Open Access Article
Open Access ArticleThe role of cytosine methylation in regulating the topology and liquid–liquid phase separation of DNA G-quadruplexes†
Mitsuki
Tsuruta
,
Sumit
Shil
,
Shinya
Taniguchi
,
Keiko
Kawauchi
and
Daisuke
Miyoshi
 *
*
Faculty of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe, Hyogo 650-0047, Japan. E-mail: miyoshi@konan-u.ac.jp
First published on 31st January 2025
Abstract
Aberrant expansion of GGGGCC DNA repeats that form G-quadruplexes (G4) is the main cause of amyotrophic lateral sclerosis (ALS). Expanded GGGGCC repeats induce liquid–liquid phase separation (LLPS) through their interaction with cellular proteins. Furthermore, GGGGCC expansion induces cytosine methylation (mC). Previous studies have shown that even slight chemical modifications of RNAs and proteins can drastically affect their LLPS ability, yet the relationship between LLPS and epigenetic DNA modifications like mC remains unexplored. As a model system, we investigated the effects of mC on LLPS induced by GGGGCC repeat DNAs and show for the first time that mC suppresses LLPS by altering the topology of G4 from being parallel to antiparallel.
Introduction
Biomolecular droplets formed in cells by liquid–liquid phase separation (LLPS), such as nucleoli and stress granules, are gaining widespread acceptance across various fields as a novel phenomenon to precisely explain spatiotemporally regulated biological functions.1 These droplets, induced by nucleic acids and proteins, regulate a variety of biological processes, including gene expression, RNA metabolism, and signal transduction.2 For example, the nucleolus, which contains rDNA and rRNA, is involved in ribosome biogenesis. Stress granules sequester mRNA and transcription factors in response to stress signals. Proteins in these droplets are typically highly charged and enriched in disordered regions. Furthermore, LLPS is often moderated by the presence of RNA and DNA through their hybridization, sequence-dependent local flexibility, and secondary structure.3–5 Consequently, it is considered that unstructured regions are essential for proteins to undergo LLPS, whereas nucleic acids require a certain secondary structure to undergo LLPS.Among the various nucleic acid structures, the G-quadruplex (G4) structure has been shown to be a key factor in facilitating LLPS, both in vitro and in cells.6,7 Nucleoli, paraspeckles, nuclear speckles, and stress granules have all been shown to contain G4-containing LLPS in cells6–9 In addition, proteins such as fused in sarcoma (FUS), zinc finger protein 706 (ZNF706), SERPINE mRNA binding protein 1 (SERBP1), and nucleolin (NCL) have all been reported to induce LLPS with G4 in vitro.10–13 G4 is a four-stranded, non-canonical, secondary structure of nucleic acids that forms in guanine-rich sequences. These G4 structures are composed of G-quartets, each of which is comprised of four guanine bases linked by Hoogsteen hydrogen bonds, and loop regions that connect the guanine repeats. The strand direction determines the folding pattern (topology) of G4, such that in a parallel topology, all of the strands are oriented in the same direction, whereas in an antiparallel topology, the neighboring strands are oriented in opposite directions. In addition, in a hybrid topology, one of the four strands is oriented in the opposite direction. The topology and stability of G4s are influenced by environmental factors such as the type and concentration of cation species, and cosolutes that promote molecular crowding conditions in living cells.14 The interactions between G4s and proteins, which often involve cationic regions like the RGG domain, play a central role in various biological processes, including replication, transcription, splicing, and translation through LLPS.12,15–17 Interestingly, LLPS is induced by G4s and proteins, linking aberrant LLPS to neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS). The most common genetic cause of ALS involves expansion of the repetitive d(GGGGCC) sequence in the first exon of C9orf72.18–20 Notably, these GGGGCC repeats can fold to form G4s and undergo LLPS through interactions with proteins. Moreover, it has been reported that expansion of the GGGGCC repeat induces hyper-cytosine methylation (mC) in both the expanded and adjacent regions,21–24 leading to silencing of the C9orf72 gene.25,26 Similarly, abnormal increases or decreases in mC in the promoter regions of cancer-related genes also contribute to the development and progression of cancer.27 It is now evident that one of the fundamental mechanisms modulating diverse biological processes involves the interplay between LLPS and molecular machinery, such as the accumulation of chemical modifications in biomolecules.28
Methylation of RNA facilitates complex formation between mRNAs and RNA-binding proteins, leading to the upregulation of LLPS.29,30 Phosphorylation of RNA-binding proteins controls LLPS by altering their binding mode, resulting in deadenylation and changes in translation.31 Phosphorylation of the C-terminal domain of RNA polymerase II promotes LLPS, facilitating transcription elongation.32 Although the effects of chemical modifications on RNA (epitranscriptomics) and proteins (post-translational modifications) on LLPS have been studied extensively, the impact of DNA modifications (epigenetics) on LLPS has received less attention.33 In this study, we investigated the impact of mC on LLPS in G4-forming DNA to obtain new insights into a combination of hypermethylation and LLPS. The findings showed that LLPS induced by G4-forming DNAs (G4-LLPS) is suppressed by mC. Structural analysis of DNAs showing different levels of mC induce a transition in G4 topology from being parallel to antiparallel, reducing the likelihood of LLPS occurrence. These results show for the first time how epigenetic modification affects G4-LLPS.
Results
Effect of mC on LLPS induced by G4
To explore the impact of mC in the loop region of G4 on the LLPS of G4s with RGG-peptide (G4-LLPS), we initially designed DNA sequences containing different levels of mC (Table 1). We selected d(GGGGCC)4 as a model template sequence. This sequence undergoes hypermethylation at the 5′-position of cytosine when the repeats are expanded.34 The 0Me, 2Me, 4Me, and 8Me have 0, 2, 4, and 8 mCs, respectively. To evaluate biological significance of (GGGGCC)4 which is used as the template sequence, we searched (GGGGCC)4 sequences on the human genome using bioinformatic tools (see Materials and methods for bioinformatic analysis). We identified 29 (GGGGCC)4 sequences on the human genome (Table S1†). The result showed that 23 of 29 (GGGGCC)4 sequences were located in the promoter regions of different genes. These bioinformatic analyses demonstrate that not only C9orf72 but also other genes are potentially controlled by (GGGGCC)4.| Abb | Sequence* | |
|---|---|---|
| a *C indicates the position of 5-methylcytosine. | ||
| DNAs | 0Me | GGGGCCGGGGCCGGGGCCGGGGCC |
| 2Me | GGGGCCGGGGCCGGGGCCGGGGCC | |
| 4Me | GGGGCCGGGGCCGGGGCCGGGGCC | |
| 8Me | GGGGCCGGGGCCGGGGCCGGGGCC | |
| Peptide | RGG-peptide | RRGDGRRRGGGGRGQGGRGRGGGF |
| KGNDDHSRGGW | ||
To quantify the LLPS of sequences using in this study, we used a model system in which LLPS can be induced by combining a G4 forming oligonucleotide with a cationic peptide, the RGG-peptide (Table 1),15 derived from the RGG domain of the Fragile X Mental Retardation Protein (FMRP), which is known for its ability to bind DNA G4.35 In this system, an increase in turbidity indicates droplet formation, with higher turbidity values corresponding to more and larger droplets.36Fig. 1A shows turbidity of a mixture of 5 μM 0Me and various concentrations of RGG-peptide (0, 1, 3, 5, 10, and 15 μM). The turbidity measured at 350 nm increased with the RGG-peptide concentration, and the methylated DNA oligonucleotides showed higher turbidity at higher RGG-peptide concentrations (Fig. S1†). The turbidity at 1800 s, that is measured 1800 seconds after the addition of RGG-peptide, was plotted against the RGG-peptide concentration (Fig. 1B). Compared to the control (0Me), the turbidity values for 2Me, 4Me, and 8Me were reduced, suggesting that mC suppresses G4-LLPS. To confirm the effect of cation concentration on G4-LLPS, we measured turbidity in the presence of various concentrations of KCl (1, 10, 100, or 1000 mM) (Fig. S2†). All sequences showed lower turbidity at higher KCl concentrations, indicating that the electrostatic interactions play a key role in G4 and RGG-peptide binding, as well as in G4-LLPS.37 To verify whether the turbidity increments are due to LLPS rather than aggregation, droplet formation was directly visualized under a confocal laser scanning microscope (Fig. 1C). As in previous reports, spherical droplets were observed in a mixtures of FAM-labeled RGG-peptide with each DNA sequence.15,38,39 The total fluorescence intensity from the droplets and droplet size under each condition were evaluated (Fig. 1D and E). The results showed that the 0Me showed the highest values in all parameters, which was consistent with the turbidity measurements shown in Fig. 1B.
