The impact of structure dimensions on initial bacterial adhesion†
Ralf
Helbig
*a,
Denise
Günther
be,
Jens
Friedrichs
a,
Florian
Rößler
be,
Andrés
Lasagni
be and
Carsten
Werner
acd
aDepartment of Biofunctional Polymer Materials – Max Bergman Center of Biomaterials, Leibniz Institute of Polymer Research, Hohe Straße 6, D-01069 Dresden, Germany. E-mail: helbig@ipfdd.de
bFraunhofer Institute for Material and Beam Technology, Winterbergstraße 28, D-01277, Dresden, Germany
cB CUBE - Center for Molecular Bioengineering, Dresden University of Technology, Arnoldstraße 18, D-01307 Dresden, Germany
dCRTD – Center of Regenerative Therapies Dresden – Center of Excellence, Fetscherstraße 105, D-01307 Dresden, Germany
eInstitute of Manufacturing Technology, University of Technology, Georg-Bähr Straße 3c, D-01069 Dresden, Germany
First published on 27th May 2016
Abstract
Substrate topography can have profound effects on initial bacterial adhesion during biofilm formation. We applied Staphylococcus epidermidis and Escherichia coli cells onto periodically structured substrates with different structure dimensions, structure types and wetting properties. We found a strong dependence of cell retention on the structure dimensions of the applied substrates. Periodicities in the range of the cell size increased, whereas smaller periodicities decreased cell retention, independent of contact time (minutes to hours) and hydrophobicity. These novel insights on the role of surface topography on bacterial retention might facilitate the development of non-fouling surfaces in the future.
Adhesion, accumulation and growth of microorganisms on man-made surfaces in contact with aerial or saline environments, designated as biofouling, can have severe negative consequences in various fields including industrial processes (e.g., food processing, textile, pulp and paper manufacturing),1,2 medicine (e.g., nosocomial infections)3–5 and in seawater-contacting equipment (e.g., pipelines, cooling and filtration systems, fishing nets, ship hulls and bridge pillars).6 Biofouling is a complex process that can generally be described by a basic sequence of events. First, a conditioning film is formed on a surface by rapid adsorption of organic molecules (mainly proteins and polysaccharides). Then, a microbial biofilm develops, which involves the attachment of bacterial cells and/or diatoms, growth and proliferation of the attached cells, formation of mature colonies and, finally, partial detachment of cell clusters. In contrast to the strongly adhering mature biofilm, the initial (minutes–hours) adhesion of bacteria onto the surfaces is generally reversible. Therefore, many antifouling strategies rely on the prevention/intervention of initial bacterial adhesion rather than on the removal of the mature biofilms.
Besides the environmental conditions, such as pH, temperature, competing organisms and nutrition, the interactions between the bacteria and the substrate are mainly influenced by interfacial properties such as chemistry, polarity, mechanical properties and structure. The structural features of substrates, such as size, spacing, aspect ratio and roughness can have both deterrent and attractive effects on the settlement of fouling organisms.7,8
On nano-rough surfaces the number of attached cells and the amount of secreted exopolysaccharides (EPS) can be strongly affected by the subtle differences in surface roughness.9–14 Bacterial biofilm formation has been observed to be more pronounced on surfaces with the root mean square roughness value of 10 nm compared to that of 5 nm and 15 nm.9 For submicron- and micron-sized structures, a permanent increase and an intermediate maximum or minimum of bacterial settlement have been observed depending on feature spacing, period or size.15–20 In some reports, a strong impact of feature sizes and/or spacing in the range of the cell size on cell–substrate interactions and colonization has been described.8,15,21–23 The structural features of substrates were also reported to influence cell orientation. The regular structures were suggested to influence settlement patterns more than irregular structures and the order of cell pattern was more pronounced on micron- than on nano-sized features, periods and spacings.7 In particular, micron-sized grooves and pillar arrays were found to guide cell orientation and proliferation.21–25
A conclusive picture of bacterial retention on structured substrates with graded structural dimensions does not currently exist.26,27 The vast knowledge bases on cell retention and orientation on materials with structures spanning the nano- and micro-scale are partially contradictory, perhaps due to the fact that different ranges of structure dimensions as well as structure types with periodically arranged or randomly distributed structures were applied. In addition, different bacterial strains and experimental conditions were used. Resolving these contradictions and exploring the role of structure types and dimensions in the initial bacterial adhesion with respect to the colonization patterns are therefore key challenges of current research.
