Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Degradation of PET microplastic particles to monomers in human serum by PETase†
Ximena Lopez-Lorenzo‡
 ab,
David Hueting‡ab,
Eliott Bosshardab and
Per-Olof Syrén
ab,
David Hueting‡ab,
Eliott Bosshardab and
Per-Olof Syrén *ab
*ab
aDepartment of Fibre and Polymer Technology, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, Stockholm, Sweden. E-mail: per-olof.syren@biotech.kth.se
bSchool of Engineering Sciences in Chemistry, Biotechnology and Health, Science for Life Laboratory, KTH Royal Institute of Technology, Stockholm, Sweden
First published on 4th March 2024
Abstract
More than 8 billion tons of plastic waste has been generated, posing severe environmental consequences and health risks. Due to prolonged exposure, microplastic particles are found in human blood and other bodily fluids. Despite a lack of toxicity studies regarding microplastics, harmful effects for humans seem plausible and cannot be excluded. As small plastic particles readily translocate from the gut to body fluids, enzyme-based treatment of serum could constitute a promising future avenue to clear synthetic polymers and their corresponding oligomers via their degradation into monomers of lower toxicity than the material they originate from. Still, whereas it is known that the enzymatic depolymerization rate of synthetic polymers varies by orders of magnitude depending on the buffer and media composition, the activity of plastic-degrading enzymes in serum was unknown. Here, we report how an engineered PETase, which we show to be generally trans-selective via induced fit docking, can depolymerize two different microplastic-like substrates of the commodity polymer polyethylene terephthalate (PET) into its non-toxic monomer terephthalic acid (TPA) alongside mono(2-hydroxyethyl)terephthalate (MHET) in human serum at 37 °C. We show that the application of PETase does not influence cell viability in vitro. Our work highlights the potential of applying biocatalysis in biomedicine and represents a first step towards finding a future solution to the problem that microplastics in the bloodstream may pose.
Introduction
400 Mt of plastic waste is generated per year,1,2 with a large amount of that plastic ending up in landfills and bodies of water. Each year, ca. 3 million tons of plastic debris enters our oceans from rivers. Every day, on average a human is exposed to hundreds of microplastic particles,3 which creates possibilities for such particles to enter our bodies upon inhalation and digestion. Small plastic particles translocate from the gut to body fluids and, further on, to organs.4,5 Recently, studies have shown that plastic particles of ≥700 nm in diameter are present in human whole blood.6 Accordingly, the way synthetic polymers are discarded could pose a health risk to humans.7Smaller plastic particles with a size < 5 mm are referred to as microplastics and are formed during, e.g., biotic processes. A further classification of microplastics is nanoplastics, which are plastic particles smaller than 1 μm. Leslie et al.,6 developed a robust analytical method which allowed for the identification and quantification of plastic particles found in human whole blood, showing that a plastic concentration of 1.6 μg mL−1 was found in their small set of analyzed donors. Polyethylene terephthalate (PET) is one of the most-used types of synthetic polymer and was found in 11 out of the 22 blood samples.6 PET is commonly used for food packaging, textiles, synthetic vascular grafts8 and other products that humans have daily contact with. Currently, there are signs that indicate that plastics have a significant impact on male fertility and on lung epithelial cells.9,10 However, there is a lack of data regarding the toxicological hazards that PET polymer particles pose, as well as data regarding human exposure and the accompanying risks. Additionally, plastic has been found in breast milk, which poses a potential threat for newborn infants.11 Despite some correlations between plastic found in bodily fluids and harmful effects, further understanding of the hazards that microplastic particles pose to human health is required to provide a full picture.5,12
Some toxicological information regarding the monomers that compose PET, terephthalic acid (TPA), bis(2-hydroxyethyl)terephthalate (BHET), mono(2-hydroxyethyl)terephthalate (MHET) and ethylene glycol (EG), is available.13 While some bacteria can metabolize TPA,14 in vivo experiments in rodents have shown that TPA is not metabolized but is rapidly excreted through urine13,15 and is non-hazardous at an uptake concentration below 1000 mg kg−1 a day (data for rats). An in vitro study of human fibroblasts showed that TPA was essentially non-toxic at concentrations up to 200 μg mL−1, while trimeric MHET and its responding monomer (MHET) were toxic at lower concentrations (25 μg mL−1 for MHET). For EG, in vivo studies showed that oral exposure of the monomer in pregnant rodents resulted in increased fetal deaths, external and skeletal malformations, and reduced body weights, when exposed to high doses of EG (≥500 mg per kg per day in mice and ≥1000 mg per kg bw per day in rats).16 However, it seems that these developmental toxicity risks associated with EG are linked to a metabolized form of EG, namely glycolic acid.16,17 As long as the accumulation of EG levels in the body does not overwhelm the oxidative enzymes responsible for converting glycolic acid further (approximately 125 mg kg−1 (ref. 16)) there should not be any developmental toxicity. The concentration of microplastics found in blood is lower than this (1.6 μg mL−1), illuminating the potential avenue to degrade microplastics into monomers with negligible toxicity. Since the possible health risks associated with microplastics can't be determined due to a lack of measured exposure data, there are no conclusions as to what extent these microplastics represent a risk.5 However, the European Environment Agency issued a report called Late Lessons from Early Warnings, highlighting how inaction regarding environmental issues due to a lack of risk data can pose a danger to society.12 For this reason and based on the recent study by Leslie et al.6 showing the occurrence of microplastics in blood, in this study, enzymatic depolymerization of microplastics in human serum was explored for the first time.
