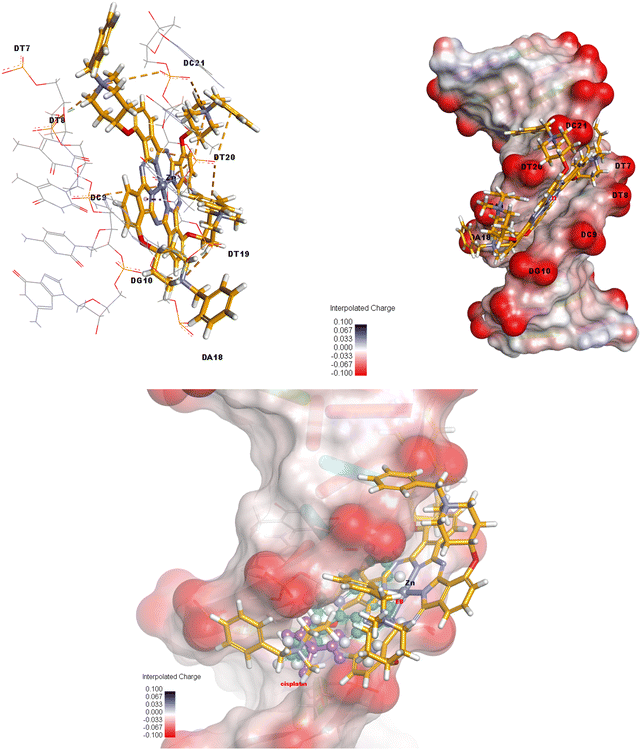
The three-dimensional molecule of the synthesized ZnPc-2 complex, which was structurally and energetically optimized with the Gaussian09 software, is presented in Fig. 7. Then, as a result of the interaction of DNA and ZnPc-2 structure, it shows a binding energy value of −13.35 kcal mol−1, as shown in Table 1.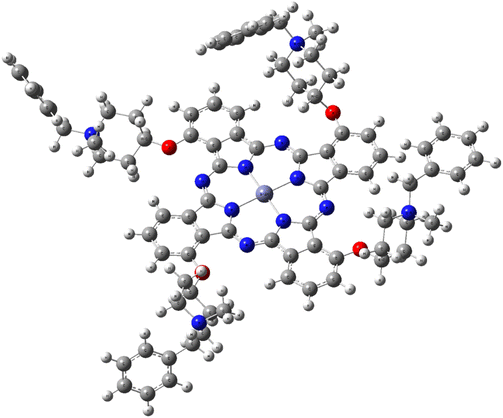 | ||
| Fig. 7 3D optimized structure of the ZnPc-2 complex on the DFT/B3LYP/LAND2DZ basis set of G09 software. | ||
| Compound | Binding energy (kcal mol−1) |
|---|---|
| ZnPc-2 | −13.35 |
| EB | −7.78 |
| Cisplatin | −6.21 |
The relevant complex showed a tendency to bind by intercalation between the base pairs of the DNA target. In other words, two electrostatic interactions occur between DC9 and DT20 nucleotides of the DNA and Zn metal of the ZnPc-2 complex, and six electrostatic interactions occur between G10, DT19, DT20, DC21, DT8 and DC21 nucleotides and N atoms of the related compound. Also, ZnPc-2 forms six hydrogen bonds with DT20, DA18, DT19, DG10 and DT7 in the target model, and forms three electrostatic and one hydrophobic interactions with the target's DT20 and DT19 nucleotides. Fig. 8 exhibits 3D interactions and the interpolated charge surface of the ZnPc-2-DNA complex. ESI Fig. 5 and 6† present 3D images of the control compounds which are ethidium bromide and cisplatin. More detailed information on the interaction distance and types also is presented in ESI Table 1 in the ESI.†
3.6. Effect of ZnPc-2 on α-amylase activity
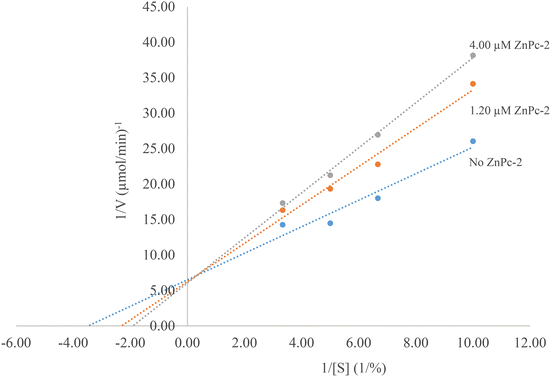
To investigate the effect of ZnPc-2 on α-amylase activity, enzyme activities were determined in the presence of different concentrations of the complex. Afterwards, relative activities were plotted against complex concentrations. The ZnPc-2 concentration causing a 50% decrease of α-amylase activity was defined as the IC50 value. IC50 values were determined to be 3.97 ± 0.32 μM and 1.64 ± 0.36 μM for ZnPc-2 and acarbose, respectively. Considering this result, it can be said that ZnPc-2 is a moderate α-amylase inhibitor.Inhibition kinetic constants and the inhibition type of ZnPc-2 were determined with two different concentrations of the complex (1.20 and 4.00 μM) using varying concentrations of soluble starch (Fig. 9). The Lineweaver–Burk plot showed that the Km value gradually increased and Vmax remained unchanged with increasing ZnPc-2 concentration indicating a competitive inhibition. Furthermore, the inhibition constant (Ki) was determined to be 6.92 μM.
