Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemical synthesis and NMR spectroscopy of long stable isotope labelled RNA†
J.
Kremser
,
E.
Strebitzer
,
R.
Plangger
,
M. A.
Juen
,
F.
Nußbaumer
,
H.
Glasner
,
K.
Breuker
and
C.
Kreutz
 *
*
Institute of Organic Chemistry and Center for Molecular Biosciences Innsbruck (CMBI), University of Innsbruck, Innrain 80/82, 6020 Innsbruck, Austria. E-mail: christoph.kreutz@uibk.ac.at
First published on 20th November 2017
Abstract
We showcase the high potential of the 2′-cyanoethoxymethyl (CEM) methodology to synthesize RNAs with naturally occurring modified residues carrying stable isotope (SI) labels for NMR spectroscopic applications. The method was applied to synthesize RNAs with sizes ranging between 60 to 80 nucleotides. The presented approach gives the possibility to selectively modify larger RNAs (>60 nucleotides) with atom-specifically 13C/15N-labelled building blocks. The method harbors the unique potential to address structural as well as dynamic features of these RNAs with NMR spectroscopy but also using other biophysical methods, such as mass spectrometry (MS), or small angle neutron/X-ray scattering (SANS, SAXS).
Solution and solid state nuclear magnetic resonance (NMR) spectroscopy have proven to be highly suitable to address structural and dynamic features of RNA.1–4 A prerequisite to apply state-of-the-art NMR experiments is the introduction of a stable isotope (SI) labelling pattern using 13C/15N labelled RNA or DNA precursors.5–8 The most wide-spread method uses labelled (2′-deoxy)-ribonucleotide triphosphates and enzymes to produce the desired RNA or DNA sequence enriched with 13C and 15N nuclei.1,5 This approach enables to produce sufficient amounts of RNA and DNA for NMR spectroscopic applications. This well-established method allows nucleotide specific labeling by mixing a SI-labeled with unlabeled d/rNTPs. Especially in larger RNAs (>60 nt) such nucleotide specific SI-labeling can still lead to significant resonance overlap. That is why, the PLOR (position-selective labelling of RNA) method was recently introduced, which holds the promise to site-specifically label RNA using SI-labelled ribonucleotide triphosphates and T7 RNA polymerase.9 An alternative method was concurrently developed making use of the synthesis of 2′-O-tri-iso-propylsilyloxymethyl (TOM)- or 2′-O-tert-butyl-dimethyl-silyl-(TBDMS)-SI-modified phosphoramidites and solid phase synthesis.10–13 The approach works well for medium sized RNAs up to 50 nts and the synthetic access to the SI-labelled building blocks is well established.10,12 Thus, the fully chemical SI-labelling protocol can be regarded as an expedient expansion to the settled enzymatic procedures to freely chose the number and positioning of SI-labeled residues into a target RNA. In our hands, however, the standard solid phase synthesis methods are not that well suited to produce larger amounts (>50 nmol) and purities higher than 95% for RNAs exceeding 60 nts. Due to this restriction, large RNAs are only accessible via enzymatic ligation strategies using T4 RNA/DNA ligase making extra optimization steps necessary or introducing new problems, such as finding the optimal ligation site or issues regarding up-scaling and yield of the ligation product.14–16 Thus, an improved synthetic procedure to directly address SI-labelling of larger RNAs (>60 nt) at amounts suitable for NMR would be highly desirable.