We further attempted to confirm the effect of mC on the fluidity of the droplets by FRAP analysis (Fig. S3†). Droplets containing FAM-tagged DNAs were photobleached, and the fluorescence recovery was examined. The recovery yields after 300 s for 0Me and 8Me were greater than 40%, which is comparable with values obtained in studies on prion or SOP1 protein-RNA LLPS.40,41 The apparent diffusion coefficients (Dapp) for these conditions were also evaluated. The Dapp value for 0Me was 0.14 ± 0.02 μm2 s−1, which is comparable with a previous studies.42,43 The Dapp value for 8Me was 0.17 ± 0.02 μm2 s−1, indicating that all droplets exhibited similar fluidity. To further demonstrate the reversibility of LLPS, we tested the resistance of these droplets to aliphatic alcohol, 1,6-hexanediol (1,6-HD), a known disrupter of droplets formed via LLPS (Fig. S4†).44 The number of droplets formed by 0Me decreased with increasing concentration of 1,6-HD (5, 10, and 20 wt%), confirming that the G4-LLPS of 0Me is reversible, a property generally associated with LLPS. Other methylated DNA sequences showed similar responses to 1,6-HD (Fig. S4†), indicating that reversibility is maintained even with mC. To further clarify the effects of mC on G4-LLPS, we employed another in vitro LLPS system, in which LLPS was induced by mixing G4 DNAs with histone H1.38 Fig. S5† shows the turbidity at 350 nm for mixtures of 3 μM G4 DNA and various concentrations of histone H1 (0.00, 0.25, 0.50, 1.00, and 1.50 μM). Similar to the results obtained for RGG-peptide shown in Fig. 1B, 0Me exhibited increased turbidity at higher histone H1 concentrations. This trend was also observed for methylated DNAs, although their turbidity values were consistently lower than those obtained for 0Me. These findings show that mC suppresses G4-LLPS, irrespective of the interacting peptide or protein, and confirm that mC reduces G4-LLPS in GGGGCC repeat sequences. Previous reports have highlighted the importance of the binding affinity between G4 and counterpart molecules for inducing LLPS.7,45 Consequently, we investigated the dissociation constant (Kd) between 0Me or 8Me, and RGG-peptide using fluorescence titration experiments. We added varying concentrations of RGG-peptide to 50 nM FAM-labeled 0Me or 8Me and observed a decrease in fluorescence intensity with an increase in RGG-peptide concentration (Fig. S6†). Fig. 1F shows the measurement results and fitting curves for fluorescence intensity for FAM-tagged 0Me or 8Me at 520 nm, plotted against the concentration of RGG-peptide. The Kd values were calculated to be 0.56 ± 0.22 and 1.62 ± 0.52 μM for 0Me and 8Me, respectively, at 25 °C (Fig. 1G). Although the LLPS behavior of 0Me and 8Me differs markedly, the Kd values showed only about a threefold difference. Biomolecular LLPS typically results from multimolecular interactions. In our LLPS model system, where the concentrations of G4 and RGG-peptide were evaluated to be 85 and 125 mM, respectively,46 if the diameter of a droplet is 1 μm, then the numbers of G4 DNA and RGG-peptide within a single droplet would be about 70 × 106 and 110 × 106, respectively. Thus, even small differences in Kd, such as those between 0 Me and 8Me with RGG-peptide, can be amplified considerably through multimolecular interactions. Next, we attempted to find whether these mC effects are just for G4. We studied the mC effects on the structure and LLPS ability of DNA duplex. We designed duplex-0Me and duplex-8Me as a DNA duplex with 0 and 8 mCs, respectively, (nucleotide sequences are listed in Table S2†). In agreement with our previous report,15 DNA duplexes with or without mC did not undergo LLPS in the concentration range used for G-quadruplex (Fig. S7†). In addition, we previously reported thermal stability of DNA duplex is increased by mCs, while mCs did not affect the duplex structure.47 These results showed that the effect of mCs on duplex properties is not significant to alter the structure and LLPS ability of DNA duplex.
Effect of mC on the structure of G4
Previous studies have shown that G4 unfolding suppresses G4-LLPS.38 Thus, we hypothesized that the suppressive effect of mC on G4-LLPS might be due to mC-induced perturbations in the G4 structure. To test this, we analyzed the structure of these oligonucleotides using UV-melting curve measurements and circular dichroism (CD) spectroscopy. The UV-melting curves, traced at 295 nm, exhibited hypochromic transitions at high temperatures for all DNA oligonucleotides (Fig. 2A), which is indicative of G4 formation.48,49 The melting temperature (Tm) derived from these melting curves was comparable across all sequences, suggesting that mCs do not alter the thermal stability of G4s (Fig. 2B). Next, to determine the G4 topology, we measured the CD spectra (Fig. 2C). The spectrum of 0Me showed positive peaks at 260 and 295 nm, consistent with either a hybrid topology or a mixture of parallel and antiparallel topologies.50,51 In contrast, the methylated sequences showed negative and positive peaks at 260 and 295 nm, respectively, are indicative of an antiparallel topology.50,51 These findings confirm that mCs induce a transition to an antiparallel G4 topology. Previous reports showed that (GGGGCC)3GGGG forms two different types of antiparallel G4s which are formed via rapid annealing in an acidic condition called AQU or slow annealing in a neutral condition called NAN.52,53 Because of the experimental condition and sample preparation, the antiparallel topology identified in this study can be similar to NAN antiparallel G4.To identify the component that shows a positive peak at 260 nm of 0Me, we focused previous reports showing that the addition of poly-ethylene glycol with molecular weight 200 (PEG200) induces parallel topology.54,55 We measured CD spectrum of 0Me in the presence of PEG200 (Fig. S8A†). Negative and positive peaks at 240 and 260 nm, respectively, were increased with increment of PEG200 concentration, indicating topology transition to parallel one. This result shows that a positive peak at 260 nm arose from parallel topology. We further superimposed the CD spectrum of 0Me in the presence of 20 wt% PEG with the CD spectra of 0Me, 2Me, 4Me, and 8Me in the buffer (Fig. S8B†). These CD spectra showed the isosbestic points at 250 and 280 nm, indicating a two-state topology transition between the parallel and the antiparallel, although further structure studies are required as shown in the previous publications for the similar oligonucleotides.52,53 To further investigate this topology transition in relation to the level of mC, we performed SVD analysis on the dataset of CD spectra at different levels of mC shown in Fig. 2C. The SVD findings showed that the first and second most significant components (i.e., SV = 115 and SV = 44, respectively) are present with and without mC (Fig. 2D). The predominant component displayed negative and positive peaks at 260 and 290 nm, respectively, indicating an antiparallel topology. The second major component showed negative and positive peaks at 240 and 260 nm, respectively, which is indicative of a parallel topology. These findings further demonstrate that G4 in 0Me consists of a mixture of parallel and antiparallel topologies, rather than a hybrid topology. Additionally, the SVD value for the most major component decreased in the presence of mC (Fig. 2E), while the second major component increased in the presence of mC. These results confirmed that mC promotes the formation of an antiparallel topology. In addition, we performed non-denaturing polyacrylamide gel electrophoresis to confirm the topological transition induced by mCs. Two distinct bands were observed in all DNAs. (Fig. S9†). Previously, it was reported that a parallel G4 migrates slower than an antiparallel G4.56 Based on these results and the CD spectra (Fig. S8†), we considered that the faster and slower bands correspond to the antiparallel topology and the parallel topology, respectively. 0Me with PEG200 showed the slower band only, confirming that the faster band and the slower band correspond to the parallel topology and the antiparallel topology, respectively. In addition, there was no other slower band (intermolecular species), indicating that DNAs used in this study do not form intermolecular structures in the experimental condition. Previous report showed that (GGGGCC)3GGGG forms inter- and intramolecular G4 structures,52 which are inconsistent with our results. However, the nucleotide sequence of 0Me is slightly different from that used in the previous study: 0Me contains two cytosines at the 3′-terminal, whereas that in the previous study does not. Although effects of the terminal cytosine nucleotides on the structure are not clear yet, it is known that a structure polymorphism is generated depending the terminal nucleotides.57 Therefore, it could be considered that structures of 0Me are partially different from those shown in the previous reports.52,53
Previous studies have reported that G4 stability and topology are influenced by mC via pKa increment and polarizability enhancement of cytosine, although the effects of mC are not consistent across all sequences.58–62 Brcic et al. demonstrated that cytosines in the loop region of the antiparallel topology formed by GGGGCC repeat DNA engage C–C base pairing and stacking interactions with the adjacent G-quartet.53 Given this information, for our sequence, the antiparallel topology appears to be the favored structure since mC promotes both the formation of C–C base pairs and their stacking interactions with adjacent G-quartets.