Previous research has emphasized the presence of so-called polyesterases (PETases) in the microbiota in saliva18 illuminating how this family of enzymes can likely be compatible with human cells. We hypothesized that PETases, when administered in serum, could enable clearance of microplastics to possibly prevent circulation of potentially harmful particles (Scheme 1a), while at the same time ideally showing low – or even abolished – activity towards semicrystalline synthetic vascular grafts. As microplastic PET shows a high degree of crystallinity due to initial biotic degradation mechanisms19–21 that favor decomposition of the amorphous parts first, such designer enzymes must have a high affinity and activity towards the crystalline parts of PET enriched in the so-called trans conformation22–24 (Scheme 1a, bottom). However, achieving hydrolysis of the crystalline parts of the polymer by PETases can constitute a challenge and often requires pre-treatment and high reaction temperatures.25–28,33 PETases are a subfamily of cutinases that were found in 2016 by Yoshida et al.,28 who discovered a bacterium, Ideonella sakaiensis, growing in plastic landfills. This bacterium can use PET as part of its carbon source thanks to its enzymes IsPETase and MHETase, which can hydrolyse the PET chain into BHET, MHET and TPA, as schematically shown in Scheme 1b. Moreover, recent studies by us have shown how an engineered IsPETase, S238A PETase, is capable of hydrolyzing microplastic PET-particles to TPA at 30 °C without the need for a second auxiliary enzyme.29 We previously found that the S238A PETase is trans selective,29 with significantly lower activity towards gauche polymer chains found in amorphous PET.22,24 This finding was further corroborated herein for model oligomers of the other important plastics polyethylene furanoate (PEF) and polylactide (PLA) (ESI Fig. 1†). Interestingly, induced-fit docking results herein highlight how S238A prefers trans substrate chain conformations, whereas WT shows gauche selectivity. These findings reinforce our hypothesis that S238A would be suitable in degrading PET microplastic substrates that are enriched in trans conformations in serum. However, PETase activity is known to be highly dependent on buffer and salt composition. For instance, Tris–HCl has been shown to inhibit enzymatic activity, as opposed to phosphate buffer,30 resulting in orders of magnitude differences in activity.31 In this study, we found that the use of WT PETase (i.e., IsPETase) and its S238A variant showed effective hydrolysis activity against a model crystalline substrate that resembles PET microplastic, in human serum and at body temperature. Investigation of a second semicrystalline PET-derived substrate with a more uniform particle size (<300 μm) and with a larger molecular weight confirmed enzymatic degradation under serum-like conditions and stresses the importance of substrate conformations and molecular weight on activity. Finally, in vitro cell-viability data shows that the application of PETase is not toxic to cells. By developing an efficient method to depolymerize microplastics in vitro, our work takes a step closer to finding a solution to the problem that microplastics in the bloodstream may pose in the future.
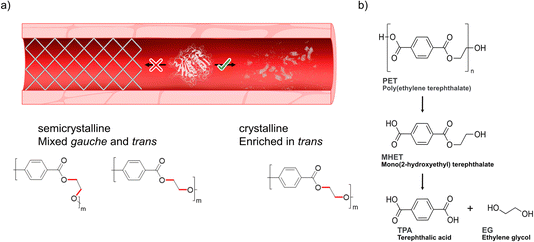 | ||
| Scheme 1 Degradation of PET. (a) Degrading microplastics in serum would ideally require a designer enzyme with high activity towards polymer substrates enriched in trans conformations22–24 (referring to the dihedral angle of the ethylene glycol moiety of the polymer) found in crystalline domains that are enriched in microplastics,19–21 while at the same time showing lower activity towards semicrystalline PET with chains adopting mixed gauche and trans forms. (b) Depolymerization steps of PET into its monomers terephthalic acid (TPA) and ethylene glycol (EG) by PETase. For simplicity, only the intermediate MHET is shown. | ||
Results and discussion
Preparation of microplastic substrate
Since PET is one of the most abundant plastics,1,2 post-consumer PET bottles were selected as a first model substrate. To prepare a microplastic-like substrate, a PET bottle was pre-treated using microwave irradiation technology (see Methods) as recently described by us.23 Microwave pre-treatment of post-consumer plastics results in hydrolysis of the material into a powder-like material, consisting of mostly polymeric chains (degree of polymerization (DP) of 12, 73% relative abundancy) and, to a lesser extent, of shorter oligomers (DP of 3, 27% relative abundancy, fraction shown in the MALDI-TOF spectrum in Fig. 1b). Detailed analysis and characterization of the material is provided in ref. 23. SEM analysis of the obtained PET powder following microwave pre-treatment showed a powder-like appearance with an average particle size of 200 μm.23 Importantly, the microwave hydrolysis occurs in the amorphous part first, resulting in a highly crystalline PET-derived substrate residing mainly in the trans conformation, reminiscent of that of microplastic particles occurring in nature, which also show enhanced crystallinity.19–21 Taken together, the model substrate falls well in the definition of a microplastic.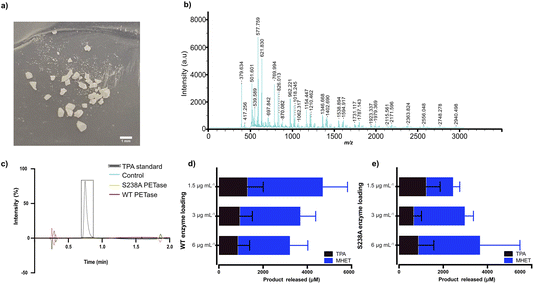 | ||