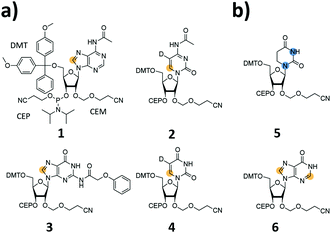
We report the synthesis of SI-labelled RNAs ranging in size between 60 to 80 nts capitalizing on the 2′-cyanoethoxymethyl (CEM) RNA synthesis method.17,18 As these CEM building blocks are not commercially available all phosphoramidites were produced in-house and we further synthesized 13C-/15N-labelled unmodified and naturally occurring modified RNA phosphoramidites (Fig. 1a and b). In detail, we focused on the synthesis of 8-13C-adenosine (1), 6-13C-5-D-cytidine (2), 8-13C-guanosine (3) and 6-13C-5-D-uridine (4) building blocks. Modified RNA building blocks include a 1,3-15N2-dihydrouridine (5) and a 2,8-13C2-inosine (6) CEM phosphoramidite. A detailed description of the synthetic procedures is given in the ESI.†
We used these monomer units to produce SI-labelled RNAs exceeding the size limitation of 60 nucleotides for NMR up to 20 nucleotides. The RNAs reported here were synthesized on a 1.3 μmol scale and on a 1000 Å controlled pore glass (CPG) solid support with 0.1 M CEM amidite solutions, i.e. a 13-fold excess of amidite was used in each coupling step. The deprotection steps followed the recommendations of Ohgi et al. but the RNA was desalted via size-exclusion chromatography and not precipitated as suggested in the original work.17 A detailed description of the chemical RNA synthesis can be found in the ESI.†
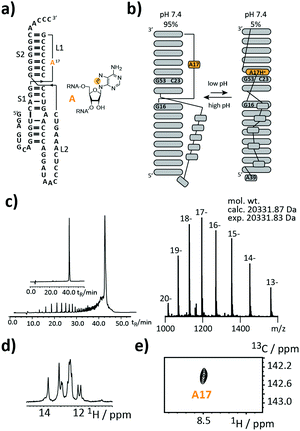
As a first target, we picked a retroviral messenger RNA, which was earlier investigated using solution NMR spectroscopy. D’Souza and co-workers found, that the murine leukaemia virus (MLV) pseudoknot (PK) is a cis-acting regulation element, which can induce alternative protein expression by the ribosomal frameshifting and the read-through mechanism.19 The MLV PK undergoes an exchange between two conformations. At low pH, A17H+ interacts with C23 and G53 leading to a release steric constraint that prevents S1–L2 tertiary interactions. At physiological pH, this state is an excited state with a population of only 6%. The pH-dependent tertiary structure transition to this excited state is a functional switch allowing the ribosome to bypass the gag stop codon (Fig. 2a and b).
We introduced a stable isotope label by replacing A17 with its 8-13C-labelled counterpart (Fig. 2a). A high-quality crude product was obtained as deduced from the anion-exchange (AIEX) chromatogram (Fig. 2c left). Characterization of the AIEX-purified MLV PK by top-down Fourier-transform ion cyclotron resonance (FT-ICR) mass spectrometry (MS)20,21 revealed a high sample homogeneity (Fig. 2c right), the correct sequence, and localized the SI label to A17 (ESI,† Fig. S1). We exploited the SI-labelling pattern to study the RNA via NMR spectroscopy (Fig. 2d and e). The imino proton spectrum is in agreement with a previously acquired spectrum.19 We also conducted a 13C CPMG relaxation dispersion experiment, but the data could not be properly fitted to give the parameters of the exchange process due to the small amplitude of the dispersion profile (data not shown).
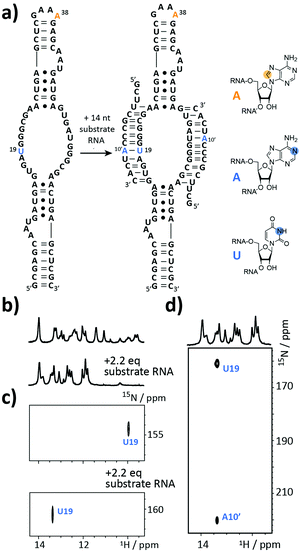
The next target is part of the box C/D ribonucleoprotein (RNP) particle, for which a structural model was recently obtained using an integrative structural biology approach.22 The RNP catalyzes the post-transcriptional modification of rRNA – the 2′-O-methylation. In the core of the complex resides a guide RNA, which recruits the substrate RNAs for processing by the fibrillarin enzyme to introduce the 2′-O-methyl group using S-adenosyl-L-methionine (SAM) as the co-factor. With 72 nt the partially symmetrized guide ssR26 represents a challenging target for the solid phase synthesis. The guide RNA was synthesized using the 8-13C-A amidite 1 but even more noteworthy a 3-15N-labelled 2′-O-TBDMS uridine amidite (Fig. 3a). The crude product gave a AIEX chromatogram with a main product peak (ESI,† Fig. S2). Thus, the CEM method tolerates a limited number of TBDMS RNA building blocks further increasing the versatility of the presented approach as many commercially available RNA modifiers use the standard 2′-O-TBDMS protecting group. So far, we introduced up to four 2′-O-TBDMS building blocks into 60 nt RNAs assembled via the CEM method.