Relationship of LLPS ability with topology of G4
The results shown in Fig. 2 suggested that the parallel topology (0Me) enhances LLPS with RGG-peptide, whereas the antiparallel topology (2Me, 4Me, and 8Me) is less favorable for undergoing LLPS. These findings suggest that different topologies may influence LLPS ability with RGG-peptide. To test this hypothesis, we used G4s with parallel, hybrid, and antiparallel topologies (Table 2 and Fig. 3A), confirmed by NMR or X-ray crystallography.63–68 Firstly, we verified the G4 topology of these DNAs in the buffer through UV-melting analysis and CD spectroscopy. The UV-melting curves for these DNAs showed hypochromicity at elevated temperatures for all DNAs, indicating that they all form G4 structures (Fig. 3B). CD spectral analysis of these DNAs further defined these topologies (Fig. 3C): P1 and P2 exhibited negative and positive peaks at 240 and 260 nm, respectively, which is indicative of a parallel topology; H1 and H2 spectra showed a negative peak at 240 nm and positive peaks at 260 and 295 nm, confirming a hybrid topology; and AP1 and AP2 showed negative and positive peaks at 260 and 295 nm, respectively, which is typical of an antiparallel topology. These results confirmed that the oligonucleotides fold to form G4s as anticipated. Using these DNAs, we analyzed the LLPS abilities associated with different topologies (Fig. 3D). It was found that P1 and P2, which form parallel topology, exhibited the highest turbidity. In contrast, the hybrid topology formed by P1 and P2 showed moderate turbidity, while AP1, with the antiparallel topology, showed very low turbidity. AP2, also the antiparallel, displayed slightly increased turbidity at higher RGG-peptide concentrations. Microscopic analysis revealed that P1, P2, H1, H2, and AP2 all formed droplets, which is consistent with the turbidity measurements (Fig. S10†). However, AP1 only formed tiny droplets. This indicates that the tendency of G4 to undergo LLPS follows the order: parallel > hybrid > antiparallel. These results demonstrated that G4-LLPS ability is dependent on the G4 topology, which in turn influences the effects of mC on G4-LLPS. It should be noted that loop orientation varies depending on the G4 topology (Fig. 3A). We hypothesize that the loop region, particularly the propeller loops in the parallel topology, is a critical factor for promoting G4-LLPS. The loop region comprises three parts: nucleosides, phosphates, and ribose moieties. To further investigate the loop effects on G4-LLPS, we designed intermolecular G4s, termed tetramer G4 (Table S3,† and Fig. 4A), which form an intermolecular parallel G4 lacking a loop region. Moreover, to identify the component of the loop region that is critical for G4-LLPS, we prepared a variant, abasic P1, modified with an abasic nucleotide (Fig. 4A). In the abasic P1 variant, all nucleotides in the loop region of P1 are modified to be abasic. Fig. 4B presents the CD spectra of 10 μM abasic P1 and 40 μM tetramer G4, which showed positive and negative peaks at 240 and 260 nm, respectively, suggesting a parallel topology. In addition, we attempted to confirm G4 formation of tetramer G4 in the presence of 100 mM K+ or Li+ by use of non-denaturing polyacrylamide gel electrophoresis (Fig. S11†). The band was observed only in the presence of K+ but not in the presence of Li+, showing that the DNA sequence forms the tetrameric G4 only in the presence of K+. In addition to gel electrophoresis, we measured fluorescence of 5 μM NMM, known as a parallel topology fluorescence probe, with 10 μM tetramer G4 in the 100 mM K+ or Li+ (Fig. S12†).69 The fluorescence increased in the presence of K+. These results further confirmed that a parallel G4 is formed in the presence of K+.The turbidity measurements for these DNAs in Fig. 4C showed that abasic P1 exhibited a lower turbidity than P1, while tetramer G4 exhibited very low turbidity, even with 15 μM of the RGG-peptide. This result showed that loop regions play a crucial role in parallel G4-LLPS. Furthermore, the lower turbidity increase observed for abasic P1 indicates that nucleobases within the propeller loops also participate in G4-LLPS. Consistent with the results shown in Fig. 1F, the Kd value for G4 with RGG-peptide appears to be a critical factor for G4-LLPS. Next, we evaluated the Kd values for G4s with different topologies using FAM-tagged P1, TAMRA-tagged H1, and TAMRA-tagged AP1. The fluorescence from these DNAs was observed to decrease as the RGG-peptide concentration increased (Fig. S13†). The fluorescence intensity was also plotted against RGG-peptide concentration to evaluate the Kd values (Fig. S14†). The Kd values for P1, H1, and AP1 with RGG-peptide at 25 °C were 0.07 ± 0.01, 0.20 ± 0.05, and 0.14 ± 0.03 μM, respectively. These values correlate with the turbidity data, suggesting that a relatively higher affinity with RGG-peptide enhances G4-LLPS. Interestingly, the parallel, hybrid, and antiparallel topologies have two, one, and zero propeller loops, respectively, which may explain the variance in LLPS ability. These results imply that propeller loops may function as scaffolds for interactions between G4 and RGG-peptide.
AP1, which was used as an antiparallel topology-forming DNA, bound to RGG-peptide with a sub-micromolar Kd value; however, it was unable to undergo LLPS on its own. We hypothesized that AP1 might engage in LLPS in the presence of preformed droplets induced by RGG-peptide (see Fig. 5A for the experimental procedure). To test this hypothesis, we mixed FAM-tagged P1 with RGG-peptide and observed droplet formation. In agreement with Fig. S10,† droplet formation of P1 and RGG-peptide was observed (Fig. 5B, top). After incubating the P1 droplets for 3600 s, we introduced TAMRA-tagged AP1 (TMR-AP1) into the mixture of P1 and RGG-peptide (Fig. 5B, second line). Interestingly, droplets containing both P1 and AP1 were observed, indicating co-localization of P1 and AP1 within the same droplets (Fig. 5C). These results demonstrate that molecules that do not independently undergo LLPS can do so in the presence of pre-formed droplets containing another sequence. This finding could be useful for isolating potential G4-forming DNA and RNA sequences that bind to RGG-peptide or proteins containing RGG domains from cells or cell lysate samples through LLPS.