In the current study, cells of a Gram-positive bacterial strain Staphylococcus epidermidis (S. epidermidis) and a Gram-negative strain of Escherichia coli (E. coli) were exposed to sets of substrates with micron- and submicron-sized structures. The substrates were prepared by Laser Interference Patterning (LIP).28–30 LIP is a mask- and moldless high throughput method that uses a standing wave pattern at the intersection of two or more coherent and collimated laser beams to create micron- and submicron-scaled structures in photo-sensitive materials like photo-resists or by ablation of polymers, ceramics and metals (Fig. S1†). The shape and dimension of the interference patterns can be adjusted by controlling the number of laser beams as well as their geometrical configuration. This method can be applied to large areas and various surface shapes and is not restricted to flat substrates. In order to comprehensively understand the impact of structure dimensions on the bacterial settlement the periodicity of the structures –Λ– was chosen to be larger, similar or smaller than the cell size of S. epidermidis (spherical with a diameter of about 1000 nm) and E. coli (rod-shaped with about 1000 nm width and 2000–3000 nm minimal length).
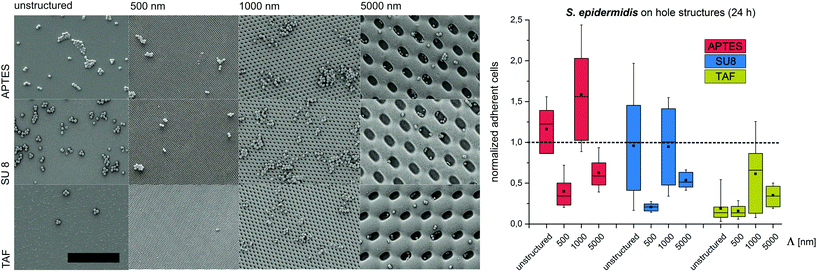
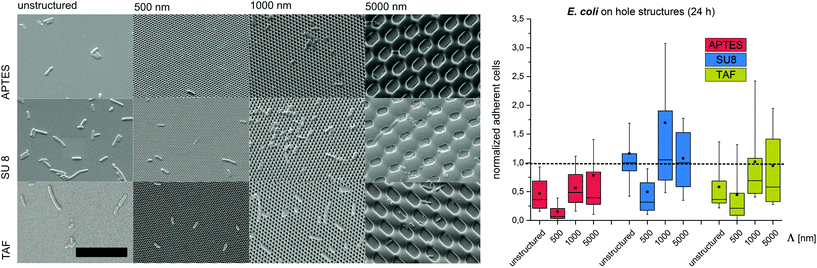
Substrates with comb-like hole patterns were incubated with the bacteria in growth medium for 24 h. Subsequently, the number of adherent cells was quantified. The highest number of S. epidermidis cells was found on substrates with a periodicity in the range of the cell size, i.e. Λ = 1000 nm (Fig. 1). Larger periodicities (Λ = 5000 nm) led to a lower number of attached cells. For E. coli no significant difference in the amount of adherent cells was detected for periodicities of 1000 nm and 5000 nm (Fig. 2). For both strains the lowest number of cells was detected on substrates with Λ = 500 nm (Fig. 1 and 2).