Only a limited number of studies have investigated the effects of phthalocyanines on α-amylase. In one of these studies, the effect of water soluble Co(II) and Zn(II) phthalocyanines (QP4P-1 and QP4P-2) on porcine pancreatic α-amylase was tested and IC50 was determined to be 128.2 μM for QP4P-2 molecule. In the study, the IC50 value for acarbose was found to be 6.9 μM.61 It was also reported that newly synthesized ZnPc, CoPc and PbPc phthalocyanine molecules inhibited α-amylase with IC50 values of 34.0 μM, 30.3 μM and 7.3 μM, respectively. The inhibition type was given as mixed-type with Ki = 4.85 μM for PbPc.62 As a result, it can be easily said that this study is one of the first studies on α-amylase inhibition by phthalocyanines.
3.7. Effect of ZnPc-2 on α-glucosidase activity
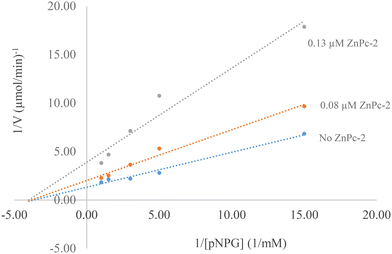
The study found that ZnPc-2 significantly inhibited the activity of α-glucosidase in a dose-dependent manner. The IC50 value of ZnPc-2 for α-glucosidase activity was determined to be 0.07 ± 0.002 μM. Also, IC50 value for acarbose was 237.241 ± 1.80 μM. Based on the results of the study, it can be concluded that ZnPc-2 is a highly effective α-glucosidase inhibitor. This suggests that ZnPc-2 has the potential to be used as a therapeutic agent for the treatment of type 2 diabetes.Inhibition kinetic constants and the inhibition type of ZnPc-2 were determined in the presence of 0.08 and 0.13 μM ZnPc-2 using varying concentrations of pNPG (Fig. 10). The Lineweaver–Burk plot indicated noncompetitive inhibition. This means that ZnPc-2 binds to a site on α-glucosidase that is different from the active site, preventing the enzyme from functioning properly. Also, the inhibition constant (Ki) was determined to be 0.07 μM.
There are several studies involving the effect of phthalocyanines on α-glucosidase activity. In one study, it was reported that IC50 values of four novel phthalocyanines complexes and acarbose were in the range of 2.23–9.10 μM and 22.80 μM, respectively, for α-glycosidase inhibition. The Ki values were in the range of 3.41–10.08 μM.63 In another study, novel peripheral phthalocyanines and their water-soluble hydrochloride derivatives had inhibitory effects on α-glycosidase. They have IC50 values in the range of 11.22–22.81 μM and Ki in the range of 9.13–24.31 μM.64 According to Güzel et al. all of the seven phthalocyanine complexes used had an inhibitory effect on α-glycosidase activity (IC50 in the range of 0.82–1.94 nM and Ki in the range of 1.56–14.78 μM) more efficient than acarbose (IC50: 0.38 μM).65
When the results of the study are wholly considered, it is seen that ZnPc-2 is a mild α-amylase and a very good α-glucosidase inhibitor. This is a desired situation. The side effects (diarrhea, abdominal pain etc.) of current α-glucosidase inhibitors used as antidiabetic drugs are thought to be caused by the fermentation of undigested carbohydrates that occurs as a result of strong α-amylase inhibition. Therefore, it is important to find new molecules having strong inhibition of α-glucosidase and low inhibition of α-amylase.66
3.8. Structure–activity relationship of ZnPc-1 and ZnPc-2
The main limitation in the applications of metallophthalocyanines is their low solubility in common organic solvents.67 To overcome this situation, we used 1-benzylpiperidin-4-ol groups containing N atoms that can be quaternized in this work. Because of the hydrophilic system of the blood, water solubility is very important in drug injection. Additionally, diamagnetic ions (such as Zn2+) contained in the structure of phthalocyanines enhance the physical and chemical properties of these compounds for various applications.30Although tekrakis-[(1-benzylpiperidin-4-yl)oxy]phthalocyaninato zinc(II) (ZnPc-1) and its water-soluble derivative (ZnPc-2) are structurally similar in terms of their basic skeletal structures, the chemical variability of the related complexes causes differences in their biological activities.