The isolated yield of full-length SI-labelled 72 nt was 95 nmol (7%). Top-down FT-ICR MS confirmed the sequence and located the SI labels to residues U19 and A38 (ESI,† Fig. S3). The structure of the 72 nt ssR26 RNA was then probed using solution NMR spectroscopy (Fig. 3b–d).
The imino proton region of the 1H-NMR spectrum indicated a well-structured RNA with several resonances in the non-standard Watson–Crick base pair chemical shift region (Fig. 3b). A 1H–15N-correlation spectrum disclosed that U19 is forming a base pairing interaction, very likely a G·U wobble base pair based on the chemical shift signature (Fig. 3c). Making use of the 8-13C-A38, an 1H–13C-HSQC spectrum of ssR26 revealed conformational heterogeneity as two 1H–13C-correlation peaks were observed (ESI,† Fig. S4). We were then interested in the consequences induced by the addition of the substrate RNA. Upon the addition of 2.2 equivalents of substrate RNA carrying a 1-15N-adenosine label, the spectra displayed significant changes. A 1H–15N-HSQC spectrum nicely reflected the structural transition of U19 to a standard A-U Watson–Crick base pair (Fig. 3c). The base pairing partner A10′ and the intermolecular A10′–U19 base pair could further be unambiguously confirmed by a HNN-COSY (Fig. 3d).23 A 1H–13C-correlation NMR experiment showed that the conformational heterogeneity of the apo ssR26 RNA was resolved and the binding of the substrate strand leads to a homogeneous folding state (ESI,† Fig. S4).
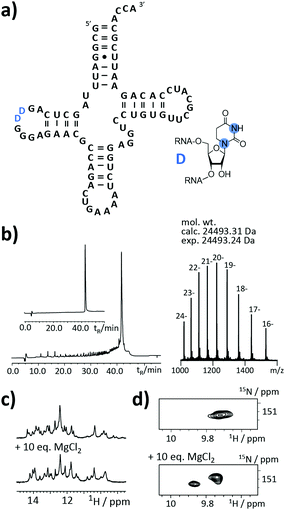
As the final example, the highly relevant class of tRNAs was selected. This RNA species fulfills a translator function by transferring the mRNA information into an amino acid sequence.24–26 But still, certain aspects of its structure/function are not clear. The functions and roles of modified residues, such as dihydrouridine (DHU) or more complex modifiers (e.g. uridine 5-oxyacetic acid), on a molecular level are not yet fully elucidated.
An as so far largely unexplored aspect of modified RNA residues is their influence on the folding landscape. SI-labelled variants of the modified RNA residues are mandatory to characterize their influence on an RNA's folding landscape by NMR.27 Two examples, the 1,3-15N2-DHU (5) and the 2,8-13C2-inosine RNA (6) amidites, are introduced here. DHU is as a dynamic hotspot as it does not form π-stacking interactions and preferentially populates the C2′-endo conformation.28 Here, we report the synthesis of a DHU modified tRNA (Fig. 4a). A high-quality crude product peak was observed and after purification, the exact mass, the sequence and the location of the SI labels (D16 and D17) were confirmed by FT-ICR MS (Fig. 4b and ESI,† Fig. S5). A 0.15 mM sample in 400 μL was obtained corresponding to a total yield of 60 nmol (4.6% yield). 1H NMR spectra and 1H–15N HSQC spectra confirmed the SI labelling (Fig. 4c and d). A folding event of the DHU modified tRNA was triggered by the addition of 10 equivalents magnesium(II) ions, which could be nicely followed via NMR spectroscopy (Fig. 4c and d).