To investigate the effect of the surface wettability/chemistry on the observed trend, the bacteria were incubated for 24 h on the structures that were hydrophilized or hydrophobized with aminosilane (APTES) or amorphous fluoropolymer TAF, respectively. The wettability, illustrated by water contact angles (Table S1†), is caused by different surface charges. APTES is positively charged,31 SU8 is slightly negatively charged (unpublished data) and TAF represents a nonpolar surface. Although the absolute number of adherent cells was different for the tested surface modifications, the relative trends were similar to the initial experiments irrespective of the surface wettability/chemistry (Fig. 1 and 2). The highest number of cells was always found on substrates with Λ = 1000 nm (both strains) and Λ = 5000 nm (E. coli only), whereas the lowest number of cells was always found on structures with Λ = 500 nm (Fig. 1 and 2). Irrespective of the structure dimensions, the experiments with S. epidermidis revealed a preference for retention on the hydrophilic APTES and a lower settlement on the hydrophobic TAF. Interestingly, E. coli cells showed a different trend. The highest number of cells was found on unmodified SU 8 surfaces, whereas few cells adhered to APTES- and TAF-modified surfaces. These results underline the fact that different bacterial strains prefer different surface chemistries, although both the strains possess an overall negative net charge.32,33
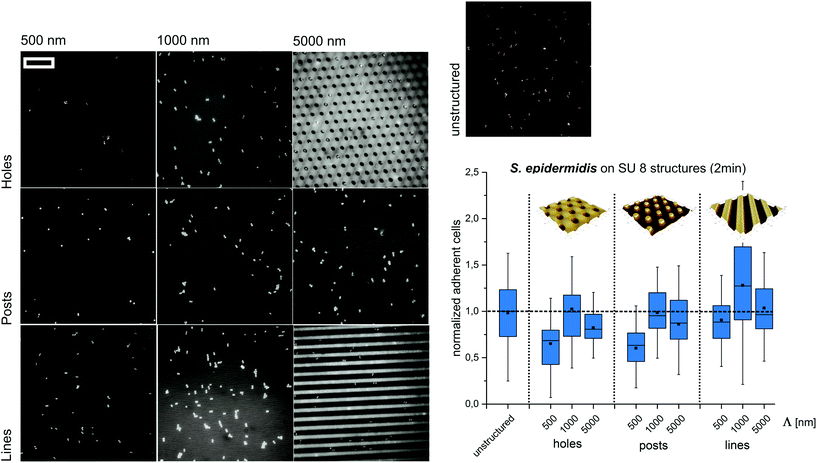
Next, we set out to test if the observed trends can be reproduced at significantly shorter contact times and different structure types. Therefore, S. epidermidis cells were incubated only for 2 min on SU8 hole structures and the attached cells were quantified. Although less pronounced, the overall trends of these experiments were comparable to the short-term adhesion experiments. The highest amount of cells was found on structures with a periodicity of 1000 nm, whereas submicron-scaled structures (Λ = 500 nm) suppress initial cell adhesion (Fig. 3).
Subsequently, we tested if the observed trends depend on the type of structure applied. Therefore, S. epidermidis cells were applied for 2 minutes to SU8 post and line structures with periods smaller, similar and larger than the cell size (wetting parameters in ESI Table S2†). Again, the highest amount of cells was observed for Λ = 1000 nm whereas the lowest amount of cells was found for Λ = 500 nm (Fig. 3).