In particular, the distribution coefficient of a drug strongly affects how easily the drug can reach its intended target in the body, how strong an effect it will have once it reaches its target, and how long it will remain in the body in its active form.68 Therefore, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of a molecule is a decision-making criterion used by medicinal chemists in preclinical drug discovery, for example, in assessing the drug-likeness of drug candidates.69 Therefore, the hydrophobicity of a compound is an important determinant of how drug-like it is.
P of a molecule is a decision-making criterion used by medicinal chemists in preclinical drug discovery, for example, in assessing the drug-likeness of drug candidates.69 Therefore, the hydrophobicity of a compound is an important determinant of how drug-like it is.
On the other hand, in the context of how the drug affects the body, the hydrophobic effect is the main driving force in the binding of drugs to their receptor targets. Hydrophobic drugs, however, tend to be more toxic because they are generally retained longer, have a wider distribution in the body, and are slightly less selective in binding to proteins. Therefore, making the drug as hydrophilic as possible is recommended while maintaining sufficient binding affinity to the therapeutic protein target. In cases where a drug reaches the target, the ideal distribution coefficient for the drug is typically intermediate (neither too lipophilic nor too hydrophilic). This situation manifests itself in both experimental and theoretical studies in our study.
4. Conclusion
In conclusion, non-peripherally tetra-substituted water soluble Zn(II) phthalocyanine was synthesized and characterized by spectral methods. The DNA binding and cleavage properties of the complex were studied by absorption spectroscopy, thermal denaturation and agarose gel electrophoresis measurements. The results of UV-Visible absorption titration and thermal denaturation measurement indicate that ZnPc-2 interacts with CT-DNA in a non-covalent manner. Gel electrophoresis results show that ZnPc-2 binds strongly to DNA molecules and exhibits effective nuclease activity. Moreover, the interaction of ZnPc-2 with DNA was investigated and evaluated by molecular docking, one of the computational methods. As a result of the calculations, it was revealed that the relevant compound exhibited a high binding tendency with the target and the reason for this was the interaction of the ligament between the helices in the DNA biomolecule. According to the docking simulation results, the complex is located in the minor groove of DNA. Both experimental and theoretical results show that the complex can interact with DNA via three binding modes: electrostatic, minor groove, and partial insertion. Consequently, due to its strong DNA binding and nuclease activity, ZnPc-2 may be suitable for antimicrobial and anticancer applications after further toxicological test systems. Additionally, studies of the antidiabetic activity of ZnPc-2 suggest that it may be a valuable new treatment for type 2 diabetes. However, more research is needed to confirm these findings in vivo and to determine the long-term safety and efficacy of ZnPc-2 in humans.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This study was supported by The Research Fund of Karadeniz Technical University (project no: FHD-2022-9976) Trabzon, Türkiye.The numerical calculations reported in this paper were partially performed at TUBITAK ULAKBIM, High Performance and Grid Computing Center (TRUBA resources).
We thank Prof. Dr Ali Osman Beldüz (Karadeniz Technical University, Faculty of Science, Department of Biology, Trabzon, Türkiye) for supplying pBR322 DNA.
References
- A. Reheman, S. Hu, L. Cao, D. Xie, G. Yan and J. Wang, Liquid-crystalline behaviour and electrorheological effect of phthalocyanine-based ionic liquid crystals, Liq. Cryst., 2021, 48, 1321–1330 CrossRef CAS
.