| Fig. 1 MALDI-TOF analysis of the microplastic substrate and initial screen of the impact of enzyme loading on degradation in Serum Body Fluid (SBF) buffer. (a) Photo of the pre-treated plastic substrate used, showcasing a diversity in particle size. (b) MALDI-TOF analysis of the microplastic substrate. (c) HPLC results of WT PETase and S238A activity against an amorphous (shredded) PET substrate with a mix of gauche and trans conformations (see ESI† for details); a standard of TPA is shown. A loading of 1.5 μg per mL WT PETase or S238A PETase was added to 2 mg mL−1 of untreated shredded PET plastic in 1 mL of SBF buffer for 1 hour of reaction time at 37 °C. Full spectra are given in the ESI.† Control refers to sample without enzyme added. (d) WT PETase loadings of 3 μg, 1.5 μg and 0.75 μg with 7.5 mg of microplastics in 500 μL of SBF buffer, for 1 hour of reaction time at 37 °C. The error bars were calculated from three independent experiments (n = 3) per enzyme loading. (e) S238A PETase loadings of 3 μg, 1.5 μg and 0.75 μg with 7.5 mg of microplastics in 500 μL of SBF buffer, for 1 hour of reaction time at 37 °C. The error bars were calculated from three independent experiments (n = 3) per enzyme loading. | ||
Enzymatic activity towards microplastic substrate in SBF buffer
Previously, we have shown the biocatalytic depolymerization activity of WT PETase and S238A PETase towards a microwaved PET-bottle-derived substrate in 50 mM phosphate buffer (pH 7.2) at 30 °C.29 However, the enzymatic activity of both WT PETase and S238A PETase in a serum-like environment and at body temperature is unknown. Previous work has shown that the choice of buffer can have a drastic effect on the PETase activity. It has been shown that Tris–HCl buffer can decrease PETase activity or even inactivate the enzyme altogether.30,32 To characterize potential PETase activity in serum, we used a Serum Body Fluid (SBF) buffer (ESI Table 1†), which resembles the pH and salinity of serum, to screen for initial activity.In a first experiment, we found that WT PETase and its S238A variant are inactive against shredded semicrystalline PET in this buffer (Fig. 1c), in accordance with previous findings.28 Turning to microplastics, the reaction conditions first needed to be optimized considering that the Tm values of PETase and S238A are approximately 40 °C.29 The reaction temperature is 37 °C to mimic body temperature. Therefore, we needed to investigate which enzyme loading leads to the best conversion of microplastic-like PET to its monomers, MHET and TPA. For the initial screen using HPLC, 1.5 μg mL−1, 3 μg mL−1 and 6 μg mL−1 of enzyme were subjected to 15 mg mL−1 of microplastic substrate. Interestingly, after one hour, the highest conversion for the WT PETase was detected with 1.5 μg mL−1 enzyme (Fig. 1d), whereas for the S238A variant, the highest activity was observed at an enzyme loading of 6 μg mL−1 (Fig. 1e). Due to the costs involved in biopharmaceutical protein production, lower enzyme concentrations would be preferred.33 At the lower enzyme loading of 1.5 μg mL−1, it was noticed that S238A had a preference for generation of TPA over MHET, which resulted in an overall similar yield of TPA for both variants. Higher product specificity towards TPA could be beneficial as this monomer is known to be rapidly excreted. Additionally, after 24 hours, the lowest enzyme loading in SBF buffer for the S238A variant gave the highest conversion to monomers (ESI Fig. 2a†). The large error bars are as expected due to the distribution of particle sizes (see Fig. 1a).
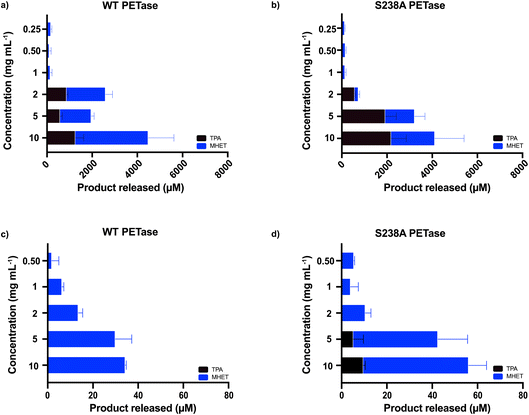
The microplastic loading in our screening experiment for determining the optimal enzyme concentration was high at 15 mg mL−1, whereas concentrations detected in whole blood are much lower. Therefore, we also investigated if using lower concentrations of microplastics (2 mg mL−1, 5 mg mL−1 and 10 mg mL−1 of microwaved PET) in 500 μL SBF buffer with 1.5 μg mL−1 enzyme would result in activity measured by HPLC. There is a clear difference in behavior between the two investigated variants at different substrate loadings (Fig. 2a and b). The WT PETase functions well at 2 mg mL−1; however, the S238A variant converts PET less efficiently, but strikingly shows a high product specificity for TPA (3.5-fold preference for TPA over MHET, whereas WT shows an inverse 2-fold preference for MHET over TPA). A reaction triplicate performed for 24 hours confirmed the specificity of S238A towards TPA and further showed that S238A was functional during this time period (ESI Fig. 2b†). At larger substrate concentrations, the two variants are comparable in overall product formation (Fig. 2a and b).6 We could observe that both enzymes were still active against lower concentrations of plastics and retained activity at 0.25 mg mL−1 (Fig. 2a and b). Besides measuring activity against shredded semicrystalline PET (Fig. 1c), we tested the enzymes against a plastic stent designed for use in applications in the bile duct (see Methods). Importantly, we could observe that the enzymes were not active against this device, as evidenced by HPLC analyses (ESI Fig. 4†).