Noteworthy, we changed the N4-acetyl group of the 2′-O-CEM-cytidine to a phenoxyacetyl moiety as the mild alkaline deprotection conditions (2 M ammonia in methanol, 37 °C, 20 h) keeping the DHU residue intact left some of the N4-acetyl groups untouched. We plan to use 15N-relaxation dispersion NMR to probe the influence of the DHU residues on the tRNA's folding landscape. In analogy, we plan to address functional dynamics induced by inosine, as a recent work reports a destabilizing effect of this modification.29
In this work, we report a synthetic access to SI-labelled RNAs using chemical solid phase synthesis. The minimal steric demand of the 2′-CEM protecting group and a clean deprotection procedure give high quality products, which can be purified using denaturing AIEX chromatography to yield RNAs up to 80 nt with the required >95% purity for NMR. We synthesized atom-specific 13C-labelled 2′-O-CEM amidites 1–4 and modified SI-labelled RNA building blocks (5, 6) and incorporated them in various RNAs. The thus obtained nucleic acids proved to be suitable for NMR investigations to probe their structure/dynamics. We foresee several areas of applications for such SI-labelled RNAs. The most obvious utilization is the unambiguous resonance assignment in larger RNAs using atom-specific SI labelled residues.30 Further, the isolated spin pair 1H–X (X = 13C/15N) topologies of the building blocks 1 to 6 minimize the scalar coupling interactions and relaxation pathways and thus make the application of relaxation based NMR experiments, such as relaxation dispersion or CEST experiments, straightforward to probe functional dynamics in RNAs.10,12,13 Besides, we anticipate a potential use of the large SI labelled RNAs in recently reported mass spectrometric methods to localize protein binding sites, which give valuable information for the 3D structure modelling of large RNP particles.31,32 We currently also focus on the synthesis of per-deuterated RNA building blocks. The building blocks will be beneficial for NMR but also for SAXS/SANS studies in an integrative structural biology approach, as the chemical RNA synthesis allows full control over segmental deuteration. This will offer the possibility to define relative domain orientations in such larger nucleic acids by contrast-matched SANS or SAXS, as recently suggested for proteins.33 To conclude, we are confident that SI-labelling of RNA via the 2′-O-CEM methodology is a competitive new approach with respect to existing chemical and enzymatic protocols to modify nucleic acids for biophysical investigations.
Conflicts of interest
There are no conflicts to declare.References
- R. P. Barnwal, F. Yang and G. Varani, Arch. Biochem. Biophys., 2017, 628, 42–56 CrossRef CAS PubMed.
- C. Dominguez, M. Schubert, O. Duss, S. Ravindranathan and F. H. T. Allain, Prog. Nucl. Magn. Reson. Spectrosc., 2011, 58, 1–61 CrossRef CAS PubMed.
- A. M. Mustoe, C. L. Brooks and H. M. Al-Hashimi, Annu. Rev. Biochem., 2014, 83, 441–466 CrossRef CAS PubMed.
- A. Marchanka, B. Simon, G. Althoff-Ospelt and T. Carlomagno, Nat. Commun., 2015, 6, 7024 CrossRef CAS PubMed.
- K. Lu, Y. Miyazaki and M. F. Summers, J. Biomol. NMR, 2009, 46, 113 CrossRef PubMed.
- C. S. Thakur and T. K. Dayie, J. Biomol. NMR, 2012, 52, 65–77 CrossRef CAS PubMed.
- L. J. Alvarado, A. P. Longhini, R. M. LeBlanc, B. Chen, C. Kreutz and T. K. Dayie, Methods Enzymol., 2014, 549, 133–162 CAS.
- M. T. Le, R. E. Brown, A. E. Simon and T. K. Dayie, Methods Enzymol., 2015, 565, 495–535 Search PubMed.
- Y. Liu, E. Holmstrom, J. Zhang, P. Yu, J. Wang, M. A. Dyba, C. De, J. Ying, S. Lockett, D. J. Nesbitt, A. R. Ferre-D’Amare, R. Sousa, J. R. Stagno and Y.-X. Wang, Nature, 2015, 522, 368–372 CrossRef CAS PubMed.
- M. A. Juen, C. H. Wunderlich, F. Nußbaumer, M. Tollinger, G. Kontaxis, R. Konrat, D. F. Hansen and C. Kreutz, Angew. Chem., Int. Ed., 2016, 55, 12008–12012 CrossRef CAS PubMed.
- S. Neuner, C. Kreutz and R. Micura, Monatsh. Chem., 2017, 148, 149–155 CrossRef CAS PubMed.
- C. H. Wunderlich, R. Spitzer, T. Santner, K. Fauster, M. Tollinger and C. Kreutz, J. Am. Chem. Soc., 2012, 134, 7558–7569 CrossRef CAS PubMed.