- B. Kadem, A. Hassan, M. Göksel, T. Basova, A. Şenocak, E. Demirbaş and M. Durmuş, High performance ternary solar cells based on P3HT:PCBM and ZnPc-hybrids, RSC Adv., 2016, 6, 93453–93462 RSC
.
- A. D. Fovo, M. Oujja, M. Sanz, A. Hernandez, M. V. Canamares, M. Castillejo and R. Fontana, Multianalytical non-invasive characterization of phthalocyanine acrylic paints through spectroscopic and non-linear optical techniques, Spectrochim. Acta, Part A, 2019, 208, 262–270 CrossRef CAS PubMed
.
- M. Benhaliliba, A. B. Ahmed and A. Ayeshamariam, Optical-electrical properties and global reactivity analysis of aluminum phthalocyanine chloride, J. Porphyrins Phthalocyanines, 2022, 26, 732–740 CrossRef CAS
.
- W. Fan, Z. Duan, W. Liu, R. Mehmood, J. Qu, Y. Cao, X. Guo and J. Zhong, Rational design of heterogenized molecular phthalocyanine hybrid single-atom electrocatalyst towards two-electron oxygen reduction, Nat. Commun., 2023, 14, 1426 CrossRef CAS PubMed
.
- E. T. Saka and K. Tekintas, Light driven photodegradation of 4-nitrophenol with novel Co and Cu phthalocyanine in aqueous media, J. Mol. Struct., 2020, 1215, 128189 CrossRef
.
- E. T. Saka, H. Yalazan, Z. Biyiklioglu, H. Kantekin and K. Tekintas, Synthesis, aggregation, photocatalytical and electrochemical properties of axially 1-benzylpiperidin-4-oxy units substituted silicon phthalocyanine, J. Mol. Struct., 2020, 1199, 126994 CrossRef CAS
.
- Z. Dong, X. Kong, Y. Wu, J. Zhang and Y. Chen, High-sensitive room-temperature NO2 sensor based on a soluble n-type phthalocyanine semiconductor, Inorg. Chem. Commun., 2017, 77, 18–22 CrossRef CAS
.
- H. Y. Yenilmez, N. Farajzadeh, G. Tollu, N. G. Kuşculu, D. Bahar, S. Özdemir and Z. A. Bayır, Silicon Phthalocyanines as Anti-infectious, Antioxidant and Anticancer Agents, ChemistrySelect, 2023, 8(19), e202300856 CrossRef CAS
.
- E. T. Saka, R. Z. Uslu Kobak, H. Alp, G. Sarkı, A. Koca and H. Kantekin, Electrochemical and spectroelectrochemical properties of new metal free, nickel(II), lead(II) and zinc(II) phthalocyanines, Synth. Met., 2016, 217, 295–303 CrossRef CAS
.
- I. Acar, E. T. Saka, S. Topçu, Z. Biyiklioglu, H. Kantekin and A. A. Kamiloglu, Synthesis and electrochemistry of new octa-substituted metal-free and metallophthalocyanines, J. Coord. Chem., 2015, 68, 1847–1858 CrossRef CAS
.
- H. Yalazan, İ. Ömeroğlu, G. Çelik, H. Kantekin and M. Durmuş, Fluorinated pyrazoline-linked axial silicon phthalocyanine, alpha (α) and beta (β) zinc phthalocyanines on photophysicochemical properties, Inorg. Chim. Acta, 2023, 551, 121480 CrossRef CAS
.
- B. Ertem, H. Yalazan, Ö. Güngör, G. Sarkı, M. Durmuş, E. T. Saka and H. Kantekin, Synthesis, structural characterization and investigation on photophysical and photochemical properties of new phthalocyanines, J. Lumin., 2018, 204, 464–471 CrossRef CAS
.
- E. Darout, K. F. McClure and V. Mascitti, Synthesis of spirofuranopyrimidine–piperidines, Tetrahedron, 2012, 68, 4596–4599 CrossRef CAS
.
- A. R. Mohite, P. R. Sultane and R. G. Bhat, Thiourea-mediated halogenation of alcohols, Tetrahedron Lett., 2012, 53, 30–35 CrossRef CAS
.
- E. Vitaku, D. T. Smith and J. T. Njardarson, Analysis of the Structural Diversity, Substitution Patterns, and Frequency of Nitrogen Heterocycles among U.S. FDA Approved Pharmaceuticals, J. Med. Chem., 2014, 57, 10257–10274 CrossRef CAS PubMed
.
