Ultrasensitive electrochemical detection and inhibition evaluation of DNA methyltransferase based on cascade strand displacement amplification†
Ruizhi
Liu
a,
Yuge
Wang
a,
Hua
Chai
bd and
Peng
Miao
 *bc
*bc
aCenter of Reproductive Medicine and Center of Prenatal Diagnosis, The First Hospital of Jilin University, Changchun 130021, China
bSuzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou 215163, China. E-mail: miaopeng@sibet.ac.cn
cShandong Laboratory of Advanced Biomaterials and Medical Devices in Weihai, Weihai 264200, China
dJinan Guoke Medical Technology Development Co., Ltd., Jinan 250103, China
First published on 15th November 2023
Abstract
An electrochemical sensing approach for ultrasensitive DNA methyltransferase (MTase) activity assay is proposed. After specific cleavage reaction in the presence of a methylated state, strand displacement polymerization (SDP) is initiated in the solution. The product of upstream SDP further triggers downstream SDP, which enriches abundant electrochemical species at the electrode. The whole process is quite convenient with shared enzymes. Due to the cascade signal amplification, ultrahigh sensitivity is promised. Inhibitor screening results are also demonstrated to be good. Besides, target MTase can be accurately determined in human serum samples, confirming excellent practical utility. This work provides a reliable approach for the analysis of MTase activity, which is of vital importance for related biological studies and clinical applications.
Introduction
DNA methylation is a critical chemical modification of DNA. It participates in controlling various biological processes like gene expression and cell differentiation.1,2 However, the exact mechanisms of the generation, maintenance and erasure of DNA methylation patterns are still not fully clarified. It is found that the balance between DNA methylation and demethylation is of great importance for human health.3,4 This epigenetic modification involves the transfer of a methyl group from the donor to acceptor, which is usually catalyzed by the DNA methyltransferase (MTase) family.5–7 MTase is regarded as an important biomarker for the study of the methylation process, which can be used to predict many genetic disorders and diseases.8,9 The fabrication of sensitive, facile and reliable tools for the analysis of MTase levels is of great importance.10,11Conventional DNA MTase assays usually rely on high-performance liquid chromatography (HPLC), high performance capillary electrophoresis (HPCE) or immunoblot analysis.12,13 However, these methods either involve radiation–hazardous contamination, or require laborious operations and high cost. Currently, several novel approaches have been well established for the analysis of DNA MTase activity based on the techniques like fluorescence, electrochemistry, SERS, mass spectrometry and colorimetry.14 For example, Yuan and coworkers fabricated a colorimetric biosensor coupling primer exchange reaction amplification and hemin/G-quadruplex DNAzyme.15 Zheng et al. developed an electrochemical and colorimetric dual modal biosensor based on the high laccase-like activity of porous organic polymer–inorganic nanocomposites.16 Liu et al. designed a cascade signal amplification approach combining strand displacement and multicomponent nucleic acid enzyme.17 To further simplify the complexity and improve the sensitivity, it remains a challenge to develop advanced sensing strategies for the determination of DNA MTase.18
To address these issues, we herein develop a novel electrochemical approach for the quantification of MTase activity. Dam is used as a model DNA MTase. Only a few DNA strands and enzymes are utilized accordingly. Cascade strand displacement polymerization (SDP) is integrated to improve the sensitivity. Common enzymes are shared in both upstream SDP in solution and downstream SDP at the electrode surface, which facilitate the operation. This biosensor circumvents the use of complicated instruments and expensive reagents. It achieves ultrahigh sensitivity due to smart amplification design and shows potential to screen Dam-targeted inhibitor drugs, which has great prospects in drug discovery and clinical diagnosis.