Moreover, we also wanted to use a commercially available PET powder (ES30-PD-000132, GoodFellow, UK, ESI Fig. 5†) as a second microplastic-reminiscent substrate. This powder shows a more uniform particle size (maximum particle size of 300 μm) whilst having a high molecular weight.29 In addition, this powder has a semi-crystalline structure, which can be compared to our microwave-generated substrate (shown in Fig. 1a) residing essentially in a fully crystalline trans-form. As expected, due to its high abundancy of gauche conformations, the activity when using the powder substrate decreased two orders of magnitude (Fig. 2c and d) compared to when using the microwave-derived trans substrate (Fig. 2a and b). Activity could clearly be monitored down to 0.5 mg mL−1 by ordinary HPLC analysis. These results indicate that enzymatic degradation of low concentrations of microplastics is possible.
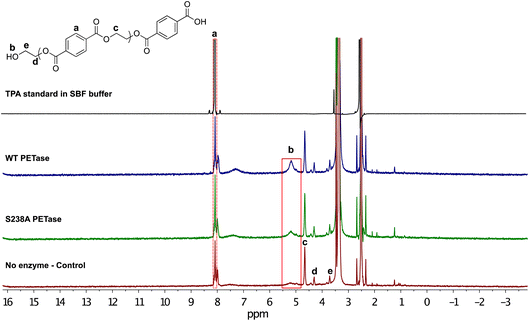
For further characterization of the depolymerized microplastic following the enzymatic reaction, the samples were subjected to 1H NMR analysis. The samples were prepared with 2.5 mg microplastic in 1.25 mL of SBF buffer and 1.5 μg mL−1 enzyme loading, for a 1 hour reaction at 37 °C. After the reaction, the samples were centrifuged and the remaining microplastic was freeze dried. The microplastic was dissolved in DMSO-d6 and the resulting 1H NMR spectra are shown in Fig. 3. The salts in the buffer and the presence of water results in a peak at 3.36 ppm, which partly overlaps with signals stemming from the internal proton peaks of the oligomeric chain of PET. The peaks at 3.71 ppm (e), 4.30 ppm (d) and 4.66 ppm (c) correspond to the inner chain protons. We observed an emerging proton peak at 5.19 ppm (b) that shows formation of MHET, as well as a benzene ring peak at 8.08 ppm (a) corresponding to aromatic protons. Hence, the NMR results support the HPLC data in Fig. 2 showing that after the enzymatic reaction, the PET-derived chain was broken down into monomers.
Depolymerization of microplastics in human serum
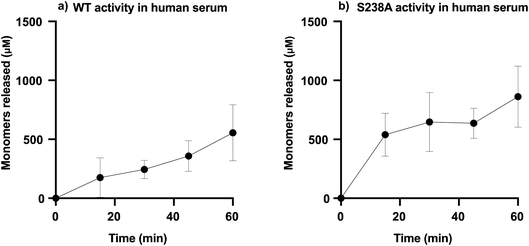
To determine the capability of WT PETase and its S238A variant in depolymerizing PET in blood, the enzymatic activity was screened using serum, which will provide an overview of the activity of the enzymes in an environment including many of the components present in whole blood. It should be noted that the solubility of TPA is different in different systems, such as SBF buffer and human serum; accordingly, the amounts of TPA monomers released were calculated according to the appropriate standard curve (ESI Fig. 6†). We performed tests in serum, measuring the activity during the initial time course of the reaction (1 h) for which both variants are still active (ESI Table 2†), using 2 mg mL−1 of microplastic substrate with 1.5 μg mL−1 enzyme, either WT PETase or its S238A variant, in a total volume of 1000 μL.Furthermore, the generated amounts of TPA and MHET from these experiments were summed as total amounts of monomers released. Fig. 4 shows the results of the enzymatic activities in human serum. It appears that the S238A variant is faster in depolymerizing PET than the WT, as it was able to release ca. 500 μM of monomers already during the first 15 minutes, representing a conversion of 5%, while the WT released only around 100 μM in the same time span. The time course in serum shows how WT and S238A are highly active and illuminates how microplastic PET is being hydrolysed into TPA and MHET, with a higher overall activity for S238A and increased preference to generate TPA (ESI Table 2†).
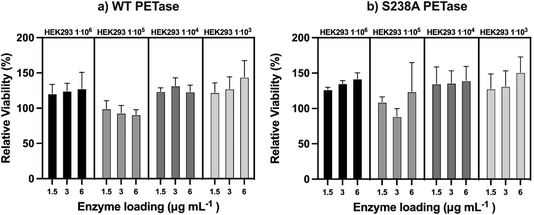
Since both WT PETase and S238A PETase have depolymerization activity in human plasma, it is important to show that the viability of human cells won't be affected by esterase activity towards membrane lipids. To determine the possible cytotoxicity of the PETases in vitro, a kidney cell line (HEK293) was subjected to an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. This colorimetric assay allows quantification of the cell metabolic activity by measuring the capacity of living cells to reduce the tetrazolium dye MTT (yellow) to formazan (purple) (ESI Fig. 7†). The cells were incubated with the enzymes for 90 minutes. This incubation time was selected as the enzymes seem to be most active up to 60 minutes at 37 °C, as shown in ESI Fig. 2.† The HEK cells were measured at four different cell densities. The densities range between 106 and 103 with a 10-fold increment. From the experiments, the viability of the cells is unaffected by the presence of the PETases. If the PETases would affect the membrane lipids, the viability would be considerably lower than 100%. However, the opposite is observed; if the HEK cells are incubated with the PETases, the cells appear to retain full relative viability (Fig. 5).