- R. D. Taylor, M. MacCoss and A. D. G. Lawson, Rings in Drugs, J. Med. Chem., 2014, 57, 5845–5859 CrossRef CAS PubMed
.
- S. S. Sajadikhah, M. T. Maghsoodlou, N. Hazeri, S. M. Habibi-Khorassani and A. C. Willis, One-pot five-component synthesis of highly functionalized piperidines using oxalic acid dihydrate as a homogenous catalyst, Chin. Chem. Lett., 2012, 23, 569–572 CrossRef CAS
.
- C. C. Jayme, I. R. Calori, E. M. F. Cunha and A. C. Tedesco, Evaluation of aluminum phthalocyanine chloride and DNA interactions for the design of an advanced drug delivery system in photodynamic therapy, Spectrochim. Acta, Part A, 2018, 201, 242–248, DOI:10.1016/j.saa.2018.05.009
.
- N. Raman, J. Dhaveethu Raja and A. Sakthivel, Synthesis, spectral characterization of Schiff base transition metal complexes: DNA cleavage and antimicrobial activity studies, J. Chem. Sci., 2007, 119(4), 303–310 CrossRef CAS
.
- N. C. López Zeballos, G. A. Gauna, M. C. García Vior, J. Awruch and L. E. Dicelio, Interaction of cationic phthalocyanines with DNA. Importance of the structure of the substituents, J. Photochem. Photobiol., B, 2014, 136, 29–33, DOI:10.1016/j.jphotobiol.2014.04.013
.
- M. Özçesmeci, Ö. B. Ecevit, S. Sürgün and E. Hamuryudan, Tetracationic fluorinated zinc(II) phthalocyanine: synthesis, characterization and DNA-binding properties, Dyes Pigm., 2013, 96, 52–58, DOI:10.1016/j.dyepig.2012.06.018
.
- M. Pitié, G. Pratviel, J. Bernadou and B. Meunier, Preferential hydroxylation by the chemical nuclease meso-tetrakis-(4-N-methylpyridiniumyl) porphyrinatomanganese III pentaacetate/KHSO5 at the 5′ carbon of deoxyriboses on both 3′ sides of three contiguous A.T base pairs in short double-stranded oligonucleotides, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 3967–3971, DOI:10.1073/pnas.89.9.3967
.
- L. Özalp, S. Sağ Erdem, B. Yüce-Dursun, Ö. Mutlu and M. Özbil, Computational insight into the phthalocyanine-DNA binding via docking and molecular dynamics simulations, Comput. Biol. Chem., 2018, 77, 87–96, DOI:10.1016/j.compbiolchem.2018.09.009
.
- O. Kurt, I. Ozcesmeci, B. S. Sesalan and M. B. Kocak, The synthesis and investigation of binding properties of a new water soluble hexadeca zinc(II) phthalocyanine with bovine serum albumin and DNA, New J. Chem., 2015, 39, 5767–5775, 10.1039/c5nj00933b
.
- M. M. Alishah, H. Y. Yenilmez, I. Ozcesmeci, B. S. Sesalan and Z. A. Bayir, Synthesis of quaternized zinc(II) and cobalt(II) phthalocyanines bearing pyridine-2-yl-ethynyl groups and their DNA binding properties, Turk. J. Chem., 2018, 42, 572–585, DOI:10.3906/kim-1707-54
.
- A. A. Kuznetsova, A. E. Lukyanets, L. I. Solovyeva, D. G. Knorre and O. S. Fedorova, DNA-binding and Oxidative Properties of Cationic Phthalocyanines and Their Dimeric Complexes with Anionic Phthalocyanines Covalently Linked to Oligonucleotides, J. Biomol. Struct. Dyn., 2008, 26(3), 307–319, DOI:10.1080/07391102.2008.10507246
.
- F. Saleem, Kanwal, K. M. Khan, S. Chigurupati, Y. Andriani, M. Solangi, S. Hameed, A. A. M. A. Hafez, F. Begum, M. A. Lodhi, M. Taha, F. Rahim and T. S. Bin Tengku Muhammad, Dicyanoanilines as potential and dual inhibitors of α-amylase and α-glucosidase enzymes: synthesis, characterization, in vitro, in silico, and kinetics studies, Arabian J. Chem., 2022, 15, 103651, DOI:10.1016/j.arabjc.2021.103651
.