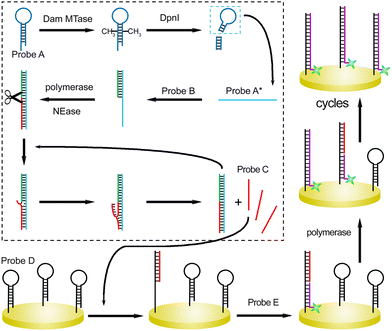
The sensing principle is described generally (Scheme 1). Seven DNA strands are first devised (Table S1†). Hairpin-structured Probe A could be recognized by Dam MTase. After the catalysis, the adenine residues in the sequences (GATC) are methylated. Next, the sites are specifically cleaved in the presence of DpnI endonuclease. The remaining sequence containing the loop segment of original Probe A is termed Probe A*. It can partially hybridize with Probe B, which acts as the primer of upstream SDP after the introduction of Klenow fragment polymerase and nicking endonuclease (NEase). With cycles of extension, nicking, and strand displacement,19 multiple Probe C strands can be generated. Probe C participates in the following SDP at the solid interface of the gold electrode. Probe D is first assembled at the substrate electrode via gold–sulfur chemistry, which also shows a hairpin structure. The loop can be opened after hybridization with Probe C. A single-stranded region is exposed, which captures the complementary sequence of Probe E. This single-stranded DNA probe acts as the downstream primer and initiates strand displacement polymerization. Probe C is then recycled for the reaction with another hairpin-structured Probe D. Therefore, a number of Probe E units can be captured at the interface. By measuring the responses from the ferrocene (Fc) labeled at the terminal of Probe E, the concentration of the reaction trigger (Dam MTase) can be evaluated.
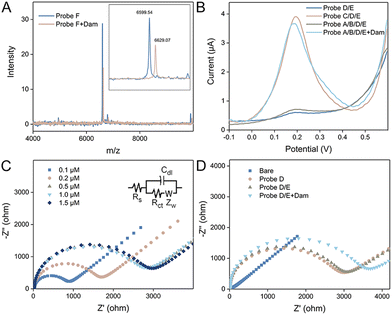
To demonstrate the feasibilities of the enzyme catalyzed reactions, polyacrylamide gel electrophoresis (PAGE) experiments are performed. After methylation of Probe A, the band does not change due to the limited resolution of PAGE to distinguish DNA methylation. However, the methylated duplex could be specifically recognized and cleaved by DpnI. Thus, after the introduction of the restriction endonuclease, Probe A is cut, reflected by the appearance of a new band at a lower position in the gel (Fig. S1A†). In contrast, in the absence of DNA methylation by Dam, DpnI does not work and the Probe A band barely changes. The gel image demonstrates the activities of Dam MTase and DpnI as expected. We then studied the SDP process in the solution by PAGE. The successful hybridization of Probes B and A* is verified by the emergence of the new band with a slower migration rate. After the introduction of Klenow polymerase, a duplex with larger molecular weight is formed and the position of the band is higher. Subsequently, with Nb·BbvCI catalyzed digestion, single-stranded Probe C is displaced, confirmed by the new band showing lower molecular weight (Fig. S1B†). Similarly, the SDP process occurring at the electrode surface is also demonstrated by PAGE. The bands of the partially formed duplex (Probe C/D) before and after extension, as well as the bands of SDP products are shown in the gel at expected positions (Fig. S1C†). Since the DNA methylation process catalyzed by Dam MTase is not directly confirmed by PAGE, we performed mass spectrometry analysis to verify the feasibility of enzyme catalysis. Considering the optimal analytical range of the instrument, we designed a shortened sequence (Probe F) for DNA methylation. As shown in Fig. 1A, after the reaction with Dam MTase, the peak position of the DNA strand shifted from 6599 to 6629 (m/z). The difference is exactly equal to m/z of two methyl groups, demonstrating that MTase successfully catalyzes the methylation process.