Previous studies suggested a dependence of bacterial cell retention and orientation on the periodicity and dimension of structured surfaces,24,25 particularly when feature sizes are in the range of the dimensions of the attaching cells.8,15,21,22 Here, we applied a systematic approach to quantify the initial adhesion of two bacterial strains on substrates with graded structure dimensions and additionally tested the effect of contact time, surface chemistry and structure type on cell retention. We suggest that, the maximized cell–substrate contact area on structures with the dimensions in the range of the cell size (Λ = 1000 nm for S. epidermidis, Λ between 1000 nm and 5000 nm for E. coli) lead to high cell retention on these structures. In contrast, topographies with structure periods below the size of individual bacterial cells (Λ = 500 nm) restrict the cell–substrate contact area and therefore lead to reduced cell retention. For topographies with structure periods bigger than the cell size (Λ = 5000 nm) cell retention strongly depends on the cell morphology. For spherical cells, cell retention was lower (compared to Λ = 1000 nm), whereas for rod-shaped cells, with the dimension of the long-axis slightly below the dimensions of the structured surface, cell retention was comparable to Λ = 1000 nm. In future studies it will be interesting to quantify the retention of E. coli on the structures with dimensions significantly bigger than the length of this bacterial strain (i.e. Λ = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm). Interestingly, the observed trend was found to be independent of the contact time between the bacteria and the substrate, the structure type and the substrate chemistry, demonstrating that surface topography is indeed a strong trigger influencing cell adhesion.
000 nm). Interestingly, the observed trend was found to be independent of the contact time between the bacteria and the substrate, the structure type and the substrate chemistry, demonstrating that surface topography is indeed a strong trigger influencing cell adhesion.
The presented results demonstrate that the structural properties of surfaces govern the initial adhesion of two important opportunistic pathogens, Staphylococcus epidermidis and Escherichia coli. Usually, a comprehensive discussion about microbial settlement and biofouling must include considerations of specific and non-specific interactions of bacteria with a substrate, such as cell appendages, specific attachment components, surface free energy, cell membrane charges, surface zeta potential and hydrophobicity. The current work proves that inhibition of bacterial colonization can be supported by sub-cell sized topographies irrespective of the physicochemical properties of the used materials and, therefore, irrespective of different nonspecific cell–substrate interactions. Nevertheless, so far this study cannot claim a long term prevention of biofilm formation which normally takes place after some days of surface colonization, but it might play an important role by inhibiting microbial dispersal in a hospital environment or in food processing areas due to the strong inhibition of the initial attachment on the devices and instruments with submicron-scaled structures. The use of sub-cell sized comb or hole structures in opposite to line and post structures should lead to applicable coatings due to the appropriate mechanical properties. Fragile line and post structures were only used to work out the underlying principal. Ongoing studies should aim at exploring the effects of similar surface characteristics on the adhesion of bacteria on respective preconditioned surfaces by hospital relevant proteins, such as albumin, fibrinogen, lysozyme, etc. as well as long term in situ assays.
Author contributions
The manuscript was written through contributions of all the authors. All the authors have given approval to the final version of the manuscript.Conflict of interest
The authors declare no competing financial interest.Funding sources
This work was supported by Deutsche Forschungsgemeinschaft (DFG WE 2539/17-1, DFG LA 2513/4-1).Acknowledgements
We are grateful to Dr Mirko Nitschke (Max Bergmann Center of Biomaterials, Leibniz Institute of Polymer Research Dresden, Germany) for support in XPS measurements for the evaluation of the amino-silanization of SU8.References
- L. V. Poulsen, Microbial Biofilm in Food Processing, LWT–Food Sci. Technol., 1999, 32, 321–326 CrossRef CAS.
- G. D. Bixler and B. Bhushan, Biofouling: Lessons from Nature, Philos. Trans. R. Soc. London, Ser. A, 2012, 370, 2381–2417 CrossRef CAS PubMed.
- I. Raad, A. Alrahwan and K. Rolston, Staphylococcus Epidermidis: Emerging Resistance and Need for Alternative Agents, Clin. Infect. Dis., 1998, 26, 1182–1187 CAS.
- C. Vuong and M. Otto, Staphylococcus Epidermidis Infections, Microbes Infect., 2002, 4, 481–489 CrossRef PubMed.
- W. Ziebuhr, S. Hennig, M. Eckart, H. Kränzler, C. Batzilla and S. Kozitskaya, Nosocomial Infections by Staphylococcus Epidermidis: How a Commensal Bacterium Turns into a Pathogen, Int. J. Antimicrob. Agents, 2006, 28(Suppl 1), S14–S20 CrossRef CAS PubMed.