Discussion
The awareness of the potential impact of microplastics on the environment and animal health has increased considerably over the years. However, no definitive proof of a negative impact for human health is yet available. As a possible future solution to combat the presence of microplastics in the human body, a first feasibility study of an enzymatic approach was chosen. As IsPETase is active at ambient temperature28 with a melting point close to 40 °C,29 this enzyme was chosen as a model system to investigate enzymatic activity under blood-like conditions.Enzymes have consolidated themselves as an essential tool in the biopharmaceutical industry34–36 and, therefore, we sought to test the potential of IsPETase (WT PETase) and its S238A variant to convert microplastic in human serum. It is well known that many enzymes have a preferred pH and salinity to operate in. This is especially relevant for PETases, which are known to be highly sensitive to reaction conditions.28 Despite earlier work showing a loss of activity for PETase over time and in Tris buffer,30 we observed activity over multiple hours for S238A PETase in SBF buffer. The buffer simulates the same pH and salinity as are present in serum. However, SBF is a simplified version of human serum without the presence of protein, antibodies, and hormones. Therefore, we sought to test the activity of PETases in human serum, still representing a relatively simple model of blood without complex clotting factors and blood cells. Our results demonstrate that PETases are highly active in serum, especially over the initial timepoints. Decreased relative activity over time might be due to protein unfolding in serum, in which the enzyme loses its functional conformation. Another potential factor that can influence enzymatic activity in serum might be the interaction with other components, especially the antibodies present. Still, in SBF buffer, S238A seems to remain active for 24 h.
It should be noted that the representative microplastic PET substrate used in this study has a particle size that is relatively large. Moreover, the substrate was added at an excessive concentration compared to what has been found in blood.6 As it is well established that substrate accessibility is key in enzymatic depolymerization23,37 and that increased surface area generally facilitates hydrolysis, our data showing relatively high depolymerization activity is encouraging. In fact, the activity of S238A PETase representing a release of ca. 1 mM monomers in blood after 1 h is similar to the activity in buffer.29 Further research is required on the interactions of PETases with other components in the blood stream. Since PETase is a bacterial enzyme, it is possible that this enzyme would be quickly neutralized when it is introduced. There are tools available that can humanize the surface of a protein to circumvent this issue38 and most likely would need to be applied to this type of enzyme for prolonged activity. Additionally, the esterase activity of PETase could be considered a problem. However, we show that the PETase does not affect viability in HEK293 cells.
Furthermore, a delivery system of PETase needs to be developed and in vivo studies are needed to verify the viability of these enzymes in removing PET and the potential of using these enzymes as therapeutic agents. Recent studies have focused on the delivery of encapsulated enzymes by erythrocytes, reducing immunogenicity and enabling a long circulation time, which could be a possible way to administer the S238A PETase enzyme.39 Nevertheless, in this study, we showed encouraging results that indicate the viability of WT PETase and S238A PETase in clearing PET microplastics from human serum, which opens avenues for enzymatic depolymerization of synthetic polymers in human blood.
Methods
Microwave pre-treatment of polyethylene terephthalate (PET)
Post-consumer PET plastic bottles were washed with 5% Tween and 50 mM sodium bicarbonate buffer (pH 7) at 30 °C and dried. The clean PET was then shredded using a SHR3D IT from 3devo. From the shredded PET, 1 g was taken in a flexiWAVE Teflon vial with 20 mL of deionized water. The vial was then placed inside a flexiWAVE microwave (Milestone, Sorisole, Italy). To control and follow the temperature, a probe was used, and the power was automatically kept to the set temperature (200 °C). A 10 minute ramp time to reach the set temperature was used and after that, the set temperature of 200 °C remained constant throughout the run of 2 h. Following the reaction, the vial was left at 4 °C for 24 hours and after that, the content of the vial was freeze-dried in a Heto LyoLab 3000 to remove water.MALDI-TOF analysis of plastic particles
The 2 hour microwaved-PET (microplastics) was dissolved in a mixture of TFA (trifluoroacetic acid) and deuterated chloroform (Sigma-Aldrich, Sweden) in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 v/v, to a final concentration of 1 mg of plastic per mL. The matrix used was 2,5-dihydroxybenzoic acid (DHB). A mixture of DHB and the dissolved PET was pipetted onto a stainless-steel plate for crystallization and then analyzed using a MALDI-TOF device (Bruker Daltonics, Germany). FlexControl and FlexAnalysis software (Bruker Daltonics, Germany) was applied for data acquisition and data analysis, respectively.
4 v/v, to a final concentration of 1 mg of plastic per mL. The matrix used was 2,5-dihydroxybenzoic acid (DHB). A mixture of DHB and the dissolved PET was pipetted onto a stainless-steel plate for crystallization and then analyzed using a MALDI-TOF device (Bruker Daltonics, Germany). FlexControl and FlexAnalysis software (Bruker Daltonics, Germany) was applied for data acquisition and data analysis, respectively.