- X. Peng, K. Liu, X. Hu, D. Gong and G. Zhang, Hesperetin-Cu(II) complex as potential α-amylase and α-glucosidase inhibitor: Inhibition mechanism and molecular docking, Spectrochim. Acta, Part A, 2023, 290, 122301, DOI:10.1016/j.saa.2022.122301
.
- G. Dilber, M. Durmus and H. Kantekin, Investigation of the photophysical and photochemical behavior of substituted zinc phthalocyanines and their water-soluble quaternized derivatives, Turk. J. Chem., 2017, 41, 917–930 CrossRef CAS
.
- E. T. Saka and Z. Biyiklioglu, Co(II) and Fe(II) phthalocyanines:
synthesis, investigation of their catalytic activity towards phenolic compounds and electrochemical behaviour, Appl. Organomet. Chem., 2015, 29, 392–399 CrossRef CAS
.
-
D. D. Perrin and W. L. F. Armarego, Purification of Laboratory Chemicals, Pergamon Press, Oxford, 2nd edn, 1989 Search PubMed
.
- A. Wolfe, G. H. Shime and T. Meehan, Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA, Biochemistry, 1987, 26, 6392–6396, DOI:10.1021/bi00394a013
.
- K. Inan, M. Kacagan, A. Ozer, A. O. Belduz and S. Canakci,
Algoriphagus trabzonensis sp. nov., isolated from freshwater, and emended description of Algoriphagus alkaliphilus, Int. J. Syst. Evol. Microbiol., 2015, 65, 2234–2240, DOI:10.1099/ijs.0.000246
.
- N. Saglam, A. Colak, K. Serbest, S. Karaböcek and S. Guner, DNA Hydrolysis by Homo- and Heteronuclear Cu(II)–Ni(II) Complexes of Two Diester-type Ligands, Monatsh. Chem., 2004, 135, 1023–1031, DOI:10.1007/s00706-004-0178-6
.
- X. Y. Meng, H. X. Zhang, M. Mezei and M. Cui, Molecular docking: a powerful approach for structure-based drug discovery, Curr. Comput.-Aided Drug Des., 2011, 7, 146–157, DOI:10.2174/157340911795677602
.
-
M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision E.01, Gaussian, Inc., Wallingford CT, 2009 Search PubMed
.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem., 2009, 30, 2785–2791, DOI:10.1002/jcc.21256
.
- L. Miller, Use of dinitrosalicylic acid reagent for determination of reducing sugar, Anal. Chem., 1959, 31, 426–428, DOI:10.1021/ac60147a030
.
- E. A. H. Mohamed, M. J. A. Siddiqui, L. F. Ang, A. Sadikun, S. H. Chan, S. C. Tan, M. Z. Asmawi and M. F. Yam, Potent α-glucosidase and α-amylase inhibitory activities of standardized 50% ethanolic extracts and sinensetin from Orthosiphon stamineus Benth as anti-diabetic mechanism, BMC Complementary Altern. Med., 2012, 12, 1–7 CrossRef PubMed
.
- H. Shen, J. Wang, J. Ao, Y. Hou, M. Xi, Y. Cai, M. Li and A. Luo, Structure-activity relationships and the underlying mechanism of α-amylase inhibition by hyperoside and quercetin: Multi-spectroscopy and molecular docking analyses, Spectrochim. Acta, Part A, 2023, 285, 121797, DOI:10.1016/j.saa.2022.121797
.
- Y. Yin, S. Sun, H. Wang, M. Guo, Z. Li, C. Lv, Z. Yang and W. Wang, Mechanism of interaction between urolithin A and α-glucosidase: Analysis by inhibition kinetics, fluorescence spectroscopy, isothermal titration calorimetry and molecular docking, J. Mol. Struct., 2023, 1286, 135567, DOI:10.1016/j.molstruc.2023.135567
.
- S. Aduri, W. R. Reddy CH and B. Sireesha, Synthesis, Characterization, DNA binding, DNA Cleavage and Antibacterial Studies of Ni(II) and Cu(II) Complexes of Pyridoxal Semicarbazone, Pharma Chem., 2017, 9(2), 90–98 CAS
.