- M. E. Callow and J. A. Callow, Marine Biofouling: A Sticky Problem, Biologiste, 2002, 49, 1–5 Search PubMed.
- M. Graham and N. Cady, Nano and Microscale Topographies for the Prevention of Bacterial Surface Fouling, Coatings, 2014, 4, 37–59 CrossRef CAS.
- K. A. Whitehead and J. Verran, The Effect of Surface Topography on the Retention of Microorganisms, Trans. Inst. Chem. Eng., Part C, 2006, 84, 253–259 CrossRef.
- A. Kerr and M. J. Cowling, The Effects of Surface Topography on the Accumulation of Biofouling, Philos. Mag., 2003, 83, 2779–2795 CrossRef CAS.
- E. P. Ivanova, V. K. Truong, J. Y. Wang, C. C. Berndt, R. T. Jones, I. I. Yusuf, I. Peake, H. W. Schmidt, C. Fluke and D. Barnes, et al., Impact of Nanoscale Roughness of Titanium Thin Film Surfaces on Bacterial Retention, Langmuir, 2010, 26, 1973–1982 CrossRef CAS PubMed.
- N. Mitik-Dineva, J. Wang, V. K. Truong, P. Stoddart, F. Malherbe, R. J. Crawford and E. P. Ivanova, Escherichia Coli, Pseudomonas Aeruginosa, and Staphylococcus Aureus Attachment Patterns on Glass Surfaces with Nanoscale Roughness, Curr. Microbiol., 2009, 58, 268–273 CrossRef CAS PubMed.
- A. V. Singh, V. Vyas, R. Patil, V. Sharma, P. E. Scopelliti, G. Bongiorno, A. Podestà, C. Lenardi, W. N. Gade and P. Milani, Quantitative Characterization of the Influence of the Nanoscale Morphology of Nanostructured Surfaces on Bacterial Adhesion and Biofilm Formation, PLoS One, 2011, 6, e25029 CAS.
- C. Satriano, G. M. L. Messina, S. Carnazza, S. Guglielmino and G. Marletta, Bacterial Adhesion onto Nanopatterned Polymer Surfaces, Mater. Sci. Eng., C, 2006, 26, 942–946 CrossRef CAS.
- M. R. Park, M. K. Banks, B. Applegate and T. J. Webster, Influence of Nanophase Titania Topography on Bacterial Attachment and Metabolism, Int. J. Nanomed., 2008, 3, 497–504 CAS.
- K. A. Whitehead, J. Colligon and J. Verran, Retention of Microbial Cells in Substratum Surface Features of Micrometer and Sub-Micrometer Dimensions, Colloids Surf., B, 2005, 41, 129–138 CrossRef CAS PubMed.
- S. E. Tebbs, a. Sawyer and T. S. Elliott, Influence of Surface Morphology on in Vitro Bacterial Adherence to Central Venous Catheters, Br. J. Anaesth., 1994, 72, 587–591 CrossRef CAS PubMed.
- S. H. Flint, J. D. Brooks and P. J. Bremer, Properties of the Stainless Steel Substrate, Influencing the Adhesion of Thermo-Resistant Streptococci, J. Food Eng., 2000, 43, 235–242 CrossRef.
- Y. Wu, J. P. Zitelli, K. S. TenHuisen, X. Yu and M. R. Libera, Differential Response of Staphylococci and Osteoblasts to Varying Titanium Surface Roughness, Biomaterials, 2011, 32, 951–960 CrossRef CAS PubMed.
- A. C. Ihnen, J.-H. Lee and W. Y. Lee, Effects of Disordered Hemispherical Micropatterns on Staphylococcus Epidermidis Biofilm Formation, Colloids Surf., B, 2010, 75, 601–607 CrossRef CAS PubMed.