Effect of cellular environmental factors on LLPS ability
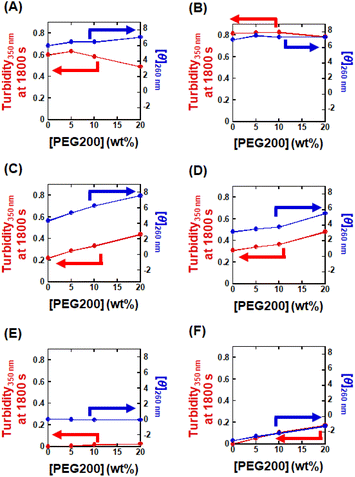
The intracellular environment, which is characterized by having numerous highly-concentrated macromolecules, differs considerably from the conditions that are typically used in biochemical research in vitro.14 Interestingly, it has been reported that molecular crowding can induce a transition in G4 topology to parallel from other forms.54 In addition, molecular crowding has also been reported to accelerate LLPS.70 Thus, to better clarify the importance of G4 topology in G4-LLPS, we investigated the effects of molecular crowding on G4-LLPS using PEG200 to reproduce molecular crowding conditions.54,71 First, we examined the molecular crowding effects on G4 topology using CD spectroscopy (Fig. S15†). For P1 and P2, which form parallel G4s, the CD spectra remained almost the same with and without PEG200, indicating that the parallel G4 topology is maintained in the crowded conditions. For H1 and H2, which form hybrid G4s, the shoulder around 295 nm and the positive peak at 260 nm decreased and increased, respectively, in the presence of PEG200. These results suggest that molecular crowding induces a hybrid to a parallel transition in topology. This finding is consistent with those of a previous NMR study which showed that molecular crowding induces the same topology transition.54 In contrast, the CD spectra of AP1 and AP2, which form antiparallel G4s, showed no significant alterations in the presence of PEG200, although there was a slight decrease in CD intensity for AP2. This suggests that the antiparallel G4 topology of AP1 and AP2 is maintained under the molecular crowding conditions induced by PEG200. Based on these structural characterizations, we then investigated the LLPS characteristics of these DNAs. We measured the turbidity of a mixture containing 5 μM of each of these DNAs, representing different G4 topologies, and 5 μM RGG-peptide in the presence of various concentrations of PEG200 (0, 5, 10, and 20 wt%) (Fig. S16†). At 1800 s, the turbidity, which is indicative of LLPS ability, and the CD intensity at 260 nm, which corresponds to the relative proportion of the parallel topology, were plotted against PEG200 concentration (Fig. 6). The turbidity for P1 decreased significantly with increasing PEG200 concentration, while P2 showed a slight decrease as PEG200 concentration increased, suggesting that G4-LLPS is suppressed by molecular crowding with PEG200. This finding contrasts with numerous reports indicating that molecular crowding typically accelerates LLPS.72,73 One possible explanation for the suppression of G4-LLPS by molecular crowding is dehydration though the G4-peptide complex formation. Molecular crowding effects can be categorized into excluded volume and hydration changes.14 The excluded volume effect, derived from the molecular crowding reagent, increases the effective concentration of solutes, thereby potentially accelerating binding reactions among multiple molecules, such as protein fibril formation.74,75 On the other hand, molecular crowding reagents such as PEG can reduce the concentration and activity of water molecules, making reactions that require more hydration unfavorable and those that involve dehydration favorable.14,55 Although molecular crowding can enhance G4-LLPS due to the multimolecular nature of these reactions, it can also inhibit G4-LLPS if complex formation is associated with dehydration. It has been reported previously that DNA G4 complex formation with a cationic ligand involves hydration, as electrostatic interactions tend to incorporate more water molecules into the complex.76 Thus, it is possible that G4-LLPS is inhibited under conditions of molecular crowding due to the formation of a more hydrated complex, although further research is necessary to elucidate the specific molecular mechanisms involved.Unlike P1 and P2, where molecular crowding suppressed LLPS, it accelerated LLPS for H1 and H2, which also underwent a topology transition from hybrid to parallel. A similar acceleration of LLPS was observed for AP2. These results suggest that the effect of molecular crowding on promoting G4-LLPS by facilitating a topology transition is more pronounced than its inhibitory effect on G4-LLPS. Conversely, in the case of AP1, whose structure remains unaffected by molecular crowding with PEG200, no significant crowding effect on LLPS was observed. This further underscores the importance of G4 topology in influencing LLPS behavior. Therefore, transitions to a parallel topology are associated with increased turbidity indicating enhanced LLPS.
Discussion
DNA and RNA G4s structures play critical roles in a variety of biological processes, including replication, transcription, splicing, and translation.16 This range of G4 functions necessitates interactions with numerous specific proteins.17 For example, ATP-dependent DNA/RNA helicases (BLM, FANCJ, and RHAU), splicing proteins (hnRNP family proteins), and RNA-transport proteins (FUS and FMRP) are all identified as G4-binding proteins.77–82 These G4 binding-proteins commonly possess nucleic acid-binding domains such as the Asp-Glu-Ala-His (DEAH) box, oligonucleotide/oligosaccharide-binding (OB) fold domain, RNA recognition motif (RRM), and RGG domain.82–84 Moreover, the specific topology of G4s is increasingly realized as being central to these G4–protein interactions. For example, the Rif1 protein, which interacts with parallel or hybrid topologies, plays a role in suppressing replication.85,86 Further combination of MS and bioinformatic studies has identified proteins that specifically bind to G4s with distinct topologies, classifying them into several biological roles.80 As shown in Fig. 3, we demonstrated that G4 topology is a critical factor in G4-LLPS, with the parallel topology being particularly favorable for undergoing G4-LLPS. A previous NMR study elucidated the complex formed between a parallel G4 topology and a short peptide (18 amino acids) derived from the RNA helicase associated with the AU-rich element (RHAU) protein.56 The hydrophilic residues of the RHAU peptide interact with G-quartets having a parallel topology via CH–π and CH3–π stacking interactions, while the positively charged residues form electrostatic interactions with phosphate groups, stabilizing the G4 structure. Thus, the selectivity of the parallel topology in G4-LLPS with RGG-peptide is attributed to both stacking and electrostatic interactions involving G-quartets and phosphate groups, respectively. Our findings suggest that the suppression of LLPS by mC can be attributed to its control of G4 topology. This suggests that mC may act as a regulatory factor influencing G4-related biological processes, not only through protein binding but also through the regulation of LLPS.Previous reports showed that the increasing repeat number of guanine-rich sequence results in tandem increment of their CD intensity, indicating that topology is retained even with multiple G4 units.87,88 Thus, it is considerable that the topology and the thermal stability of intramolecular G-quadruplexes formed by GGGGCC repeats are not affected by the repeat number. On the other hand, the larger number of repeats of GGGGCC may induce more intermolecular associations, resulting in aggregation. In fact, it was reported that longer GGGGCC repeats tend to aggregate by themselves. The self-association is governed by hydrogen bonds for base-pairing, stacking interactions for end-to-end stacking of G4, as well as electrostatic interactions.89–91 Further studies are required to investigate the effects of mC on GGGGCC aggregation.
This study provides new insights into how the combination of chemical modifications and LLPS of nucleic acids synergistically regulates gene expression, highlighting the role of small modified functional groups, such as the methyl group on cytosine, in amplifying these regulatory signals.