- F. Arjmand and M. Aziz, Synthesis and characterization of dinuclear macrocyclic cobalt(II), copper(II) and zinc(II) complexes derived from 2,2,20,20-S,S[bis(bis-N, N-2-thiobenzimidazolyloxalato-1,2-ethane)]: DNA binding and cleavage studies, Eur. J. Med. Chem., 2009, 44, 834–844, DOI:10.1016/j.ejmech.2008.05.006
.
- V. Uma, M. Kanthimathi, T. Weyhermuller and B. U. Nair, Oxidative DNA cleavage mediated by a new copper(II) terpyridine complex: Crystal structure and DNA binding studies, J. Inorg. Biochem., 2005, 99, 2299–2307, DOI:10.1016/j.jinorgbio.2005.08.011
.
- L. Hassani, F. Hakimian, E. Safaei and Z. Fazeli, Antibacterial effect of cationic porphyrazines and anionic phthalocyanine and their interaction with plasmid DNA, J. Mol. Struct., 2013, 1052, 221–227, DOI:10.1016/j.molstruc.2013.07.054
.
- N. C. López Zeballos, G. A. Gauna, M. C. García Vior, J. Awruch and L. E. Dicelio, Interaction of cationic phthalocyanines with DNA. Importance of the structure of the substituents, J. Photochem. Photobiol., B, 2014, 136, 29–33, DOI:10.1016/j.jphotobiol.2014.04.013
.
- S. Demir and F. Yuksel, Novel highly water soluble zinc(II) phthalocyanines: Synthesis, photochemistry and DNA binding behaviours, Inorg. Chim. Acta, 2023, 548, 121373, DOI:10.1016/j.ica.2022.121373
.
- A. Arslantas and M. S. Agirtas, Investigation of DNA Binding Activities of Peripherally 2,10,16,24-Tetrakis Dimethyl 5-(Phenoxy)-Isophthalate-Substituted Ni(II) Phthalocyanine Complex, ChemistrySelect, 2018, 3, 3155–3160, DOI:10.1002/slct.201800572
.
- H. Baş, B. Barut, Z. Biyiklioglu and A. Özel, Synthesis, DNA interaction, topoisomerase I, II inhibitory and cytotoxic effects of water soluble silicon(IV) phthalocyanine and napthalocyanines bearing 1-acetylpiperazine units, Dyes Pigm., 2019, 160, 136–144, DOI:10.1016/j.dyepig.2018.08.005
.
- B. Wang, Z. Y. Yang and T. R. Li, Synthesis, characterization, and DNA-binding properties of the Ln(III) complexes with 6-hydroxy chromone-3-carbaldehyde-(20-hydroxy)benzoyl hydrazone, Bioorg. Med. Chem., 2006, 14, 6012–6021, DOI:10.1016/j.bmc.2006.05.015
.
- P. Mendu, C. Gyana Kumari and R. Ragi, Synthesis, Characterization, DNA Binding, DNA Cleavage and Antimicrobial Studies of Schiff Base Ligand and its Metal Complexes, J. Fluoresc., 2015, 25, 369–378, DOI:10.1007/s10895-015-1520-6
.
- H. Yalazan, B. Barut, B. Ertem, C. Ö. Yalçın, Y. Ünver, A. Özel, I. Ömeroglu, M. Durmus and H. Kantekin, DNA interaction and anticancer properties of new peripheral phthalocyanines carrying tosylated 4-morpholinoaniline units, Polyhedron, 2020, 177, 114319, DOI:10.1016/j.poly.2019.114319
.
- J. Mergny and L. Lacroix, Analysis of thermal melting curves, Oligonucleotides, 2003, 13, 515–537, DOI:10.1089/154545703322860825
.
- T. K. Goswami, S. Gadadhar, A. A. Karande and A. R. Chakravarty, Photocytotoxic ferrocene-appended (L-tyrosine) copper(II)complexes of phenanthroline bases, Polyhedron, 2013, 52, 1287–1298, DOI:10.1016/j.poly.2012.06.018
.
- M. R. Rodríguez, M. J. Lavecchia, B. S. Parajon-Costa, A. C. Gonzalez-Baro, M. R. Gonzalez-Baro and E. R. Cattaneo, DNA cleavage mechanism by metal complexes of Cu(II), Zn(II) and VO(IV) with a schiff-base ligand, Biochimie, 2012, 186, 43–50, DOI:10.1016/j.biochi.2021.04.002
.