The feasibility of the proposed method is then evaluated by electrochemical techniques. First, square wave voltammetry characterization was performed and the results are depicted in Fig. 1B. The electrochemical species of Fc is labeled at the 5′ terminal of Probe E. Its commentary sequence immobilized at the electrode is hidden in the hairpin. Therefore, after incubating Probe E with the Probe D immobilized electrode, no SWV peak can be observed. In contrast, in the presence of Probe C, the loop of Probe D is opened and Probe E can be captured. As a result, a significant SWV peak appears around 0.2 V. Since Probe C can be generated by the SDP process of Probe A/B with the methylated state, the SWV curves are compared without and with Dam MTase. It is clear that only with the enzyme catalyzed reaction, the SWV peak is retained, indicating that square wave voltammetry is suitable for the analysis of Dam MTase activity. We then utilized EIS to probe the electrode interface. Negatively charged DNA strands immobilized at the electrode can repel electrochemical species intensely. As a result, the charge transfer resistance is highly related to the density of DNA strands. As depicted in Fig. 1C, with the increase of Probe D concentration, the semicircular domain is increased and nearly saturated with a concentration of 0.5 μM. Therefore, this critical value is used for the following experiments. Charge transfer resistance (Rct) can be calculated according to the Nyquist diagrams. We modified seven independent electrodes with Probe D (0.5 μM), and the values are quite consistent (average: 2642 ± 37 Ω), demonstrating good reproducibility. We then compared the EIS curves of the electrodes with different treatments. The bare electrode shows a straight line without any semicircular domain. A semicircle appears for the Probe D modified electrode, which is nearly the same compared with the electrode after Probe E incubation. This is because Probe E cannot hybridize with Probe D directly. After the SDP of Probe A/B in the methylated state, Probe C strands are generated, which assist in the immobilization of Probe E. Therefore, the semicircular domain of the Nyquist diagram is further enlarged. The EIS results further confirm the feasibility of the proposed sensing mechanism (Fig. 1D).
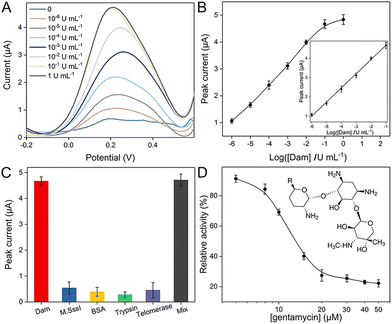
Before electrochemical quantification of Dam MTase, several critical parameters are optimized, including the template concentration of Probe A for SDP and reaction time. With a larger amount of Probe A strand and longer incubation time, the obtained SWV peaks are larger. Optimized conditions are selected (80 nM and 120 min) when the peak intensities reach saturation (Fig. S2†). Under these experimental conditions, SWV curves are compared corresponding to a series of initial Dam MTase concentrations. As shown in Fig. 2A, the peak increases with more Dam MTase, indicating that larger amounts of Fc labeled Probe E are immobilized. The relationship between the Dam concentration and electrochemical response is summarized in Fig. 2B. A linear range is established from 10−6 to 10−1 U mL−1. The linear equation is as follows:
| i = 5.41 + 0.74cDam (n = 4, R2 = 0.997). |
The limit of detection (LOD) is 10−6 U mL−1 (S/N = 3). The low LOD and wide analytical range are superior to those of most recently reported methods (Table S2†).
The selectivity of the proposed electrochemical biosensor is explored by employing different enzymes, including other unspecific MTase. After introducing M·SssI, BSA, trypsin and telomerase in the reaction system, respectively, the electrochemical signals are recorded with the same operation. As shown in Fig. 2C, the peak intensities are limited due to insufficient localization of Fc labeled Probe E. However, after blending these proteins with Dam MTase, the enzyme in the mixture can still transfer methyl groups and initiate further cleavage and SDP reactions, which is verified by the significantly larger electrochemical response. We further introduced human serum samples to test the potential practical utility. These real samples are first collected and diluted 10 fold with 10 mM phosphate buffered saline (PBS). Afterward, different concentrations of Dam MTase are spiked (0.1, 0.2, 0.3, 0.5, and 1 U mL−1) in these samples, which are transferred into the reaction system. The recorded electrochemical responses are referred to the established linear equation for calculation. The estimated concentrations are in good accordance with original spiked values (Table S3†). The tiny relative errors and satisfactory recoveries are due to the negligible interference of diluted serum samples on this method. The potential of this method for clinical analysis is thus demonstrated. Screening of the inhibitors of MTase is highly desired for the purpose of biological studies and drug discovery.20,21 To evaluate the potential use of this method for the study of inhibitors, we have introduced gentamycin as a model inhibitor. Although there are several different enzymes involved in this system, previous studies have proved that gentamycin does not inhibit the activities of DpnI, Klenow and Nb·BbvCI.22,23 Different concentrations of gentamycin are added to Probe A solution with DNA Dam MTase before the subsequent reaction procedures. The dose-dependent inhibitory effect of Dam MTase is then studied. As shown in Fig. 2D, the relative activity (RA) decreases with more gentamycin (from 5 to 50 μM), indicating an elevated inhibitory effect with larger concentration. A half-maximum inhibitory concentration (IC50) of 13.1 μM is estimated, which well matches with those in a previous study.24 It also reveals that this approach has great potential application for the inhibitor screening applications.