Materials and Methods
Materials
All DNA strands used in this study were acquired from Sigma-Aldrich Japan K. K. (Tokyo, Japan) and Hokkaido System Science Co., Ltd (Hokkaido, Japan). Extinction coefficients for single-stranded DNA oligonucleotides were calculated from mono- and dinucleotide data using the nearest-neighbor approximation model.92 Absorption of the oligonucleotides was measured at 260 nm and 95 °C using an UV spectrophotometer (UV-1800; Shimadzu Co., Ltd, Kyoto, Japan) connected to a temperature controller (TMSPC-8; Shimadzu Co., Ltd). The concentration of high-performance liquid chromatography (HPLC)-grade peptides (Genscript Japan Inc., Tokyo, Japan) was measured at 280 nm as tryptophan (Trp) absorbance using a UV spectrophotometer (UV-1800; Shimadzu Co., Ltd). Chemical reagents were purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan) and FUJIFILM Wako Pure Chemical Co. (Osaka, Japan).Fluorescence spectroscopy
Fluorescence spectra were measured using a spectrofluorometer (FP-8200; JASCO, Tokyo, Japan) equipped with a temperature controller and a 0.3 cm × 0.3 cm quartz cell, maintained at 25 °C. Before measurement, each sample was heated to 90 °C for 5 min and then gently cooled to 25 °C at a rate of 0.5 °C min−1 for annealing. The fluorescence spectra of 6-carboxyfluorescein (FAM)- or 5-carboxytetramethylrhodamine (TAMRA)-tagged DNA were measured over the ranges of 510 to 610 nm or 560 to 660 nm and excited at 495 nm and 550 nm, respectively. All experiments were conducted in a buffer composed of 100 mM KCl, 10 mM K2HPO4 (pH 7.0), and 1 mM K2EDTA at 25 °C. For measurement of NMM fluorescence, we prepared sample containing 40 μM tetramer G4 and 5 μM NMM in a K+ and Li+ (100 mM LiCl, 10 mM LiH2PO4 (pH 7.0), and 1 mM Li2EDTA) buffer. The fluorescence intensity at 520 nm for FAM-tagged DNA or at 580 nm for TAMRA-tagged DNA was plotted against the concentration of the RGG-peptide. These values were fitted to eqn (1) using KaleidaGraph (Synergy Software, Reading, PA) to evaluate the dissociation constants (Kd) at 25 °C: | (1) |
Thermal analysis
The melting curves of the DNA oligonucleotides were measured by monitoring the absorption at 260 and 295 nm using a UV spectrophotometer (UV-1800; Shimadzu Co., Ltd) connected to a temperature controller (Shimadzu Co., Ltd). DNA samples in a buffer containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0), and 1 mM K2EDTA were heated at a rate of 0.5 °C min−1 from 25 to 95 °C to trace the thermal denaturation curves. Before measurements were initiated, each sample was heated to 95 °C for 5 min and gently cooled to 25 °C at −0.5 °C min−1.Circular dichroism spectroscopy
Circular dichroism (CD) spectra of 10 μM oligonucleotides were measured in a buffer containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0), and 1 mM K2EDTA at 25 °C using a J-820 spectropolarimeter (JASCO Co., Ltd, Tokyo, Japan) equipped with a JASCO PTC-424L temperature controller using a quartz cell with 0.1 cm path length under N2 gas flow. The spectrum was averaged by at least three scans. Before measurement, each sample was heated to 95 °C for 5 min and gently cooled to 25 °C at a rate of −0.5 °C min−1 for annealing.Singular value decomposition analysis
For the singular value decomposition (SVD) analysis, a dataset of CD spectra from DNA oligonucleotides with various levels of mC was utilized. SVD was performed using the KinTek Explorer software package (KinTek Corp., Snow Shoe, PA). The mathematical details of the SVD analysis have been reported previously.93 The significant components (spectra) were determined based on a statistical parameter, the relative magnitude of the singular values (SV). Higher SVs correspond to more significant components.Turbidity measurement
Turbidity was measured at 350 nm using a UV spectrophotometer (UV-1800; Shimadzu Co., Ltd) connected to a temperature controller Shimadzu Co., Ltd The measurements were performed using a solution of DNA oligonucleotide, RGG-peptide, and mixtures of both using a quartz cell with a 1 cm path length. Various concentrations of RGG-peptide solution (1, 3, 5, 10, or 15 μM) were added to the DNA solution (final concentration of 5 μM) after 300 s incubation following the initiation of measurement. All measurements were conducted in a KCl buffer (100 mM KCl, 10 mM K2HPO4 (pH 7.0), and 1 mM K2EDTA) at 25 °C. Both DNA and peptide samples were separately heated to 95 °C for 5 min and then gently cooled to 25 °C at a rate of −0.5 °C min−1.Droplet imaging
Images were captured using a confocal laser scanning microscope (A1R, Nikon Co., Tokyo, Japan) using a 60× oil-immersion objective lens. Samples consisting of mixture of FAM- or TAMRA-labeled DNA and RGG-peptide were imaged in a buffer containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0), and 1 mM K2EDTA after incubation for 30 min at room temperature. Before imaging, DNA and peptide samples were separately heated to 95 °C for 5 min and then gently cooled to 25 °C at a rate of −0.5 °C min−1. To measure the droplet area, the fluorescence intensities inside the droplets were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD). The average fluorescence intensity values within each droplet were calculated by averaging these values on a per-droplet basis after subtracting the background.Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were conducted on a mixture of 5 μM fluorescence-conjugated DNA and 5 μM RGG-peptide in a buffer containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0), and 1 mM K2EDTA under a confocal microscope (A1R, Nikon Co.) using a 60× oil-immersion objective lens. Photobleaching was performed at 488 nm and fluorescence intensity at the bleached spot was measured with ImageJ. Before measurements, each sample was heated to 90 °C for 5 min and then gently cooled to 25 °C at a rate of −0.5 °C min−1. After subtracting the background, the intensity recovery traces were normalized to the prebleach level and fitted to an exponential curve using eqn (2) to estimate the apparent recovery time, τ,| Y = a(1 − e−t/τ) | (2) |
| Dapp = r2/τ | (3) |
The main text of the article should appear here with headings as appropriate.
Non-denaturing polyacrylamide gel electrophoresis
0Me, 2Me, 4Me, and 8Me in the absence or presence of 20 wt% PEG200 were diluted to be 10 μM in a K+ buffer containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0), and 1 mM K2EDTA. 40 μM tetramer G4 was mixed with the K+ buffer or a Li+ buffer (100 mM LiCl, 10 mM LiH2PO4 (pH 7.0), and 1 mM Li2EDTA). The samples were heated to 95 °C for 5 min and cooled to −0.5 °C min−1. The samples were loaded onto an 18% polyacrylamide gel at room temperature. After electrophoresis, the gels were stained with SYBR Gold (Thermo Fisher Scientific, Inc., Tokyo, Japan) and fluorescent bands were imaged by FLA-7000 (Fujifilm, Tokyo, Japan).Bioinformatic analysis
To determine where the [(GGGGCC)4] sequences map to in the genome sequence, analyses were performed using R: Version-4.4.1, Rstudio: Version-2024.4.2.764, and R scripts (Biostrings: Version-2.72.1, BS genome.Hsapiens.UCSC.hg38: Version-1.4.5) to extract the BED file that show location of [(GGGGCC)4]. In order to investigate promoter region which contain [(GGGGCC)4], ENSEMBL Gene IDs were obtained using R scripts (GenomicRanges: Version-1.56.2, rtracklayer: Version-1.64.0). The promoter region in this study was defined as upstream 2000 bp to downstream 200 bp from the transcription start site (TSS). Gene Symbols were converted from ENSEMBL Gene ID to Gene symbol converter using Biotools (https://www.biotools.fr).Data availability
All experimental supporting data are available in the ESI.†Author contributions
Conceptualization, D. M.; methodology, D. M.; formal analysis, M. T. S. S. and S. T.; investigation, M. T. S. S. and S. T.; data curation, M. T. and S.S.; writing—original draft preparation, M. T.; writing—review and editing, D. M.; visualization, D. M.; supervision, D. M.; project administration, D. M.; funding acquisition, M. T., K. K., and D. M. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported by Grant-in-Aid for JSPS KAKENHI (Grant Numbers 24K21801, 23H03020, 21H02062, 20K21259, and 17H06351) and for JSPS Fellowship (Grant Number 22J21167). Additional support was provided by research grants from the Asahi Glass Foundation, Japan, the Junzo Tateno Foundation and the Hirao Taro Foundation, Konan Gakuen for Academic Research, Japan.References
- A. A. Hyman, C. A. Weber and F. Julicher, Liquid-liquid phase separation in biology, Annu. Rev. Cell Dev. Biol., 2014, 30, 39–58 CrossRef CAS PubMed.
- S. Boeynaems, S. Chong, J. Gsponer, L. Holt, D. Milovanovic, D. M. Mitrea, O. Mueller-Cajar, B. Portz, J. F. Reilly, C. D. Reinkemeier, B. R. Sabari, S. Sanulli, J. Shorter, E. Sontag, L. Strader, J. Stachowiak, S. C. Weber, M. White, H. Zhang, M. Zweckstetter, S. Elbaum-Garfinkle and R. Kriwacki, Phase Separation in Biology and Disease; Current Perspectives and Open Questions, J. Mol. Biol., 2023, 435, 167971 CrossRef CAS PubMed.
- A. Shakya and J. T. King, DNA Local-Flexibility-Dependent Assembly of Phase-Separated Liquid Droplets, Biophys. J., 2018, 115, 1840–1847 CrossRef CAS PubMed.
- H. Zhang, S. Elbaum-Garfinkle, E. M. Langdon, N. Taylor, P. Occhipinti, A. A. Bridges, C. P. Brangwynne and A. S. Gladfelter, RNA Controls PolyQ Protein Phase Transitions, Mol. Cell, 2015, 60, 220–230 CrossRef CAS PubMed.