- S. G. K.R., B. B. Mathew, C. N. Sudhamani and H. S. Bhojya Naik, Mechanism of DNA binding and cleavage, biomedicine and biotechnology, Biomed. Biotechnol., 2014, 2(1), 1–9, DOI:10.12691/bb-2-1-1
.
- Z. Yu and J. A. Cowan, Metal complexes promoting catalytic cleavage of nucleic acids d biochemical tools and therapeutics, Curr. Opin. Chem. Biol., 2018, 43, 37–42, DOI:10.1016/j.cbpa.2017.10.029
.
- S. Dülger, N. Saglam, A. O. Beldüz, S. Güner and S. Karaböcek, DNA cleavage by homo- and heterotetranuclear Cu(II) and Mn(II) complexes with tetrathioether-tetrathiol moiety, BioMetals, 2000, 13, 261–265, DOI:10.1023/a:1009222705659
.
- N. Saglam, A. Colak, K. Serbest, S. Dülger, S. Güner, S. Karaböcek and A. O. Beldüz, Oxidative cleavage of DNA by homo- and heteronuclear Cu(II)-Mn(II) complexes of an oxime-type ligand, BioMetals, 2002, 15, 357–365 CrossRef CAS PubMed
.
- E. T. Saka, U. Cakmak, C. Akkol and Z. Biyiklioglu, An investigation on photocatalytic and α-amylase inhibitory activities of Co(II) and Zn(II) phthalocyanines, Polyhedron, 2023, 243, 116522, DOI:10.1016/j.poly.2023.116522
.
- I. Değirmencioğlu, F. O. Tuncay, U. Cakmak and Y. Kolcuoglu, The synthesis of novel piperazine-benzodioxole substituted phthalocyanines and investigation of their α-amylase and tyrosinase inhibition properties, J. Organomet. Chem., 2021, 951, 122012, DOI:10.1016/j.jorganchem.2021.122012
.
- A. Günsel, P. Yıldırım, Y. Taslimi, T. Erden, T. Taskin-Tok, H. Pişkin, A. T. Bilgiçli, I. Gülçin and M. N. Yarasir, Cytotoxicity effects and biochemical investigation of novel tetrakis-phthalocyanines bearing 2-thiocytosine moieties with molecular docking studies, Inorg. Chem. Commun., 2022, 138, 109263, DOI:10.1016/j.inoche.2022.109263
.
- A. Günsel, P. Taslimi, G. Y. Atmaca, A. T. Bilgicli, H. Pişkin, Y. Ceylan, A. Erdoğmuş, M. N. Yarasir and I. Gülçin, Novel potential metabolic enzymes inhibitor, photosensitizer and antibacterial agents based on water-soluble phthalocyanine bearing imidazole derivative, J. Mol. Struct., 2021, 1237, 130402, DOI:10.1016/j.molstruc.2021.130402
.
- E. Güzel, U. M. Koçyiğit, P. Taslimi, S. Erkan and O. S. Taskin, Biologically active phthalocyanine metal complexes: Preparation, evaluation of α–glycosidase and anticholinesterase enzyme inhibition activities, and molecular docking studies, J. Biochem. Mol. Toxicol., 2021, 35, 1–9, DOI:10.1002/jbt.22765
.
- M. Santoso, L. L. Ong, N. P. Aijijiyah, F. A. Wati, A. Azminah, R. M. Annuur, A. Fadlan and Z. M. Judeh, Synthesis, α-glucosidase inhibition, α-amylase inhibition, and molecular docking studies of 3, 3-di (indolyl) indolin-2-ones, Heliyon, 2022, 8, e09045 CrossRef CAS PubMed
.
- J. Obirai and T. Nyokong, Synthesis, spectral and electrochemical characterization of mercaptopyrimidine-substituted cobalt, manganese and Zn(II) phthalocyanine complexes, Electrochim. Acta, 2005, 50, 3296–3304, DOI:10.1016/j.electacta.2004.12.003
.
-
N. Bodor and P. Buchwald, Chapter 2.2: Pharmacokinetic Phase: ADME, in Retrometabolic Drug Design and Targeting, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2nd edn, 2012, ISBN 978-1-118-40776-9 Search PubMed
.
- P. D. Leeson and B. Springthorpe, The influence of drug-like concepts on decision-making in medicinal chemistry, Nat. Rev. Drug Discovery, 2007, 6(11), 881–890, DOI:10.1038/nrd2445
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dt01138d |
| This journal is © The Royal Society of Chemistry 2024 |