Conclusions
In summary, we have fabricated a sensitive biosensor to detect Dam MTase activity by combining upstream and downstream strand displacement reactions after Dam MTase triggered methylation and DpnI assisted digestion. The finally recorded electrochemical response is highly related to the activity of Dam MTase. Due to the cascade signal amplification, exceptional sensitivity is achieved. The LOD of 10−6 U mL−1 is superior to those of most previous biosensors. This method is also applicable for the screening of MTase inhibitors. We further achieved the analysis of target MTase in biological samples, demonstrating its potential for biological investigations and clinical diagnosis.Author contributions
Ruizhi Liu: conceptualization, data curation, and writing – original draft. Yuge Wang: investigation and formal analysis. Hua Chai: investigation and validation. Peng Miao: project administration and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the Project of Shandong Provincial Laboratory (SYS202207) and the Natural Science Foundation of Shandong Province (ZR2021YQ54 and ZR2023QB106).Notes and references
- T. Y. Sun, Y. Y. Xu, Y. Xiang, J. H. Ou, E. J. Soderblom and Y. R. Diao, Nat. Genet., 2023, 55, 1324–1335 CrossRef.
- K. Y. Li, C. H. T. Tam, H. B. Liu, S. Day, C. K. P. Lim, W. Y. So, C. Huang, G. Z. Jiang, M. Shi, H. M. Lee, H. y. Lan, C. C. Szeto, R. L. Hanson, R. G. Nelson, K. Susztak, J. C. N. Chan, K. Y. Yip, R. C. W. Ma and T. Consortium, Nat. Commun., 2023, 14, 2543 CrossRef PubMed.
- T. T. Wang, P. L. Li, Q. C. Qi, S. J. Zhang, Y. Xie, J. Wang, S. B. Liu, S. H. Ma, S. J. Li, T. T. Gong, H. T. Xu, M. Q. Xiong, G. H. Li, C. G. You, Z. F. Luo, J. Li, L. T. Du and C. X. Wang, Nat. Commun., 2023, 14, 4724 CrossRef PubMed.
- L. McAllan, D. Baranasic, S. Villicaña, S. Brown, W. H. Zhang, B. Lehne, M. Adamo, A. Jenkinson, M. Elkalaawy, B. Mohammadi, M. Hashemi, N. Fernandes, N. Lambie, R. Williams, C. Christiansen, Y. W. Yang, L. Zudina, V. Lagou, S. L. Tan, J. Castillo-Fernandez, J. W. D. King, R. Soong, P. Elliott, J. Scott, I. Prokopenko, I. Cebola, M. Loh, B. Lenhard, R. L. Batterham, J. T. Bell, J. C. Chambers, J. S. Kooner and W. R. Scott, Nat. Commun., 2023, 14, 2784 CrossRef PubMed.
- Y. Q. Chang, L. D. Sun, K. J. Kokura, J. R. Horton, M. Fukuda, A. Espejo, V. Izumi, J. M. Koomen, M. T. Bedford, X. Zhang, Y. Shinkai, J. Fang and X. D. Cheng, Nat. Commun., 2011, 2, 533 CrossRef PubMed.