- J. R. Vieregg, M. Lueckheide, A. B. Marciel, L. Leon, A. J. Bologna, J. R. Rivera and M. V. Tirrell, Oligonucleotide-Peptide Complexes: Phase Control by Hybridization, J. Am. Chem. Soc., 2018, 140, 1632–1638 CrossRef CAS PubMed.
- I. Pavlova, M. Iudin, A. Surdina, V. Severov and A. Varizhuk, G-Quadruplexes in Nuclear Biomolecular Condensates, Genes, 2023, 14, 1076 CrossRef CAS PubMed.
- S. Asamitsu, Y. Yabuki, K. Matsuo, M. Kawasaki, Y. Hirose, G. Kashiwazaki, A. Chandran, T. Bando, D. O. Wang, H. Sugiyama and N. Shioda, RNA G-quadruplex organizes stress granule assembly through DNAPTP6 in neurons, Sci. Adv., 2023, 9, eade2035 CrossRef CAS PubMed.
- A. Ghosh, S. P. Pandey, D. C. Joshi, P. Rana, A. H. Ansari, J. S. Sundar, P. Singh, Y. Khan, M. K. Ekka, D. Chakraborty and S. Maiti, Identification of G-quadruplex structures in MALAT1 lncRNA that interact with nucleolin and nucleophosmin, Nucleic Acids Res., 2023, 51, 9415–9431 CrossRef CAS PubMed.
- E. A. J. Simko, H. Liu, T. Zhang, A. Velasquez, S. Teli, A. R. Haeusler and J. Wang, G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA, Nucleic Acids Res., 2020, 48, 7421–7438 CAS.
- A. R. Haeusler, C. J. Donnelly, G. Periz, E. A. Simko, P. G. Shaw, M. S. Kim, N. J. Maragakis, J. C. Troncoso, A. Pandey, R. Sattler, J. D. Rothstein and J. Wang, C9orf72 nucleotide repeat structures initiate molecular cascades of disease, Nature, 2014, 507, 195–200 CrossRef CAS PubMed.
- A. Ishiguro, J. Lu, D. Ozawa, Y. Nagai and A. Ishihama, ALS-linked FUS mutations dysregulate G-quadruplex-dependent liquid-liquid phase separation and liquid-to-solid transition, J. Biol. Chem., 2021, 297, 101284 CrossRef CAS PubMed.
- X. Liu, Y. Xiong, C. Zhang, R. Lai, H. Liu, R. Peng, T. Fu, Q. Liu, X. Fang, S. Mann and W. Tan, G-Quadruplex-Induced Liquid-Liquid Phase Separation in Biomimetic Protocells, J. Am. Chem. Soc., 2021, 143, 11036–11043 CrossRef CAS PubMed.
- B. R. Sahoo, V. Kocman, N. Clark, N. Myers, X. Deng, E. L. Wong, H. J. Yang, A. Kotar, B. B. Guzman, D. Dominguez, J. Plavec and J. C. A. Bardwell, Protein G-quadruplex interactions and their effects on phase transitions and protein aggregation, Nucleic Acids Res., 2024, 52, 4702 CrossRef CAS PubMed.
- S. Nakano, D. Miyoshi and N. Sugimoto, Effects of molecular crowding on the structures, interactions, and functions of nucleic acids, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS PubMed.
- M. Tsuruta, T. Torii, K. Kohata, K. Kawauchi, H. Tateishi-Karimata, N. Sugimoto and D. Miyoshi, Controlling liquid-liquid phase separation of G-quadruplex-forming RNAs in a sequence-specific manner, Chem. Commun., 2022, 58, 12931–12934 RSC.
- D. Varshney, J. Spiegel, K. Zyner, D. Tannahill and S. Balasubramanian, The regulation and functions of DNA and RNA G-quadruplexes, Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS PubMed.
- L. Oganesian and T. M. Bryan, Physiological relevance of telomeric G-quadruplex formation: a potential drug target, Bioessays, 2007, 29, 155–165 Search PubMed.
- P. E. Ash, K. F. Bieniek, T. F. Gendron, T. Caulfield, W. L. Lin, M. Dejesus-Hernandez, M. M. van Blitterswijk, K. Jansen-West, J. W. Paul, 3rd, R. Rademakers, K. B. Boylan, D. W. Dickson and L. Petrucelli, Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS, Neuron, 2013, 77, 639–646 CrossRef CAS PubMed.
- A. Jain and R. D. Vale, RNA phase transitions in repeat expansion disorders, Nature, 2017, 546, 243–247 CrossRef CAS PubMed.
- K. M. Green, M. R. Glineburg, M. G. Kearse, B. N. Flores, A. E. Linsalata, S. J. Fedak, A. C. Goldstrohm, S. J. Barmada and P. K. Todd, RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response, Nat. Commun., 2017, 8, 2005 Search PubMed.
- P. O. Bauer, Methylation of C9orf72 expansion reduces RNA foci formation and dipeptide-repeat proteins expression in cells, Neurosci. Lett., 2016, 612, 204–209 CrossRef CAS PubMed.
- A. Bird, DNA methylation patterns and epigenetic memory, Genes Dev., 2002, 16, 6–21 CrossRef CAS PubMed.
- J. S. Sutcliffe, D. L. Nelson, F. Zhang, M. Pieretti, C. T. Caskey, D. Saxe and S. T. Warren, DNA methylation represses FMR-1 transcription in fragile X syndrome, Hum. Mol. Genet., 1992, 1, 397–400 Search PubMed.
- M. Pieretti, F. P. Zhang, Y. H. Fu, S. T. Warren, B. A. Oostra, C. T. Caskey and D. L. Nelson, Absence of expression of the FMR-1 gene in fragile X syndrome, Cell, 1991, 66, 817–822 CrossRef CAS PubMed.
- E. Y. Liu, J. Russ, K. Wu, D. Neal, E. Suh, A. G. McNally, D. J. Irwin, V. M. Van Deerlin and E. B. Lee, C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD, Acta Neuropathol., 2014, 128, 525–541 CrossRef CAS PubMed.
- S. Kumar, V. Chinnusamy and T. Mohapatra, Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond, Front. Genet., 2018, 9, 640 CrossRef CAS PubMed.
- Z. Jin and Y. Liu, DNA methylation in human diseases, Genes Dis., 2018, 5, 1–8 CrossRef CAS PubMed.
- J. Li, M. Zhang, W. Ma, B. Yang, H. Lu, F. Zhou and L. Zhang, Post-translational modifications in liquid-liquid phase separation: a comprehensive review, Mol. Biomed., 2022, 3, 13 CrossRef PubMed.
- R. J. Ries, S. Zaccara, P. Klein, A. Olarerin-George, S. Namkoong, B. F. Pickering, D. P. Patil, H. Kwak, J. H. Lee and S. R. Jaffrey, m(6)A enhances the phase separation potential of mRNA, Nature, 2019, 571, 424–428 CrossRef CAS PubMed.
- X. Wang, M. Wang, X. Dai, X. Han, Y. Zhou, W. Lai, L. Zhang, Y. Yang, Y. Chen, H. Wang, Y.-L. Zhao, B. Shen, Y. Zhang, Y. Huang and Y.-G. Yang, RNA 5-methylcytosine regulates YBX2-dependent liquid-liquid phase separation, Fundam. Res., 2022, 2, 48–55 CrossRef CAS PubMed.
- T. H. Kim, B. Tsang, R. M. Vernon, N. Sonenberg, L. E. Kay and J. D. Forman-Kay, Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation, Science, 2019, 365, 825–829 CrossRef CAS PubMed.
- H. Lu, D. Yu, A. S. Hansen, S. Ganguly, R. Liu, A. Heckert, X. Darzacq and Q. Zhou, Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II, Nature, 2018, 558, 318–323 CrossRef CAS PubMed.
- Y. Liu, W. Feng, Y. Wang and B. Wu, Crosstalk between protein post-translational modifications and phase separation, Cell Commun. Signaling, 2024, 22, 110 CrossRef PubMed.