- J. Verran, A. Packer, P. Kelly and K. A. Whitehead, The Retention of Bacteria on Hygienic Surfaces Presenting Scratches of Microbial Dimensions, Lett. Appl. Microbiol., 2010, 50, 258–263 CrossRef CAS PubMed.
- A. K. Epstein, A. I. Hochbaum, P. Kim and J. Aizenberg, Control of Bacterial Biofilm Growth on Surfaces by Nanostructural Mechanics and Geometry, Nanotechnology, 2011, 22, 494007–494014 CrossRef CAS PubMed.
- A. I. Hochbaum and J. Aizenberg, Bacteria Pattern Spontaneously on Periodic Nanostructure Arrays, Nano Lett., 2010, 10, 3717–3721 CrossRef CAS PubMed.
- J. Valle, S. Burgui, D. Langheinrich, C. Gil, C. Solano, A. Toledo-Arana, R. Helbig, A. Lasagni and I. Lasa, Evaluation of Surface Microtopography Engineered by Direct Laser Interference for Bacterial Anti-Biofouling, Macromol. Biosci., 2015, 15, 1060–1069 CrossRef CAS PubMed.
- C. Diaz, P. L. Schilardi, R. C. Salvarezza and M. F. L. Mele de, Nano/Microscale Order Affects the Early Stages of Biofilm Formation on Metal Surfaces, Langmuir, 2007, 23, 11206–11210 CrossRef CAS PubMed.
- C. Díaz, P. Schilardi and M. F. L. de Mele, Influence of Surface Sub-Micropattern on the Adhesion of Pioneer Bacteria on Metals, Artif. Organs, 2008, 32, 292–298 CrossRef PubMed.
- K. Anselme, P. Davidson, A. M. Popa, M. Giazzon, M. Liley and L. Ploux, The Interaction of Cells and Bacteria with Surfaces Structured at the Nanometre Scale, Acta Biomater., 2010, 6, 3824–3846 CrossRef CAS PubMed.
- K. Bazaka, R. J. Crawford and E. P. Ivanova, Do Bacteria Differentiate between Degrees of Nanoscale Surface Roughness?, Biotechnol. J., 2011, 6, 1103–1114 CrossRef CAS PubMed.
- M. Campbell, D. Sharp, M. Harrison, R. Denning and A. Turberfield, Fabrication of Photonic Crystals for the Visible Spectrum by Holographic Lithography, Nature, 2000, 404, 53–56 CrossRef CAS PubMed.
- H. Misawa, T. Kondo, S. Juodkazis, V. Mizeikis and S. Matsuo, Holographic Lithography of Periodic Two- and Three-Dimensional Microstructures in Photoresist SU-8, Opt. Express, 2006, 14, 7943–7953 CrossRef PubMed.
- A. F. Lasagni and B. S. Menéndez-Ormaza, Two- and Three-Dimensional Micro- and Sub-Micrometer Periodic Structures Using Two-Beam Laser Interference Lithography, Adv. Eng. Mater., 2010, 12, 54–60 CrossRef CAS.
- T. Osaki, R. Zimmermann, T. Kratzmüller, R. Schweiss and C. Werner, Polyanion Protection of Silane Bonds to Silicon Oxide Revealed by Electrokinetic Measurements, Langmuir, 2004, 20, 524–527 CrossRef CAS PubMed.
- M. G. Katsikogianni and Y. F. Missirlis, Interactions of Bacteria with Specific Biomaterial Surface Chemistries under Flow Conditions, Acta Biomater., 2010, 6, 1107–1118 CrossRef CAS PubMed.
- K. A. Soni, A. K. Balasubramanian, A. Beskok and S. D. Pillai, Zeta Potential of Selected Bacteria in Drinking Water When Dead, Starved, or Exposed to Minimal and Rich Culture Media, Curr. Microbiol., 2008, 56, 93–97 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6bm00078a |
| This journal is © The Royal Society of Chemistry 2016 |