- Z. Yang, J. R. Horton, L. Zhou, X. J. Zhang, A. P. Dong, X. Zhang, S. L. Schlagman, V. Kossykh, S. Hattman and X. D. Cheng, Nat. Struct. Mol. Biol., 2003, 10, 849–855 CrossRef CAS PubMed.
- A. L. Furst and J. K. Barton, Chem. Biol., 2015, 22, 938–945 CrossRef CAS PubMed.
- B. van Steensel and S. Henikoff, Nat. Biotechnol., 2000, 18, 424–428 CrossRef CAS.
- N. Luo, M. J. Nixon, P. I. Gonzalez-Ericsson, V. Sanchez, S. R. Opalenik, H. L. Li, C. A. Zahnow, M. L. Nickels, F. Liu, M. N. Tantawy, M. E. Sanders, H. C. Manning and J. M. Balko, Nat. Commun., 2018, 9, 248 CrossRef.
- Y. Long, K. Ubych, E. Jagu and R. K. Neely, Bioconjugate Chem., 2021, 32, 192–198 CrossRef CAS PubMed.
- S. Rauf, L. Zhang, A. Ali, J. Ahmad, Y. Liu and J. H. Li, Anal. Chem., 2017, 89, 13252–13260 CrossRef CAS PubMed.
- S. Pradhan, A. Bacolla, R. D. Wells and R. J. Roberts, J. Biol. Chem., 1999, 274, 33002–33010 CrossRef CAS.
- L. Lennard and H. J. Singleton, J. Chromatogr. B: Biomed. Sci. Appl., 1994, 661, 25–33 CrossRef CAS PubMed.
- R. Z. Chen, H. Y. Shi, X. Y. Meng, Y. Y. Su, H. Y. Wang and Y. He, Anal. Chem., 2019, 91, 3597–3603 CrossRef.
- W. X. Yuan, K. T. Xiao, X. X. Liu, Y. M. Lai, F. Z. Luo, W. Xiao, J. J. Wu, P. Pan, Y. K. Li and H. Xiao, Anal. Chim. Acta, 2023, 1273, 341559 CrossRef PubMed.
- Z. K. Zheng, T. T. Liu, H. Y. Zhao, L. Cui and X. M. Zhang, Sens. Actuators, B, 2022, 372, 132650 CrossRef.
- S. C. Liu, M. He, B. B. Chen, X. Yin, Q. Kang, Y. Xu and B. Hu, Sens. Actuators, B, 2022, 362, 131758 CrossRef.
- L. Schermelleh, F. Spada, H. P. Easwaran, K. Zolghadr, J. B. Margot, M. C. Cardoso and H. Leonhardt, Nat. Methods, 2005, 2, 751–756 CrossRef PubMed.
- Y. Jiang, H. Chai, N. H. Feng and P. Miao, ACS Sustainable Chem. Eng., 2021, 9, 9257–9263 CrossRef.
- D. Greiner, T. Bonaldi, R. Eskeland, E. Roemer and A. Imhof, Nat. Chem. Biol., 2005, 1, 143–145 CrossRef PubMed.
- J. J. Jiang, J. Liu, D. Sanders, S. M. Qian, W. D. Ren, J. K. Song, F. Q. Liu and X. H. Zhong, Nat. Plants, 2021, 7, 184–197 CrossRef PubMed.
- J. Li, H. F. Yan, K. M. Wang, W. H. Tan and X. W. Zhou, Anal. Chem., 2007, 79, 1050–1056 CrossRef PubMed.
- M. Qing, Y. Fan, S. L. Chen, H. Q. Luo and N. B. Li, Biosens. Bioelectron., 2021, 192, 113487 CrossRef PubMed.
- L. J. Wang, X. Han, C. C. Li and C. Y. Zhang, Chem. Sci., 2018, 9, 6053–6061 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Details of experimental information, Fig. S1, S2 and Tables S1–S3. See DOI: https://doi.org/10.1039/d3an01780j |
| This journal is © The Royal Society of Chemistry 2024 |