Expression, production, and purification of enzymes
pET21b+ vectors encoding the WT PETase and S238A PETase genes were transformed into E. coli C43(DE3) and inoculated on a Nutrient Agar plate with 100 μg per mL ampicillin (Sigma-Aldrich, Sweden). Following overnight culture at 37 °C, single colonies containing WT PETase and S238A PETase were pre-cultured in 10 mL of 2xYT liquid medium (5 g of NaCl, 10 g of yeast extract, 16 g of Tryptone per liter, pH 7.0) supplemented with 100 μg per mL ampicillin in a 100 mL flask at 37 °C (200 rpm). Each culture was then transferred into 2 L flasks with 400 mL fresh 2xYT liquid medium containing ampicillin and was incubated at 37 °C (200 rpm). 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added as the OD600 reached 0.6, and following 20 hours of cultivation at 20 °C (160 rpm), the cell pellets were collected by centrifugation at 4500 rpm for 20 minutes. The collected cells were lysed with B-PER complete lysis buffer (Thermo Fisher Scientific, USA), and the collected protein was purified using an ÄKTA protein purification system (GE Healthcare Life Science, Sweden) with a Ni-NTA His-tag column (Qiagen, USA). 10 mM sodium phosphate buffer, pH 7.4, with 500 mM NaCl and 40 mM or 300 mM imidazole were used as the equilibrium buffer and elution buffer, respectively. The purified proteins were desalted with a PD-10 desalting column and finally stored in 10 mM Tris–HCl buffer, pH 7.4. The protein purification was confirmed with sodium dodecyl sulphate-polyacrylamide gel electrophoresis with a gradient of 4–15% (Mini-PROTEAN TGX Stain-Free, BioRad, Sweden). The SeeBlueTM Plus2 (Thermo Fisher Scientific, USA) pre-stained protein standard was used as a marker. The protein concentration was determined using a Nanodrop.Simulated body fluid (SBF) buffer preparation
The SBF buffer was prepared according to Kokubo et al. (1990).40 This buffer contains ion concentrations of Na+, K+, Mg2+, Ca2+, Cl−, HCO3−, HPO42−, and SO2− that resemble those that can be found in human blood plasma. It was later buffered to pH 7.4 using 50 mM trishydroxymethylaminomethane (Tris) and 45 mM hydrochloric acid (HCl) and dissolved at 36.5 °C. The ion concentration (mM) is as shown in ESI Table 1.†Analysis of enzymatic activity and sample preparation in SBF buffer and human serum
For both SBF and human serum samples, the different microplastic amounts (either 15 mg mL−1, 10 mg mL−1, 5 mg mL−1 or 2 mg mL−1) were weighed into 2 mL glass vials. For the SBF samples, the different amounts of enzyme loading (either 6 μg mL−1, 3 μg mL−1 or 1.5 μg mL−1) were added to 500 μL of the SBF buffer (pH 7.4), which was then added to the 2 mL vials with the microplastic substrate. Before the addition of the enzyme, the buffer with plastic was incubated at 37 °C for 10 minutes. After addition of the enzyme to the SBF buffer, samples were taken at t = 0, 1 h, 3 h and 24 h. The reaction was quenched with 500 μL methanol and centrifuged for 1 minute at 7000 rpm at room temperature. 150 μL of the supernatant was transferred to HPLC vials. In the serum experiments, 1 mL of the serum (Human Serum (H4522), sterile filtered, Merck, USA) was added to the 2 mL glass vials with pre-weighted plastic. The samples were incubated at 37 °C for 10 minutes before addition of the enzymes. After the enzyme was added, 200 μL samples were taken at t = 0, 15, 30, 45 and 60 minutes. The reaction was quenched at 95 °C for 1 minute according to safety protocols. The sample was then mixed with 200 μL methanol and centrifuged at 7000 rpm for 10 minutes. 150 μL supernatant was carefully transferred to an HPLC vial for analysis.The plastic stent used was an Advanix Biliary Stent with a NaviFlex RX Delivery system (Boston Scientific) containing a polyethylene-based material, a polyether-amide and Nylon.
HPLC analysis
High-performance liquid chromatography (HPLC, Agilent 1200 Series, Agilent Technologies, USA) with an ACE C8 column (50 × 3 mm, 3 μm) was used to analyze the products released from the PET pre-treatment and the monomeric products released following enzymatic reaction either in SBF buffer or human serum. The flow rate was 1 mL min−1, at 50 °C with an acetonitrile gradient from 10% to 97%, and 0.1% trifluoroacetic acid/H2O. The amount of TPA and MHET that can be attributed to the microwave pretreatment (reaction time 0) was measured at reaction time 0 in control experiments and these values were subtracted in the calculation of enzyme activity for accurate determination of the enzymatic activity. The concentrations of the released products were calculated using a standard curve of TPA ranging from 0.016 mM to 2 mM in SBF buffer or 0.016 mM to 1 mM in human plasma (ESI Fig. 6†).MTT cytotoxicity assay
A colorimetric kit for cell survival and proliferation (lot no. 4030556m, Millipore, USA) was used to analyze the samples. HEK293 cells were grown in DMEM + 10% fetal bovine serum. The cells were split using TrypLE™ (ThermoFisher, USA) and DMEM + 10% fetal bovine serum (ThermoFisher, USA) to a cell density of 103 to 106 cells per mL. 100 μL of the cell suspension was added per well in a 96-well plate. The cells were incubated for 90 minutes after addition of enzyme (1.5 μg mL−1, 3 μg mL−1 and 6 μg mL−1). After 90 minutes, 10 μL of AB Solution (MTT) was added. The solution was mixed by gentle tapping on the plate, after which the plate was incubated for 4 hours at 37 °C. After the incubation period, 100 μL of isopropanol with 0.04 M HCl was added and thoroughly mixed. The absorbance was measured at 570 nm after 20 minutes on an ELISA plate reader. The viability was calculated using the formula: 100 × ((fluorometric signal of cells + enzyme)/(fluorometric signal of cells without enzyme)).Proton nuclear magnetic resonance (1H NMR)