- Z. Xi, M. Zhang, A. C. Bruni, R. G. Maletta, R. Colao, P. Fratta, J. M. Polke, M. G. Sweeney, E. Mudanohwo, B. Nacmias, S. Sorbi, M. C. Tartaglia, I. Rainero, E. Rubino, L. Pinessi, D. Galimberti, E. I. Surace, P. McGoldrick, P. McKeever, D. Moreno, C. Sato, Y. Liang, J. Keith, L. Zinman, J. Robertson and E. Rogaeva, The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients, Acta Neuropathol., 2015, 129, 715–727 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, J. C. Darnell, A. Serganov, A. Majumdar, S. Ilin, T. Raslin, A. Polonskaia, C. Chen, D. Clain, R. B. Darnell and D. J. Patel, Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction, Nat. Struct. Mol. Biol., 2011, 18, 796–804 CrossRef CAS PubMed.
- S. Alberti, S. Saha, J. B. Woodruff, T. M. Franzmann, J. Wang and A. A. Hyman, A User's Guide for Phase Separation Assays with Purified Proteins, J. Mol. Biol., 2018, 430, 4806–4820 CrossRef CAS PubMed.
- D. M. Mitrea, B. Chandra, M. C. Ferrolino, E. B. Gibbs, M. Tolbert, M. R. White and R. W. Kriwacki, Methods for Physical Characterization of Phase-Separated Bodies and Membrane-less Organelles, J. Mol. Biol., 2018, 430, 4773–4805 CrossRef CAS PubMed.
- M. Mimura, S. Tomita, Y. Shinkai, T. Hosokai, H. Kumeta, T. Saio, K. Shiraki and R. Kurita, Quadruplex Folding Promotes the Condensation of Linker Histones and DNAs via Liquid-Liquid Phase Separation, J. Am. Chem. Soc., 2021, 143, 9849–9857 CrossRef CAS PubMed.
- A. Patel, H. O. Lee, L. Jawerth, S. Maharana, M. Jahnel, M. Y. Hein, S. Stoynov, J. Mahamid, S. Saha, T. M. Franzmann, A. Pozniakovski, I. Poser, N. Maghelli, L. A. Royer, M. Weigert, E. W. Myers, S. Grill, D. Drechsel, A. A. Hyman and S. Alberti, A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation, Cell, 2015, 162, 1066–1077 CrossRef CAS PubMed.
- A. Agarwal, S. K. Rai, A. Avni and S. Mukhopadhyay, An intrinsically disordered pathological prion variant Y145Stop converts into self-seeding amyloids via liquid-liquid phase separation, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2100968118 CrossRef CAS PubMed.
- W. Qu, Z. Wang and H. Zhang, Phase separation of the C. elegans Polycomb protein SOP-2 is modulated by RNA and sumoylation, Protein Cell, 2020, 11, 202–207 CrossRef CAS PubMed.
- T. Kaur, I. Alshareedah, W. Wang, J. Ngo, M. M. Moosa and P. R. Banerjee, Molecular Crowding Tunes Material States of Ribonucleoprotein Condensates, Biomolecules, 2019, 9, 71 CrossRef CAS PubMed.
- I. Alshareedah, T. Kaur, J. Ngo, H. Seppala, L. D. Kounatse, W. Wang, M. M. Moosa and P. R. Banerjee, Interplay between Short-Range Attraction and Long-Range Repulsion Controls Reentrant Liquid Condensation of Ribonucleoprotein-RNA Complexes, J. Am. Chem. Soc., 2019, 141, 14593–14602 CrossRef CAS PubMed.
- S. Kroschwald, S. Maharana and A. Simon, Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments, Matters, 2017, 3, e201702000010 Search PubMed.
- S. Shil, M. Tsuruta, K. Kawauchi and D. Miyoshi, Factors Affecting Liquid-Liquid Phase Separation of RGG Peptides with DNA G-Quadruplex, ChemMedChem, 2024, e202400460, DOI:10.1002/cmdc.202400460.
- K. Yokosawa, M. Tsuruta, S. Kajimoto, S. Naoki, D. Miyoshi and T. Nakabayashi, Quantification of the concentration in a droplet formed by liquid–liquid phase separation of G-quadruplex-forming RNA, Chem. Phys. Lett., 2023, 826, 140634 CrossRef CAS.
- M. Tsuruta, Y. Sugitani, N. Sugimoto and D. Miyoshi, Combined Effects of Methylated Cytosine and Molecular Crowding on the Thermodynamic Stability of DNA Duplexes, Int. J. Mol. Sci., 2021, 22, 947 CrossRef CAS PubMed.
- J.-L. Mergny, A.-T. Phan and L. Lacroix, Following G-quartet formation by UV-spectroscopy, FEBS Lett., 1998, 435, 74–78 CrossRef CAS PubMed.
- J. L. Mergny, J. Li, L. Lacroix, S. Amrane and J. B. Chaires, Thermal difference spectra: a specific signature for nucleic acid structures, Nucleic Acids Res., 2005, 33, e138 CrossRef PubMed.
- M. Vorlíčková, I. Kejnovská, K. Bednářová, D. Renčiuk and J. Kypr, Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes, Chirality, 2012, 24, 691–698 Search PubMed.
- A. Randazzo, G. P. Spada and M. W. da Silva, Circular dichroism of quadruplex structures, Top. Curr. Chem., 2013, 330, 67–86 CrossRef CAS PubMed.
- J. Brčić and J. Plavec, Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution, Nucleic Acids Res., 2015, 43, 8590–8600 CrossRef PubMed.
- J. Brcic and J. Plavec, NMR structure of a G-quadruplex formed by four d(G4C2) repeats: insights into structural polymorphism, Nucleic Acids Res., 2018, 46, 11605–11617 CAS.
- D. Miyoshi, A. Nakao and N. Sugimoto, Molecular crowding regulates the structural switch of the DNA G-quadruplex, Biochemistry, 2002, 41, 15017–15024 CrossRef CAS PubMed.
- B. Heddi and A. T. Phan, Structure of human telomeric DNA in crowded solution, J. Am. Chem. Soc., 2011, 133, 9824–9833 CrossRef CAS PubMed.
- B. Heddi, V. V. Cheong, H. Martadinata and A. T. Phan, Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: solution structure of a peptide-quadruplex complex, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9608–9613 Search PubMed.
- Y. Ma, K. Iida and K. Nagasawa, Topologies of G-quadruplex: Biological functions and regulation by ligands, Biochem. Biophys. Res. Commun., 2020, 531, 3–17 Search PubMed.
- J. Lin, J. Q. Hou, H. D. Xiang, Y. Y. Yan, Y. C. Gu, J. H. Tan, D. Li, L. Q. Gu, T. M. Ou and Z. S. Huang, Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: implications for epigenetic regulation, Biochem. Biophys. Res. Commun., 2013, 433, 368–373 CrossRef CAS PubMed.
- K. Tsukakoshi, S. Saito, W. Yoshida, S. Goto and K. Ikebukuro, CpG Methylation Changes G-Quadruplex Structures Derived from Gene Promoters and Interaction with VEGF and SP1, Molecules, 2018, 23, 944 CrossRef PubMed.
- J. J. Fox and D. Shugar, Spectrophotometric studies of nucleic acid derivatives and related compounds as a function of pH. II. Natural and synthetic pyrimidine nucleosides, Biochim. Biophys. Acta, 1952, 9, 369–384 CrossRef CAS PubMed.
- L. C. Sowers, B. R. Shaw and W. D. Sedwick, Base stacking and molecular polarizability: effect of a methyl group in the 5-position of pyrimidines, Biochem. Biophys. Res. Commun., 1987, 148, 790–794 CrossRef CAS PubMed.
- K. J. Miller and J. Savchik, A new empirical method to calculate average molecular polarizabilities, J. Am. Chem. Soc., 1979, 101, 7206 CrossRef CAS.
- R. I. Mathad, E. Hatzakis, J. Dai and D. Yang, c-MYC promoter G-quadruplex formed at the 5'-end of NHE III1 element: insights into biological relevance and parallel-stranded G-quadruplex stability, Nucleic Acids Res., 2011, 39, 9023–9033 CrossRef CAS PubMed.