- H. Zhou, I. J. Kimsey, E. N. Nikolova, B. Sathyamoorthy, G. Grazioli, J. McSally, T. Bai, C. H. Wunderlich, C. Kreutz, I. Andricioaei and H. M. Al-Hashimi, Nat. Struct. Mol. Biol., 2016, 23, 803–810 CAS.
- R. Rieder, C. Höbartner and R. Micura, in Riboswitches: Methods and Protocols, ed. A. Serganov, Humana Press, Totowa, NJ, 2009, pp. 15–24, DOI:10.1007/978-1-59745-558-9_2.
- B. Chen, R. LeBlanc and T. K. Dayie, Angew. Chem., Int. Ed., 2016, 55, 2724–2727 CrossRef CAS PubMed.
- O. Duss, N. Diarra dit Konté and F. H. T. Allain, Methods Enzymol., 2015, 565, 537–562 Search PubMed.
- Y. Shiba, H. Masuda, N. Watanabe, T. Ego, K. Takagaki, K. Ishiyama, T. Ohgi and J. Yano, Nucleic Acids Res., 2007, 35, 3287–3296 CrossRef CAS PubMed.
- T. Ohgi, Y. Masutomi, K. Ishiyama, H. Kitagawa, Y. Shiba and J. Yano, Org. Lett., 2005, 7, 3477–3480 CrossRef CAS PubMed.
- B. Houck-Loomis, M. A. Durney, C. Salguero, N. Shankar, J. M. Nagle, S. P. Goff and V. M. D’Souza, Nature, 2011, 480, 561–564 CAS.
- H. Glasner, C. Riml, R. Micura and K. Breuker, Nucleic Acids Res., 2017, 45, 8014–8025 CrossRef PubMed.
- M. Taucher and K. Breuker, Angew. Chem., Int. Ed., 2012, 51, 11289–11292 CrossRef CAS PubMed.
- A. Lapinaite, B. Simon, L. Skjaerven, M. Rakwalska-Bange, F. Gabel and T. Carlomagno, Nature, 2013, 502, 519–523 CrossRef CAS PubMed.
- A. J. Dingley, L. Nisius, F. Cordier and S. Grzesiek, Nat. Protoc., 2008, 3, 242–248 CrossRef CAS PubMed.
- M. Helm, Nucleic Acids Res., 2006, 34, 721–733 CrossRef CAS PubMed.
- J. E. Jackman and J. D. Alfonzo, Wiley Interdiscip. Rev.: RNA, 2013, 4, 35–48 CrossRef CAS PubMed.
- V. Ramakrishnan, Cell, 2014, 159, 979–984 CrossRef CAS PubMed.
- J. R. Bothe, E. N. Nikolova, C. D. Eichhorn, J. Chugh, A. L. Hansen and H. M. Al-Hashimi, Nat. Methods, 2011, 8, 919–931 CrossRef CAS PubMed.
- N. Dyubankova, E. Sochacka, K. Kraszewska, B. Nawrot, P. Herdewijn and E. Lescrinier, Org. Biomol. Chem., 2015, 13, 4960–4966 CAS.
- A. Dallmann, A. V. Beribisky, F. Gnerlich, M. Rübbelke, S. Schiesser, T. Carell and M. Sattler, Chem. – Eur. J., 2016, 22, 15350–15359 CrossRef CAS PubMed.
- A. C. Wolter, A. K. Weickhmann, A. H. Nasiri, K. Hantke, O. Ohlenschläger, C. H. Wunderlich, C. Kreutz, E. Duchardt-Ferner and J. Wöhnert, Angew. Chem., Int. Ed., 2017, 56, 401–404 CrossRef CAS PubMed.
- G. Dorn, A. Leitner, J. Boudet, S. Campagne, C. von Schroetter, A. Moursy, R. Aebersold and F. H. T. Allain, Nat. Methods, 2017, 14, 487–490 CrossRef CAS PubMed.
- E.-M. Schneeberger and K. Breuker, Angew. Chem., Int. Ed., 2017, 56, 1254–1258 CrossRef CAS PubMed.
- M. Sonntag, P. K. A. Jagtap, B. Simon, M.-S. Appavou, A. Geerlof, R. Stehle, F. Gabel, J. Hennig and M. Sattler, Angew. Chem., Int. Ed., 2017, 56, 9322–9325 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc06747j |
| This journal is © The Royal Society of Chemistry 2017 |