A Bruker Avance AM 400 NMR instrument was used to record the 1H NMR spectra of the samples before and after enzymatic reaction using DMSO-d6 as the solvent.Induced fit docking (IFD)
The X-ray crystal structure of IsPETase (PDB ID: 6EQE41) was downloaded from the RCSB Protein Data Bank. The structure of the variant S238A was generated by in silico mutagenesis using the ‘mutate residue’ tool in Maestro. All protein structures were prepared for IFD using the protein preparation wizard in the software Schrödinger Maestro Suite (Schrödinger, LCC, USA). All crystallographic water molecules were removed, and the missing hydrogens were added at a set pH of 7.5. The polymer substrates – polyethylene furanoate and polylactide – were represented by an oligomer prepared in Maestro using the LigPrep wizard and were all energy-minimized using the OPLS4 force field. Finally, both polymers were used to generate IFD calculations of both of the enzymes.Ethical statement
The purchased sterile-filtered human serum (H4522) sample represents a randomized anonymous donor. No personal or patient data is disclosed, or traceable. Prior to the execution of this study, the project and its setup were discussed with the Research Ethics function of the KTH Research Support Office. No ethical permit was required for the study. A risk assessment on working with serum for this project was approved by the lab safety coordinator according to implemented safety routines at KTH/Science for Life Laboratory. The use of the plastic stent in this study was approved by Boston Scientific.Data availability
All raw data is available upon reasonable request and was submitted alongside the manuscript.Author contributions
P. O. S., X. L. L. and D. H. designed the research; X. L. L., D. H. and E. B. performed experiments and analyzed the data; X. L. L. and D. H. prepared the manuscript; P. O. S. supervised the research and revised the manuscript. All authors approved the final version of the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The authors want to thank Prof. Minna Hakkarainen for assistance with microwave pre-treatment and fruitful discussions. Funding from the NovoNordisk Foundation (project EazyPlast grant no. NNF20OC0064972) and the Swedish research council FORMAS (project BioPlastEco grant no. 2021-02509) is greatly acknowledged. Computations were enabled by resources provided by the National Academic Infrastructure for Supercomputing in Sweden (NAISS), partially funded by the Swedish Research Council through grant agreement no. 2022-06725.References
- C. Jönsson, et al., Biocatalysis in the Recycling Landscape for Synthetic Polymers and Plastics towards Circular Textiles, ChemSusChem, 2021, 14, 4028–4040 CrossRef PubMed.
- R. Geyer, J. R. Jambeck and K. L. Law, Production, use, and fate of all plastics ever made, Sci. Adv., 2017, 3, e1700782 CrossRef PubMed.
- K. D. Cox, et al., Human Consumption of Microplastics, Environ. Sci. Technol., 2019, 53, 7068–7074 CrossRef CAS PubMed.
- C. Q. Yong, S. Valiyaveettil and B. L. Tang, Toxicity of Microplastics and Nanoplastics in Mammalian Systems, Int. J. Environ. Res. Public Health, 2020, 17, 1509 CrossRef CAS PubMed.
- A. D. Vethaak and J. Legler, Microplastics and human health, Science, 2021, 371, 672–674 CrossRef CAS PubMed.
- H. A. Leslie, et al., Discovery and quantification of plastic particle pollution in human blood, Environ. Int., 2022, 163, 107199 CrossRef CAS PubMed.
- L. J. J. Meijer, T. van Emmerik, R. van der Ent, C. Schmidt and L. Lebreton, More than 1000 rivers account for 80% of global riverine plastic emissions into the ocean, Sci. Adv., 2021, 7, eaaz5803 CrossRef PubMed.
- D. Wang, Y. Xu, Q. Li and L.-S. Turng, Artificial small-diameter blood vessels: materials, fabrication, surface modification, mechanical properties, and bioactive functionalities, J. Mater. Chem. B, 2020, 8, 1801–1822 RSC.
- C.-D. Dong, et al., Polystyrene microplastic particles: In vitro pulmonary toxicity assessment, J. Hazard. Mater., 2020, 385, 121575 CrossRef CAS PubMed.
- S. D'Angelo and R. Meccariello, Microplastics: A Threat for Male Fertility, Int. J. Environ. Res. Public Health, 2021, 18, 2392 CrossRef PubMed.
- A. Ragusa, et al., Raman Microspectroscopy Detection and Characterisation of Microplastics in Human Breastmilk, Polymers, 2022, 14, 2700 CrossRef CAS PubMed.
- H. A. Leslie and M. H. Depledge, Where is the evidence that human exposure to microplastics is safe?, Environ. Int., 2020, 142, 105807 CrossRef CAS PubMed.
- G. L. Ball, C. J. McLellan and V. S. Bhat, Toxicological review and oral risk assessment of terephthalic acid (TPA) and its esters: a category approach, Crit. Rev. Toxicol., 2012, 42, 28–67 CrossRef CAS PubMed.
- R. A. Wilkes, et al., Complex regulation in a Comamonas platform for diverse aromatic carbon metabolism, Nat. Chem. Biol., 2023, 19, 651–662 CrossRef CAS PubMed.
- A. Hoshi and K. Kuretani, Metabolism of terephthalic acid. III. Absorption of Terephthalic Acid from Gastrointestinal Tract and Detection of Its Metabolites, Chem. Pharm. Bull., 1967, 15, 1979–1984 CrossRef CAS PubMed.