- S. T. Hsu, P. Varnai, A. Bugaut, A. P. Reszka, S. Neidle and S. Balasubramanian, A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics, J. Am. Chem. Soc., 2009, 131, 13399–13409 CrossRef CAS PubMed.
- J. Dai, D. Chen, R. A. Jones, L. H. Hurley and D. Yang, NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region, Nucleic Acids Res., 2006, 34, 5133–5144 Search PubMed.
- M. Marusic, P. Sket, L. Bauer, V. Viglasky and J. Plavec, Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops, Nucleic Acids Res., 2012, 40, 6946–6956 CrossRef CAS PubMed.
- P. Schultze, R. F. Macaya and J. Feigon, Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG), J. Mol. Biol., 1994, 235, 1532–1547 CrossRef CAS PubMed.
- Y. Wang and D. J. Patel, Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex, J. Mol. Biol., 1995, 251, 76–94 Search PubMed.
- A. Yett, L. Y. Lin, D. Beseiso, J. Miao and L. A. Yatsunyk, N-methyl mesoporphyrin IX as a highly selective light-up probe for G-quadruplex DNA, J. Porphyrins Phthalocyanines, 2019, 23, 1195–1215 Search PubMed.
- M. Poudyal, K. Patel, L. Gadhe, A. S. Sawner, P. Kadu, D. Datta, S. Mukherjee, S. Ray, A. Navalkar, S. Maiti, D. Chatterjee, J. Devi, R. Bera, N. Gahlot, J. Joseph, R. Padinhateeri and S. K. Maji, Intermolecular interactions underlie protein/peptide phase separation irrespective of sequence and structure at crowded milieu, Nat. Commun., 2023, 14, 6199 Search PubMed.
- C. H. Spink and J. B. Chaires, Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA, Biochemistry, 1999, 38, 496–508 CrossRef CAS PubMed.
- J. Wang, J. M. Choi, A. S. Holehouse, H. O. Lee, X. Zhang, M. Jahnel, S. Maharana, R. Lemaitre, A. Pozniakovsky, D. Drechsel, I. Poser, R. V. Pappu, S. Alberti and A. A. Hyman, A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins, Cell, 2018, 174, 688–699.e616 CrossRef CAS PubMed.
- A. Molliex, J. Temirov, J. Lee, M. Coughlin, A. P. Kanagaraj, H. J. Kim, T. Mittag and J. P. Taylor, Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization, Cell, 2015, 163, 123–133 CrossRef CAS PubMed.
- L. A. Munishkina, E. M. Cooper, V. N. Uversky and A. L. Fink, The effect of macromolecular crowding on protein aggregation and amyloid fibril formation, J. Mol. Recognit., 2004, 17, 456–464 CrossRef CAS PubMed.
- Q. Ma, J. B. Fan, Z. Zhou, B. R. Zhou, S. R. Meng, J. Y. Hu, J. Chen and Y. Liang, The contrasting effect of macromolecular crowding on amyloid fibril formation, PLoS One, 2012, 7, e36288 CrossRef CAS PubMed.
- H. Yaku, T. Murashima, D. Miyoshi and N. Sugimoto, In vitro assays predictive of telomerase inhibitory effect of G-quadruplex ligands in cell nuclei, J. Phys. Chem. B, 2014, 118, 2605–2614 CrossRef CAS PubMed.
- H. Sun, J. K. Karow, I. D. Hickson and N. Maizels, The Bloom's syndrome helicase unwinds G4 DNA, J. Biol. Chem., 1998, 273, 27587–27592 CrossRef CAS PubMed.
- Y. Wu, K. Shin-ya and R. M. Brosh, Jr., FANCJ helicase defective in Fanconi anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability, Mol. Cell. Biol., 2008, 28, 4116–4128 CrossRef CAS PubMed.
- J. P. Vaughn, S. D. Creacy, E. D. Routh, C. Joyner-Butt, G. S. Jenkins, S. Pauli, Y. Nagamine and S. A. Akman, The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates, J. Biol. Chem., 2005, 280, 38117–38120 CrossRef CAS PubMed.
- A. Pipier, A. Devaux, T. Lavergne, A. Adrait, Y. Couté, S. Britton, P. Calsou, J. F. Riou, E. Defrancq and D. Gomez, Constrained G4 structures unveil topology specificity of known and new G4 binding proteins, Sci. Rep., 2021, 11, 13469 CrossRef CAS PubMed.
- R. Yagi, T. Miyazaki and T. Oyoshi, G-quadruplex binding ability of TLS/FUS depends on the beta-spiral structure of the RGG domain, Nucleic Acids Res., 2018, 46, 5894–5901 CrossRef CAS PubMed.
- K. Takahama, A. Miyawaki, T. Shitara, K. Mitsuya, M. Morikawa, M. Hagihara, K. Kino, A. Yamamoto and T. Oyoshi, G-Quadruplex DNA- and RNA-Specific-Binding Proteins Engineered from the RGG Domain of TLS/FUS, ACS Chem. Biol., 2015, 10, 2564–2569 CrossRef CAS PubMed.
- R. L. Flynn and L. Zou, Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians, Crit. Rev. Biochem. Mol. Biol., 2010, 45, 266–275 CrossRef CAS PubMed.
- E. K. S. McRae, E. P. Booy, G. P. Padilla-Meier and S. A. McKenna, On Characterizing the Interactions between Proteins and Guanine Quadruplex Structures of Nucleic Acids, J. Nucleic Acids, 2017, 2017, 9675348 Search PubMed.
- H. Masai, R. Fukatsu, N. Kakusho, Y. Kanoh, K. Moriyama, Y. Ma, K. Iida and K. Nagasawa, Rif1 promotes association of G-quadruplex (G4) by its specific G4 binding and oligomerization activities, Sci. Rep., 2019, 9, 8618 CrossRef PubMed.
- Y. Kanoh, S. Matsumoto, R. Fukatsu, N. Kakusho, N. Kono, C. Renard-Guillet, K. Masuda, K. Iida, K. Nagasawa, K. Shirahige and H. Masai, Rif1 binds to G quadruplexes and suppresses replication over long distances, Nat. Struct. Mol. Biol., 2015, 22, 889–897 CrossRef CAS PubMed.
- H. Q. Yu, D. Miyoshi and N. Sugimoto, Characterization of structure and stability of long telomeric DNA G-quadruplexes, J. Am. Chem. Soc., 2006, 128, 15461–15468 CrossRef CAS PubMed.
- H. Yu, X. Gu, S. Nakano, D. Miyoshi and N. Sugimoto, Beads-on-a-string structure of long telomeric DNAs under molecular crowding conditions, J. Am. Chem. Soc., 2012, 134, 20060–20069 CrossRef CAS PubMed.
- Y. Teng, H. Tateishi-Karimata and N. Sugimoto, RNA G-Quadruplexes Facilitate RNA Accumulation in G-Rich Repeat Expansions, Biochemistry, 2020, 59, 1972–1980 CrossRef CAS PubMed.
- F. Raguseo, Y. Wang, J. Li, M. Petrić Howe, R. Balendra, A. Huyghebaert, D. M. Vadukul, D. A. Tanase, T. E. Maher, L. Malouf, R. Rubio-Sánchez, F. A. Aprile, Y. Elani, R. Patani, L. Di Michele and M. Di Antonio, The ALS/FTD-related C9orf72 hexanucleotide repeat expansion forms RNA condensates through multimolecular G-quadruplexes, Nat. Commun., 2023, 14, 8272 CrossRef CAS PubMed.
- S. Wang and Y. Xu, RNA structure promotes liquid-to-solid phase transition of short RNAs in neuronal dysfunction, Commun. Biol., 2024, 7, 137 CrossRef CAS PubMed.
- N. Sugimoto, M. Nakano and S. Nakano, Thermodynamics-structure relationship of single mismatches in RNA/DNA duplexes, Biochemistry, 2000, 39, 11270–11281 CrossRef CAS PubMed.
- R. D. Gray and J. B. Chaires, Analysis of multidimensional G-quadruplex melting curves, Curr. Protoc. Nucleic Acid Chem., 2011, 17.4.1–17.4.16 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc06959e |
| This journal is © The Royal Society of Chemistry 2025 |