- National Toxicology Program, NTP-CERHR Monograph on the Potential Human Reproductive and Developmental Effects of Styrene, NTP CERHR MON, 2006 Search PubMed.
- D. Jacobsen and K. E. McMartin, Methanol and Ethylene Glycol Poisonings: Mechanism of Toxicity, Clinical Course, Diagnosis and Treatment, Med. Toxicol., 1986, 1, 309–334 CrossRef CAS PubMed.
- M. F. M. White and S. Wallace, A New PETase from the Human Saliva Metagenome and Its Functional Modification via Genetic Code Expansion in Bacteria, Angew. Chem., Int. Ed., 2023, 62, e202216963 CrossRef CAS PubMed.
- S. Miri, et al., Biodegradation of microplastics: better late than never, Chemosphere, 2022, 286, 131670 CrossRef CAS PubMed.
- M. Dilara Hatinoglu and F. Dilek Sanin, Fate and effects of polyethylene terephthalate (PET) microplastics during anaerobic digestion of alkaline-thermal pretreated sludge, Waste Manage., 2022, 153, 376–385 CrossRef CAS PubMed.
- A. L. Andrady, The plastic in microplastics: a review, Mar. Pollut. Bull., 2017, 119, 12–22 CrossRef CAS PubMed.
- J. S. Becker, R. D. Brown, D. R. Killelea, H. Yuan and S. J. Sibener, Comparative surface dynamics of amorphous and semicrystalline polymer films, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 977–982 CrossRef CAS PubMed.
- B. Guo, et al., Fast Depolymerization of PET Bottle Mediated by Microwave Pre-treatment and an Engineered PETase, ChemSusChem, 2023, 16, e202300742 CrossRef CAS PubMed.
- R. Wei, et al., Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation, Nat. Commun., 2019, 10, 5581 CrossRef CAS PubMed.
- V. Tournier, et al., An engineered PET depolymerase to break down and recycle plastic bottles, Nature, 2020, 580, 216–219 CrossRef CAS PubMed.
- P. Blázquez-Sánchez, et al., Antarctic Polyester Hydrolases Degrade Aliphatic and Aromatic Polyesters at Moderate Temperatures, Appl. Environ. Microbiol., 2022, 88, e01842 CrossRef PubMed.
- H. Lu, et al., Machine learning-aided engineering of hydrolases for PET depolymerization, Nature, 2022, 604, 662–667 CrossRef CAS PubMed.
- S. Yoshida, et al., A bacterium that degrades and assimilates poly(ethylene terephthalate), Science, 2016, 351, 1196–1199 CrossRef CAS PubMed.
- B. Guo, et al., Conformational Selection in Biocatalytic Plastic Degradation by PETase, ACS Catal., 2022, 3397–3409 CrossRef CAS.
- J. Schmidt, et al., Effect of Tris, MOPS, and phosphate buffers on the hydrolysis of polyethylene terephthalate films by polyester hydrolases, FEBS Open Bio, 2016, 6, 919–927 CrossRef CAS PubMed.
- K. Świderek, et al., Mechanistic studies of a lipase unveil effect of pH on hydrolysis products of small PET modules, Nat. Commun., 2023, 14, 3556 CrossRef PubMed.
- A. Maurya, A. Bhattacharya and S. K. Khare, Enzymatic Remediation of Polyethylene Terephthalate (PET)–Based Polymers for Effective Management of Plastic Wastes: An Overview, Front. Bioeng. Biotechnol., 2020, 8, 602325 CrossRef PubMed.
- J. Puetz and F. M. Wurm, Recombinant Proteins for Industrial versus Pharmaceutical Purposes: A Review of Process and Pricing, Processes, 2019, 7, 476 CrossRef CAS.
- N. M. Hendrikse, et al., Exploring the therapeutic potential of modern and ancestral phenylalanine/tyrosine ammonia-lyases as supplementary treatment of hereditary tyrosinemia, Sci. Rep., 2020, 10, 1315 CrossRef CAS PubMed.
- N. M. Hendrikse, et al., Ancestral lysosomal enzymes with increased activity harbor therapeutic potential for treatment of Hunter syndrome, iScience, 2021, 24, 102154 CrossRef CAS PubMed.
- M. T. Reetz, G. Qu and Z. Sun, Engineered enzymes for the synthesis of pharmaceuticals and other high-value products, Nat. Synth., 2024, 3, 19–32 CrossRef.
- R. K. Brizendine, et al., Particle Size Reduction of Poly(ethylene terephthalate) Increases the Rate of Enzymatic Depolymerization But Does Not Increase the Overall Conversion Extent, ACS Sustain. Chem. Eng., 2022, 10, 9131–9140 CrossRef CAS.
- Y. Choi, C. Hua, C. L. Sentman, M. E. Ackerman and C. Bailey-Kellogg, Antibody humanization by structure-based computational protein design, mAbs, 2015, 7, 1045–1057 CrossRef CAS PubMed.
- L. Rossi, F. Pierigè, A. Bregalda and M. Magnani, Preclinical developments of enzyme-loaded red blood cells, Expert Opin. Drug Delivery, 2021, 18, 43–54 CrossRef CAS PubMed.
- T. Kokubo, H. Kushitani, S. Sakka, T. Kitsugi and T. Yamamum, Surface-Structure Changes in Bioactive, J. Biomed. Mater. Res., 1990, 24, 721–734 CrossRef CAS PubMed.
- H. Austin, et al., Characterization and engineering of a plastic degrading aromatic polyesterase, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E4350–E4357 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fd00014e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |