A robust metabolomics approach for the evaluation of human embryos from in vitro fertilization†
Cecilia Beatriz
Figoli
 a,
Marcelo
Garcea
b,
Claudio
Bisioli
b,
Valeria
Tafintseva
c,
Volha
Shapaval
c,
Mariana
Gómez Peña
b,
Luz
Gibbons
a,
Marcelo
Garcea
b,
Claudio
Bisioli
b,
Valeria
Tafintseva
c,
Volha
Shapaval
c,
Mariana
Gómez Peña
b,
Luz
Gibbons
 d,
Fernando
Althabe
d,
Osvaldo Miguel
Yantorno
a,
Marcos
Horton
b,
Jürgen
Schmitt
e,
Peter
Lasch
d,
Fernando
Althabe
d,
Osvaldo Miguel
Yantorno
a,
Marcos
Horton
b,
Jürgen
Schmitt
e,
Peter
Lasch
 f,
Achim
Kohler
*c and
Alejandra
Bosch
f,
Achim
Kohler
*c and
Alejandra
Bosch
 *a
*a
aLaboratorio de Bioespectrosocpia, CINDEFI-CONICET, CCT La Plata, Facultad de Ciencias Exactas, UNLP, 1900 La Plata, Argentina. E-mail: bosch@quimica.unlp.edu.ar
bPREGNA Medicina Reproductiva, C1425 AYV Ciudad Autónoma de Buenos Aires, Argentina
cFaculty of Science and Technology, Norwegian University of Life Sciences, 1432 Ås, Norway. E-mail: achim.kohler@nmbu.no
dIECS, Instituto de Efectividad Clínica y Sanitaria, C1414 Ciudad Autónoma de Buenos Aires, Argentina
eSynthon GmbH, 69198 Schriersheim, Germany
fCentre for Biological Threats and Special Pathogens (ZBS) Proteomics and Spectroscopy Unit, Robert Koch-Institut, 13353 Berlin, Germany
First published on 25th August 2021
Abstract
The identification of the most competent embryos for transfer to the uterus constitutes the main challenge of in vitro fertilization (IVF). We established a metabolomic-based approach by applying Fourier transform infrared (FTIR) spectroscopy on 130 samples of 3-day embryo culture supernatants from 26 embryos that implanted and 104 embryos that failed. On examining the internal structure of the data by unsupervised multivariate analysis, we found that the supernatant spectra of nonimplanted embryos constituted a highly heterogeneous group. Whereas ∼40% of these supernatants were spectroscopically indistinguishable from those of successfully implanted embryos, ∼60% exhibited diverse, heterogeneous metabolic fingerprints. This observation proved to be the direct result of pregnancy's multifactorial nature, involving both intrinsic embryonic traits and external characteristics. Our data analysis strategy thus involved one-class modelling techniques employing soft independent modelling of class analogy that identified deviant fingerprints as unsuitable for implantation. From these findings, we could develop a noninvasive Fourier-transform-infrared-spectroscopy–based approach that represents a shift in the fundamental paradigm for data modelling applied in assisted-fertilization technologies.
1. Introduction
Infertility—a multifactorial disorder that affects around 15% of the reproductive couples worldwide—is a markedly increasing health problem due to the postponement of parenthood.1 Since the first successful in vitro fertilization (IVF) birth in 1978, more than eight million children have been born with the help of assisted reproduction techniques.2,3 Although IVF is widely used to treat infertile couples, in many instances that approach does not resolve infertility problems because of its low success rate.4 Several conditions lead to implantation failure, including reduced endometrial receptivity,5,6 embryonic defects such as genetic abnormalities, the overall clinical status of the mother, faults in the embryo-transfer technique, and/or other multifactorial causes.7,8 One of the most crucial steps for a successful IVF treatment is definitively the selection of a competent embryo(s) for transfer. The evaluation of embryos’ development morphology by light microscopy still represents the usual clinically established method for assessing embryo viability.9,10 This technique constitutes a fast, easy, and affordable evaluation and has been considered as the universally accepted method of choice for embryo selection.9,11 Nevertheless, owing to the significant interobserver variability and subjectivity reported in the literature, together with the low capability of morphological evaluation by light microscopy in predicting the implantation rate of an embryo (i.e., below 30%), that approach represents an inefficient methodology for embryo selection.12–16 Alternatively, invasive methods, such as preimplantation genetic testing (PGT) used to determine the genetic profiling of embryos before implantation, involve certain risks, since biopsy might negatively influence further embryo development.17–21 A noninvasive and rapid evaluation of the embryo-implantation potential before transfer, therefore, constitutes one of the most crucial challenges in IVF treatments.In the last decade, different techniques have been proposed as alternative noninvasive technologies to evaluate the embryo implantation potential.22 Human embryos, while developing in the culture media, consume available nutrients and release metabolites, thus modifying culture supernatants. Therefore, a detailed chemical analysis of the spent supernatant of an embryo's culture medium provides information reflecting cellular metabolic activities and the overall developmental status of the embryo. The relationship between the metabolic parameters and embryo viability was reported for the first time in 1980 by Renard and collaborators.23 A number of proof-of-principle studies related to the chemical composition of an embryo's culture supernatants and the subsequent embryo-implantation outcome reported that embryos achieving implantation were different in metabolomic profile from those that failed in implantation.24,25
The application of several technologies such as vibrational spectroscopy (near infrared, NIR, and mid-infrared, MIR, plus Raman spectroscopy), nuclear-magnetic resonance (NMR), and matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF) for the assessment of culture supernatants provides a picture of an embryo's metabolism and genetic-expression patterns. Therefore, these methodologies have been broadly applied for the evaluation of embryo metabolomics.26,27 In the last decade, studies on metabolomic profiling in spent culture media and the subsequent embryo viability were carried out through the use of different spectroscopy-based technologies.24,28–34 NIR spectroscopy combined with supervised mathematical models was established to estimate the reproductive potential of embryos.24,28–34 Different fertility centers were included in those trials, with the number of recruited patients ranging from 30 to 417. Nevertheless, none of those NIR-based metabolomics models were able to improve clinical-pregnancy rates when compared to the results obtained by analyzing embryo morphology by light microscopy.29,30,32 In addition, a preliminary MIR-spectroscopy assay demonstrated the great potential of Fourier transform infrared (FTIR) spectroscopy in the screening of the embryonic-implantation potential. However, only 7 samples of 26-hour-embryo-culture supernatants from 5 patients were studied and no additional publications with larger cohorts of patients have appeared so far.35 Finally, Bracewell-Milnes and collaborators (2017), by reviewing the potential of the metabolomics technologies as applied to IVF, concluded that the metabolomic profiling of embryo supernatants, as studied to date, has not evidenced any improvement in the prediction of embryonic viability in clinical practice.26
FTIR spectroscopy is a noninvasive analytical physicochemical technique providing information about the total biochemical composition of the analyzed material and has the remarkable advantage of involving a straightforward form of sample preparation and a short spectral-data-acquisition time. FTIR spectroscopy has been successfully used as an analytical tool in a wide range of fields including food, biotechnology, and microbiological and medical diagnostics.36–46,90 The potential of FTIR analysis of blood components (e.g., serum and plasma) and other biofluids (e.g., bile, urine and sputum) for diagnostic purposes has been widely investigated and recognized.47–55 The great ability of this spectroscopic technology to detect small changes in different types of samples has led to its application in other fields such as the study of extracellular and intracellular metabolites in bacterial,41 fungal,56 and mammalian-cell cultures.39,40 Furthermore, FTIR spectroscopy has been applied as a valuable tool in metabolomics studies since it is highly sensitive for the simultaneous detection of carbohydrates, lipids, proteins, nucleic acids, amino acids, fatty acids, sugars and many other small molecules.57 In particular, glucose, glycerol, and acetic acid were measured in Escherichia coli cultures,41 while glucose and lactate concentrations were evaluated in mammalian cell line culture supernatants.39
In view of this strong background, the aim of the present study was to test FTIR spectroscopy combined with multivariate data analysis as a means for a noninvasive assessment of human embryo metabolomics. For this purpose, we characterized 3-day-embryo-culture supernatants by FTIR spectroscopy and evaluated whether changes in the infrared patterns could be associated with the outcome of IVF. We also expected to understand how different clinical characteristics of the mother may impact embryos’ metabolomics and implantation rates.
2. Experimental
2.1. Ethical approval
Participants were recruited and provided written consent according to section IRB00001745-IORG 0001315 of the protocols approved by a national ethics committee for medical education and clinical research [Centro de Educación Médica e Investigaciones Clínicas Norberto Quirno (CEMIC), Argentina].2.2. Patients
The patients participating in the study were recruited at the fertility center “PREGNA-Medicina Reproductiva”, Buenos Aires, Argentina during a 3-year period. All women below 42 years treated and undergoing IVF treatments were considered for participation in the study, but patients with more than two previous IVF attempts were excluded. The diagnosis of the cause of female or male infertility, the protocol applied for ovarian-stimulation, and whether conventional IVF or intracytoplasmic sperm injection was employed were not considered as exclusion criteria. The stimulation protocols consisted of the application of gonadotropins and the gonadotrophin-releasing–hormone antagonist in combination with recombinant and/or highly purified urinary gonadotropins.2.3. Construction of a database platform for research-data management—“OpenClinica”
This research was performed under strict international biosafety regulations, applying the “Best Practices for Research Data Management”. For this purpose, we used an open-source software OpenClinica, based on electronic forms and protocols for storage, classification, analysis, and data visualization.58 For the construction of this database, information such as patients’ personal details, hormonal treatments, embryo morphology, pregnancy outcome, and results from the quality tests of the acquired spectra. This platform is currently operating effectively on the web site https://www.openclinica.com/. For data analyses some of the clinical data registered at the OpenClinica database such as mother's age, BMI, and smoking habits were converted from numerical variables into qualitative ones (see ESI† Converting numerical into qualitative variables).2.4. Embryo culture
Upon oocyte retrieval, cumulus–oocyte complexes were placed in four-well plates containing 500 μL of G-IVF Plus medium covered with mineral oil (OVOIL-Culture Oil) to avoid evaporation (all the culture media used in this study were from Vitrolife, Göteborg, Sweden). In parallel, semen samples were processed through discontinuous 90–50% density gradients (Spermgrad) and double washed in a sperm preparation medium. The sperm suspension was adjusted to 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 motile sperm per mL in that medium and kept at room temperature (20–23 °C) until insemination. For conventional IVF, insemination was carried out 4 h after oocyte retrieval in the G-IVF Plus culture medium. For intracytoplasmic sperm injection, cumulus cells and the corona radiata of oocytes were removed by a brief exposure to 80 IU mL−1 of hyaluronidase (Hyase-10×) 3–4 h after collection. This fertilization procedure was performed according to the standard protocol.59,60
000 motile sperm per mL in that medium and kept at room temperature (20–23 °C) until insemination. For conventional IVF, insemination was carried out 4 h after oocyte retrieval in the G-IVF Plus culture medium. For intracytoplasmic sperm injection, cumulus cells and the corona radiata of oocytes were removed by a brief exposure to 80 IU mL−1 of hyaluronidase (Hyase-10×) 3–4 h after collection. This fertilization procedure was performed according to the standard protocol.59,60
After 18–20 h of insemination, fertilization was checked and two pronuclei-stage embryos were cultured individually in 40 μL droplets of overnight-equilibrated G-1 Plus medium covered with culture oil in an IVF Tri-gas Incubator Model G185 (K-Systems, Birkerød, Denmark). For each cohort of embryos, a drop of G1 Plus medium was incubated under the same conditions as a control.
Embryos incubated for 3 days were individually observed by optical microscopy for morphological grading according to the Istanbul-consensus criteria (European Society of Human Reproduction and Embryology and the Alpha Scientists’ Special-Interest Group).9 The embryos were classified as grade 1 (good quality), grade 2 (fair quality), grade 3 (poor quality) and grade 4 (arrested or undeveloped).
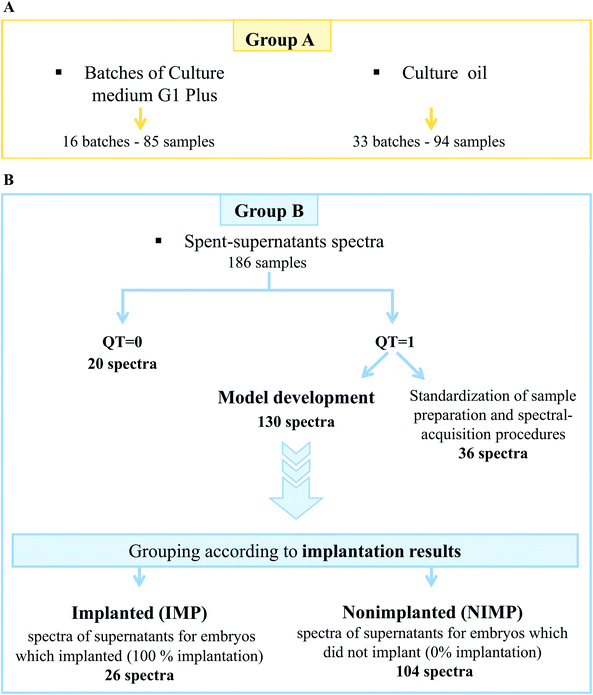
The highest quality embryos from each cohort (one or two) were selected to be transferred. All the single embryos were retrieved from their supernatants. In all instances, only fresh embryos were transferred as described elsewhere after placing them in G-2 Plus culture medium.61 The spent supernatants, drops of control culture media, samples of different batches of the G-1 Plus culture medium, and samples of the culture oil used were registered and stored under liquid nitrogen for further FTIR spectral analyses.
2.5. Sample preparation and FTIR spectral acquisition
The dried-film FTIR technology55,62–64 was applied to analyze the 3-day-embryo-culture supernatants. For this purpose, a protocol for sample preparation was optimized. Cryopreserved supernatants were thawed at room temperature (25 °C) and centrifuged for 5 min at 1690g to separate possible remnants of the culture oil in the samples. Different volumes of supernatants (15, 20, and 30 μL) were then pipetted onto each well of a ZnSe 96-well microtiter plate. The samples were dried under moderate vacuum (0.1 bar) or until transparent films were formed.53,64 FTIR absorption spectra were recorded using a Vertex 70 FTIR spectrometer coupled to the high-throughput HTS-XT automatic module under dried-air circulation (Bruker Optics GmbH, Ettlingen, Germany). The spectra were recorded in the transmission mode within the spectral range 650 and 4000 cm−1 with a 6 cm−1 spectral resolution by taking 64 scans that were subsequently averaged. Before each sample measurement, the background spectra of the ZnSe substrate were recorded in order to account for the variation in water vapor and CO2, with the OPUS spectroscopy software (version 7.0; Bruker Optics GmbH, Ettlingen, Germany) being used for automatic spectral acquisition. The FTIR spectra of both the different batches of the culture medium and of the culture oil, used for the different reproducibility studies here performed, were likewise measured according to the procedure described above.2.6. Spectral data analyses
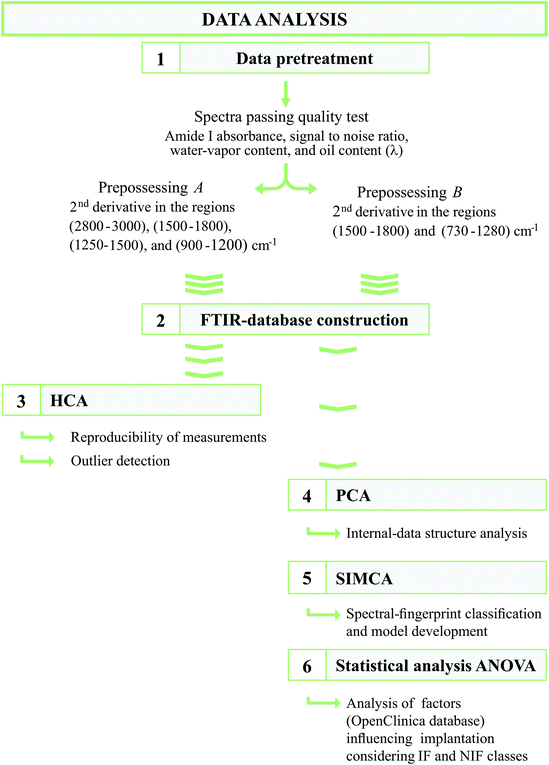
The spectral data analysis flow sheet applied in this study was specifically developed for the optimized processing of embryo-supernatant FTIR spectra and comprised the following routines: (1) data pretreatment, (2) construction of the FTIR database, (3) hierarchical-cluster analysis (HCA), (4) principal-component analysis (PCA), (5) soft independent modelling of class analogy (SIMCA), and (6) statistical analysis (see Fig. 1).Data quality test is an important step in spectral data analysis and is known to improve data mining and data modelling results.66 Therefore, as a first step in data analysis, all raw spectra were subjected to a quality test of our own design through the use of the OPUS-spectroscopy software version 7.0 (Bruker Optics, Ettlingen, Germany). This test involved checking the following parameters: (i) absorbance in the amide I region (1600–1700 cm−1) with acceptable values being between 0.125 and 1.20 absorbance units, (ii) the signal-to-noise ratio (calculated from the first derivatives of the spectra between 2000 and 2100 cm−1) with admissible values being lower than 1.5 × 10−4 and, (iii) water-vapor content (determined from the first derivatives of the absorbance values between 1837 and 1847 cm−1) with acceptable values being lower than 3 × 10−4.53,64 Because of the small quantities of culture oil that were always left in the supernatants after the centrifugation, which could further interfere with the FTIR spectral signals, an additional parameter in the quality test, the so-called “λ”, was defined. Parameter λ indicates the level of contamination of the supernatant with the culture oil in the infrared-absorbance spectra as follows: the rate of the intensity of the peak at 2933 cm−1 assigned as the C–H stretching of >CH2 groups (lipids)—used as a marker band of oil content (I2933)—and the intensity of the peak at 1655 cm−1 assigned to amide I—used as an internal standard of the total biomass (I1655, where λ = I2933/I1655; ESI Fig. S1†). Only spectra with λ ≤ 0.33 were included in the FTIR database.
For spectra that passed the quality test, two different types of spectral preprocessing were developed: one for technical reproducibility analysis (preprocessing A) which was applied among (i) the different wells of the ZnSe optical plate, (ii) the samples of the different culture medium batches, and (iii) the samples recovered within each culture medium batch. Another preprocessing (preprocessing B) was applied to the spectra of the supernatant samples before the PCA and SIMCA analyses. In the first type, the so-called preprocessing A, the spectra were preprocessed by calculating the second derivative (Savitzky–Golay, 17 windows size) in the regions 2800–3000, 1500–1800, 1250–1500, and 900–1200 cm−1; in the second, the so-called preprocessing B, the data were preprocessed by taking the second derivatives (Savitzky–Golay, 3rd-degree polynomial, 17 windows size) in the regions 1500–1800 and 730–1280 cm−1, followed by normalization by extended multiplicative signal correction (EMSC) with linear term.67,91,92
Out of the 186 FTIR-embryo-supernatants acquired, 130 were used for modelling, 20 were discarded due to their high amount of culture oil content (λ > 0.33), and 36 were purely used for the optimization of the different experimental procedures (such as sample preparation and the type of drying process, see Fig. 2).
The supernatant spectra of the IMP group pertained to embryos whose morphologies were 30.8% grade 1 and 69.2% grade 2. Whereas embryos of grade 3 were not found, the supernatants of the NIMP group contained 33.7%, 50.0%, and 16.4% of embryos with morphologies of grades 1, 2, and 3, respectively.
2.7. Statistical analysis
An one-way analysis of variance (ANOVA) was performed on the maternal metadata, with the IF class being considered separately, and on the IF versus the NIF class obtained by SIMCA modelling. The different parameters, such as the maternal age, body-mass index (BMI), and smoking habits, were analyzed as a single parameter both separately by ANOVA and altogether by the multivariate analysis of variance (MANOVA).Chi2 analysis was performed to evaluate the embryo distribution within the IF and NIF classes.
3. Results & discussion
3.1. A robust FTIR experimental approach for the metabolomics analysis of 3-day-embryo-culture supernatants
In the development of a novel method for the evaluation and selection of embryo-implantation potential based on FTIR vibrational spectroscopy, we established a 3 h protocol for sample preparation and spectral acquisition (Fig. 3). The protocol stated in brief is as follows: supernatants are recovered from individual embryo cultures and centrifuged to remove the culture oil. Then 30 μL is transferred to a 96-multiwell ZnSe optical plate. Next, the samples are dried under moderate vacuum (0.1 bar) for 45 min until transparent films are obtained.53,64 Finally, FTIR transmission measurements are carried out in the spectral range 650–4000 cm−1 at a 6 cm−1 spectral resolution. | ||
| Fig. 3 Flow sheet of the standardized approach for the FTIR spectral measurements of embryo supernatants. | ||
Because FTIR is an extremely sensitive analytical technique, a strict reproducibility analysis was performed to assure the construction of a robust infrared spectral database for further data analysis (Methods). We studied the reproducibility level of the measurements obtained for the 96 positions of the ZnSe optical plate using the same batch of fresh culture medium. This analysis demonstrated that the spectral quality was not affected by the desiccation of the samples observed during the lengthy time required for the measurement of 96 samples (ESI Fig. S2†). A high level of reproducibility was also observed among the samples from each batch and among different batches of the G1 Plus culture medium (ESI Fig. S3 and S4†).
The selected spectral pretreatment approaches carried out here provided an increase in the resolution and an easier interpretation of the spectra. It allowed the detection of outliers and helped to deal with the interference from the culture oil in the FTIR spectra. Overall, the experimental protocol proposed here improved the FTIR-based method previously reported,35 facilitated the subsequent data analyses and allowed achieving robust and reliable classification models.
3.2. FTIR spectral characterization of spent embryo-culture medium
As the vibrational spectra provide the biochemical information of embryo-culture supernatants, they provide a picture of embryonic metabolomics. Particularly, NIR spectroscopy has represented a widespread analytical technique for carrying out a direct study of embryo supernatants.24,29,30,32–34,72 The NIR technology produces wide bands due to the absorption of few signals from the molecular overtones along with a combination of the stretching-bending vibrations of atomic groups such as O–H, C–H, and N–H.73 In contrast, MIR spectroscopy provides much more information about the biochemical composition of the biological materials.64 The MIR spectra arise from the stretching and bending vibrations resulting from all bonds that exhibit a transition dipole moment, such as C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–O, O–H, N–H, and C–N, among others. Fig. 4, panel A, depicts a representative and exemplary FTIR absorption spectrum of a 3-day-embryo-culture supernatant recovered from a successfully implanted embryo (class IMP in this study), while Fig. 4, panel B lists the assigned spectral bands and their respective functional groups. The main spectral windows (W1–W5) associated with the molecular building blocks of complex biological samples64 could be identified: the spectral region associated preferentially with lipids (W1) between 2800 and 3000 cm−1 exhibits bands assigned to the symmetric and antisymmetric C–H stretching modes of the methyl groups (–CH3) detected at 2874 and 2969 cm−1, respectively, and the antisymmetric C–H stretching mode of methylene residues (>CH2) at 2933 cm−1. The infrared spectral region associated with protein absorptions (W2) evidenced the typical amide-I and amide-II bands at 1655 and 1545 cm−1, respectively. The mixed region (W3) between 1200 and 1500 cm−1 represents the absorptions of the stretching and bending vibrations from fatty acids, polysaccharides, nucleic acids, and proteins. A characteristic band is observed around 1400 cm−1, the absorbance of which may be attributed to the symmetric stretching vibrations of the –COO− functional groups of amino-acid side chains or free fatty acids. In this region, a typical amide-III band at 1315 cm−1 was also observed along with bands of different >P
O, C–O, O–H, N–H, and C–N, among others. Fig. 4, panel A, depicts a representative and exemplary FTIR absorption spectrum of a 3-day-embryo-culture supernatant recovered from a successfully implanted embryo (class IMP in this study), while Fig. 4, panel B lists the assigned spectral bands and their respective functional groups. The main spectral windows (W1–W5) associated with the molecular building blocks of complex biological samples64 could be identified: the spectral region associated preferentially with lipids (W1) between 2800 and 3000 cm−1 exhibits bands assigned to the symmetric and antisymmetric C–H stretching modes of the methyl groups (–CH3) detected at 2874 and 2969 cm−1, respectively, and the antisymmetric C–H stretching mode of methylene residues (>CH2) at 2933 cm−1. The infrared spectral region associated with protein absorptions (W2) evidenced the typical amide-I and amide-II bands at 1655 and 1545 cm−1, respectively. The mixed region (W3) between 1200 and 1500 cm−1 represents the absorptions of the stretching and bending vibrations from fatty acids, polysaccharides, nucleic acids, and proteins. A characteristic band is observed around 1400 cm−1, the absorbance of which may be attributed to the symmetric stretching vibrations of the –COO− functional groups of amino-acid side chains or free fatty acids. In this region, a typical amide-III band at 1315 cm−1 was also observed along with bands of different >P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O asymmetric stretching vibrations at around 1230 cm−1. The vibrational modes of the carbohydrate region (W4) between 900 and 1200 cm−1 is generally dominated by the symmetric stretching vibrations of the PO2− groups (1090 cm−1) in nucleic acids and a complex sequence of peaks mainly attributed to the C–O–C and C–O–P stretching vibrations of various oligo- and polysaccharides.64 That region also contains bands assigned to the C–O stretching vibrations in carboxylic groups. Finally, the region between 650 and 900 cm−1 (W5) contains weakly expressed bands arising from the aromatic-ring vibrations of phenylalanine, tyrosine, tryptophan, and the various nucleotides. With the exception of only a few peaks (e.g., a band near 720 cm−1, resulting from the >CH2-rocking modes of the fatty-acid chains), valid assignments can hardly be achieved. W5 exhibits a variety of extremely characteristic features superimposed on an underlying broad spectral contour. Therefore, we refer to this spectral domain as the true fingerprint region.64
O asymmetric stretching vibrations at around 1230 cm−1. The vibrational modes of the carbohydrate region (W4) between 900 and 1200 cm−1 is generally dominated by the symmetric stretching vibrations of the PO2− groups (1090 cm−1) in nucleic acids and a complex sequence of peaks mainly attributed to the C–O–C and C–O–P stretching vibrations of various oligo- and polysaccharides.64 That region also contains bands assigned to the C–O stretching vibrations in carboxylic groups. Finally, the region between 650 and 900 cm−1 (W5) contains weakly expressed bands arising from the aromatic-ring vibrations of phenylalanine, tyrosine, tryptophan, and the various nucleotides. With the exception of only a few peaks (e.g., a band near 720 cm−1, resulting from the >CH2-rocking modes of the fatty-acid chains), valid assignments can hardly be achieved. W5 exhibits a variety of extremely characteristic features superimposed on an underlying broad spectral contour. Therefore, we refer to this spectral domain as the true fingerprint region.64
The dried-film FTIR spectroscopy approach proposed here for the analyses of embryo supernatants improves the sensitivity of metabolomics analyses by providing much more information about the biochemical composition than other spectroscopic methodologies such as NIR. Moreover, it guarantees that the strong spectroscopic features of water absorption that interfere in FTIR spectra are avoided.55,62–64 This approach increases the benefits of the vibrational spectroscopy technologies that are simple, rapid, and inexpensive for assessing biological samples.
3.3. Development of a predictive embryo-implantation model based on the FTIR metabolomic profile of embryo-supernatants
Although depending on many different parameters,54 the crucial step of the IVF process is clearly the selection of fully competent embryo(s) for transfer. Accordingly, obtaining and selecting an embryo with the highest implantation potential is still the key objective within the state of the art in IVF laboratories. In this respect, different noninvasive proteomics and metabolomics technologies have constantly been proposed allowing differentiation between the embryos that appear morphologically identical and have the potential to identify the embryo ploidy status.11,26,27 Nevertheless, we are convinced that the limited success of the discrimination results is not due to the technologies but rather the modelling approach selected to perform the task. We believe that the statistical models for embryo differentiation can be improved if the embryo data are reconsidered (restructured) and the modelling approach is selected appropriately to solve the classification problem.We first studied the internal structure of the spectral data by the unsupervised multivariate method PCA. This analysis aims at transforming the original variables, which here in infrared spectroscopy referred to wavenumbers, into smaller numbers of new variables or principal components that describe the main variation patterns.67 The PCA-score plot obtained (Fig. 5, panel A) revealed that the spectral fingerprints of the supernatants from the implanted embryos (IMP, black squares) were quite similar among themselves and formed a relatively homogeneous cluster in the principal-component space. In contrast, the spectra recorded from the supernatants of the NIMP embryos (green squares) were distributed over a much wider range, did not follow any specific pattern, thus exhibiting a larger spectral heterogeneity (see also ESI Heterogeneity analysis and ESI Fig. S5†). However, within this context, we also need to mention that a significant fraction of spectral fingerprints from the NIMP embryos were indistinguishable from the footprints of the supernatants of the implanted embryos (see NIMP fingerprints overlapping with IMP footprints in Fig. 5, panel A). This observation and the higher heterogeneity level of the NIMP fingerprints suggested the presence of altered metabolic states in the embryos that did not implant. We were thus tempted to speculate that some of those altered states may be responsible for the nonimplantation. In contrast, among the NIMP group of samples, a significant number of embryos with biochemical implantation fingerprints similar to or overlapping with those of IMP might belong to embryos that could be good candidates on the basis of their metabolomic patterns but failed to implant owing to the other parameters not associated with metabolomics and thus were not reflected in the spectroscopic methods (e.g., maternal features like overweight, age, and smoking habits, among other). Those samples were therefore falsely grouped in the wrong class from the metabolomics point of view.
In most of the metabolomic-based investigations carried out thus far, the available data are first preclassified according to the implantation outcomes in two classes: (i) samples belonging to embryos that implanted or resulted in a live birth and (ii) samples belonging to embryos that failed to do either. Then, these two classes serve as reference data for training different supervised learning algorithms.55,64 Supervised data processing methods such as genetic algorithms or least-squares regressions use these spectra a priori assigned to classes such as teaching information data to build models that are later used to predict the outcomes of unknown samples.24,28–33,35 Under our working hypothesis, there is a certain group of NIMP embryos that express metabolomic biomarkers for embryo implantation but fail to implant for other reasons. Consequently, the use of the fingerprints of nonimplanted embryos as one of the references to establish a model disregards the reality that implantation is a multifactorial event.
To test our hypothesis, we employed the SIMCA method,70 a one-class modelling technique that involves only a single class of objects to establish a discrimination model. The model can then be used to classify any new subject as to either belonging to the class or being outside of the class. This approach is particularly well suited for the example of embryo-supernatant data as embryos with implantation results (IMP) are quite similar among themselves and can be used to establish such a model, whereas the embryos with nonimplantation results (NIMP) are more disparate. Thus, a SIMCA model was established with the spectra of the IMP group. The SIMCA model is represented by a class border (see Fig. 5, panel B, curved line) that separates samples that belong to the class from those falling outside the class. The model then, upon challenged with the NIMP samples, separated that class into two groups. The first contained the so-called “implantation fingerprints” (IF)—i.e., comprising spectra from samples with a metabolic fingerprint similar to IMP (Fig. 5, panel B, orange dots). The second class, formed by putative “nonimplantation fingerprints” (NIF), contained spectra from supernatants with altered metabolic states (Fig. 5, panel B, blue dots). We need to note here that the NIMP spectra are exclusively—but no IMP samples—plotted in Fig. 5, panel B. From this plot, we can clearly recognize that some NIMP fingerprints belong to the model space characterized by implanted-embryo fingerprints (IF, orange dots) while others fall outside that model space (NIF, blue dots). This situation is also reflected in the logic diagram in Fig. 5, panel C; which illustrates that the class IF was composed of all 26 of the IMP spectra along with 40 of the NIMP spectra having features of the metabolic-implantation fingerprints (the 40 orange dots in Fig. 5, panel B), altogether totalling 66 samples. The NIF class contained the remaining 64 fingerprints from the NIMP spectra (blue dots in Fig. 5, panel B). Interestingly, the classification by the SIMCA model provided an almost perfect balance between the IF and NIF classes (i.e., 66 versus 64). This observation suggested that, according to our hypothesis, roughly half of the embryos had implantation potential. As the restructuring of data involved redistributing the NIMP group into two classes according to the implantation metabolomic fingerprint (IF and NIF), it was also worthy to study whether the spectral data heterogeneity was modified (ESI Heterogeneity analysis†). As expected, we found that the IF spectra were quite homogeneous yielding a D value and a standard deviation similar to the ones obtained for the IMP group (D ± SD = 10.88 ± 7.24 for IF group vs. D ± SD = 11.70 ± 7.00 for IMP). Interestingly, the NIF group was highly heterogeneous, giving even higher D and standard deviation values than the NIMP class (D ± SD = 33.35 ± 25.72 for NIF class vs. D ± SD = 21.98 ± 16.97 for NIMP group). This high deviation or variation in the NIF class might indicate the diversity of the abnormal metabolic profiles and/or cellular processes that may occur in abnormal embryo development in contrast to the IF class characterized by a defined metabolic pathway.
An in-depth analysis of the second derivatives of the spectra belonging to the NIF and IF groups revealed certain spectral differences mainly in the spectral regions W2 and W3 (ESI Fig. S6†). A decrease in the intensities of the 1530 and 1515 cm−1 peaks associated with amino acids was observed in the IF spectra in comparison with those of the NIF spectra. In addition, in accordance with a Raman based study,74 these spectra exhibited differences in the region 1330–1440 cm−1 (W3) that could be attributed to the symmetric stretching vibrations of the –COO− functional groups of amino-acid side chains. These spectral variances could be indicative of a difference in the amino-acid turnover between the two groups of embryos, as previously inferred by other technologies. Moreover, these changes in the spent culture medium are in accord with previous studies indicating that amino acid turnover could be used to evaluate embryonic viability.22,75,76 Particularly, the turnover of three amino acids, Asn, Gly and Leu, was previously reported to be significantly correlated with clinical pregnancy and live birth.77,78 Furthermore, importantly it was demonstrated that those correlations were independent of known predictors, such as female age and embryo morphological grade.77 These results revealed that SIMCA analysis could in fact discriminate between embryos presenting different metabolomic profiles.
Another important finding was that among the spectra in IF class, we encountered a proportion of samples that although presenting metabolomic fingerprints (IF class) did not implant (green squares in IF group, Fig. 5, panel C). Aiming to identify the potential causes for those embryos with IF to have failed to implant, different statistical analyses were performed using the patients’ clinical data registered in the OpenClinica database (cf. the Methods section and ESI† for converting numerical into qualitative variables). This database contained certain parameters obtained from patients that are known to be associated with implantation outcomes such as women's age (between 27 to 42 years classified into three categories, <35, from 35 to 40, and >40), women's BMI, divided into 4 classes from normal weight to obesity, and smoking habits (categorized into 5 groups, from nonsmoker to smoker at more than 20 cigarettes per day). Each of these parameters was analyzed independently (ANOVA) and in combination (MANOVA). Through this approach, we could confirm that woman's age was the main factor significantly associated (p < 0.05) with the nonimplantation of embryos within the class NIMP classified by SIMCA as the IF class (comprising those having implantation potential; ESI Fig. S7, panel A†). Accordingly, upon analyzing the distribution of the mothers’ ages, we found that within the IF class the IMP group (those same 26 samples) did not contain women older than 40 years, while the NIMP group (40 samples) included women of all three age-categories (age groups 1, 2 and 3; ESI Fig. S8, panel A†).
Another question that we wanted to address was if any of the external parameters analyzed here could have triggered the embryos to express either IF or NIF metabolomics. In an ANOVA comparison of the classes IF (26 + 40 samples) and NIF (64 samples), we found that the samples predicted as NIF were typical of women of a statistically relevant higher age (ESI Fig. S7, panel B†). In fact, a comparison of the distribution of the mothers’ ages within IF and NIF revealed that the NIF class contained a higher number of women older than 40 years (ESI Fig. S8, panel B†). It is well known that age has the strongest influence on a woman's chance to become pregnant,79–85 with advanced age causing a reduction in the ovarian follicular pool, perturbations in ovulation, and an increase in meiotic errors within the oocyte.86,87 This example is one among several different clinical predispositions that could induce an embryo to express an implantation or nonimplantation metabolomic fingerprint. Another element to consider at this point is that in the last decades it has widely been studied that the metabolic profile of biological specimens is affected by numerous factors, such as age, ethnicity, environment, lifestyle and other factors.88 In our study, the metabolomic fingerprinting of embryo supernatants may present the contribution of biological markers associated with mothers’ age together with the signatures owing to embryos implantation potential. Nevertheless, the classification model by SIMCA has been built using only IMP class which happened to be just young mothers (age 0–2 grade). However, when the model was applied to the rest of the data (NIMP class), we observed that the implantation fingerprint (IF samples) was detected for mothers of the older group too (grade 3) (ESI Fig. S7, panel B†). This demonstrated that the model does not depend on the mother's age but rather focuses on the embryo's differences to detect whether the embryos are of good quality to be implanted. Besides, we also showed that amino acid turnover represents one of those metabolomic markers that contribute to the discrimination between IF and NIF (ESI Fig. S6†). We can therefore address that the discriminative signatures between IF and NIF are dominated by features emerging from implantation potential and not from women's age.
By combining patients’ clinical data stored at OpenClinica and the results of our SIMCA model, we could show how the successful implantation of an embryo relies on intricate and multiple contributions. As it has been previously reported, embryo implantation depends not only on eggs and sperm, which determine the quality of resulting embryos, but also on the endometrium, ovarian-stimulation regimes, laboratory conditions, and many other external parameters.89
Of further interest to us was to compare embryo morphology within the IF and NIF classes (Fig. 5, panel D). A statistical analysis (Chi2) demonstrated that the morphological distribution of the embryos (grades 1, 2, and 3) observed within the IF and the NIF classes was not significantly different (p = 0.125). In accordance with this result, previous studies have demonstrated that other technique grading systems based on the embryo-cleavage rate and morphological features were found not to be correlated with the different metabolomic patterns.75 Thus, embryo morphology seems not to be correlated with its phenotypic traits.
In a retrospective analysis examining the combined results of FTIR and SIMCA, we observed that within the NIMP group more than 50% (64 out of 104 embryos) presented an NIF pattern (Fig. 5, panel C). Thus, these NIMP-NIF embryos would not have been transferred to mothers if they had first been analyzed by the approach employing both FTIR and SIMCA. We need, however, to remark that the criterion for selecting the embryos for transfer was based largely on their morphological appearance (86% of these embryos presented the highest morphology grade according to the Istanbul consensus). Our results indicate, therefore, that reconstruction of the data including SIMCA in the IF and NIF will certainly be helpful as a practical adjunct since this additional information would decrease the implantation rate failures and significantly improve the overall IVF outcome. Nonetheless, these results definitely have to be verified experimentally in a follow-up study.
Different strategies can be described for the process of selecting the embryo with the highest probability of implantation within an embryo cohort. The first represents the development of models that identify embryo biomarkers associated with embryo “quality”. In this regard, we can apply different approaches—namely, those based on embryonic morphology (microscopic morphological analysis and time-lapse imaging techniques), on the quality of the embryo's genetic material (noninvasive PGT), or on embryo metabolomics (NIR, FTIR, Raman, and proteomics methods). Then, if a large cohort of patients were available, by combining more than one of these different approaches to assess embryonic viability, a further improvement in developing robust multifactorial implantation models for selecting the best embryo for transfer would also be possible. Nevertheless, even then, we still could not ensure that such embryos with the best “score” would implant. We have demonstrated here that suitable candidates from the metabolomic point of view may fail in their implantation owing to other parameters, of equal consequence, associated with patients’ life-style, habits, and/or external characteristics such as smoking habits, BMI, age, and stress—in addition to the embryo-transfer quality—just to mention a few. Consequently, a margin of uncertainty involving the patients’ profile and external conditions will always exist, the elements of which are often difficult to be included in a model. Therefore, we believe that to enhance the efficiency of IVF procedures, efforts should focus particularly on two aspects (i) combining different approaches for embryo assessment (morphokinetics, genetics, proteomics, and metabolomics) and (ii) improving dataset classification and data analysis. The combination of different embryo-assessment methods could enable the evaluation of an embryo's complete status with respect to implantation outcome. We have here established the basis for how embryo data should be analyzed in order to develop novel models for embryo selection in IVF treatments. This research, therefore, constitutes a significant contribution in the area of assisted fertilization by offering a novel approach for embryo assessment and data treatment in IVF.
4. Conclusions
In this work, we have demonstrated for the first time the potential of FTIR spectroscopy combined with multivariate analysis for gaining insights into the fundamental aspects associated with embryo metabolomics and for improving implantation outcomes. Our findings have significant implications for the understanding of how embryo data should be classified and analyzed in order to develop new models for embryo selection in IVF treatments. This research has also allowed us to understand the impact of the different clinical characteristics of the mother—such as mother's age—on embryonic metabolomics and implantation rates, demonstrating the importance of assuming that implantation represents a highly complex multifactorial process when developing novel IVF models. Although this study was focused on FTIR spectroscopy of 3-day embryo culture supernatants, the data analysis strategy proposed here can be applied to future research analyzing data from 5 or 6-day embryo culture as well as different kinds of data obtained from genetics, proteomics, or metabolomics.Author contributions
C. B. F.: conceptualization, formal analysis, investigation, methodology, software, validation, visualization, writing – original draft, and writing – review & editing. M. G.: conceptualization, formal analysis, resources, and writing – review & editing. C. B.: conceptualization, resources, and writing – review & editing. V. T.: conceptualization, formal analysis, investigation, methodology, software, validation, writing – original draft, and writing – review & editing. M. G. P.: resources. L. G.: data curation. F. A.: data curation. V. S.: conceptualization, methodology, software, visualization, writing – original draft, and writing – review & editing. J. S.: conceptualization, methodology, and software. P. L.: conceptualization, supervision, visualization, and writing – review & editing. O. M. Y.: conceptualization, funding acquisition, project administration, and writing – review & editing. M. H.: funding acquisition, project administration, and supervision. A. K.: conceptualization, methodology, software, writing – original draft, and writing – review & editing. A. B.: conceptualization, investigation, funding acquisition, project administration, supervision, visualization, writing – original draft, and writing – review & editing.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
Dr Donald F. Haggerty, a retired academic career investigator and native English speaker, edited the final version of the manuscript.This research was supported by a Merck Serono award, “Grant for Fertility Innovation” GFI-2012-1; by an award from Comisión de Investigaciones Científicas de la Provincia de Buenos Aires, (CIC-PBA), “PREMIO CIENCIA Y COMUNIDAD 2013”; by an award from Ministerio de Ciencia, Tecnología e Innovación Productiva (MINCyT), “PREMIO INNOVAR 2015”; and from a grant of the Fondo para la Investigación Científica y Tecnológica (FONCYT), ANR-800-183/11. C. B. F. was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (CONICET); A. B. was supported by CIC PBA.
References
- M. N. Mascarenhas, S. R. Flaxman, T. Boerma, S. Vanderpoel and G. A. Stevens, PLoS Med., 2012, 9, e1001356 CrossRef PubMed.
- C. De Geyter, Best Pract. Res., Clin. Endocrinol. Metab., 2019, 33, 3–8 CrossRef PubMed.
- J. de Mouzon, G. M. Chambers, F. Zegers-Hochschild, R. Mansour, O. Ishihara, M. Banker, S. Dyer, M. Kupka and G. David Adamson, Hum. Reprod., 2020, 35, 1900–1913 CrossRef PubMed.
- G. D. Adamson, J. de Mouzon, G. M. Chambers, F. Zegers-Hochschild, R. Mansour, O. Ishihara, M. Banker and S. Dyer, Fertil. Steril., 2018, 110, 1067–1080 CrossRef PubMed.
- A. Kasius, J. G. Smit, H. L. Torrance, M. J. C. Eijkemans, B. W. Mol, B. C. Opmeer and F. J. M. Broekmans, Hum. Reprod. Update, 2014, 20, 530–541 CrossRef PubMed.
- O. Lebovitz and R. Orvieto, Gynecol. Endocrinol., 2014, 30, 409–414 CrossRef CAS PubMed.
- E. J. Margalioth, A. Ben-Chetrit, M. Gal and T. Eldar-Geva, Hum. Reprod., 2006, 21, 3036–3043 CrossRef CAS PubMed.
- S. A. Roberts, M. Hann and D. R. Brison, Reprod. BioMed. Online, 2016, 32, 197–206 CrossRef PubMed.
- Alpha and ESHRE, Hum. Fertil., 2011, 26, 1270–1283 CrossRef PubMed.
- L. Nel-Themaat and Z. P. Nagy, Placenta, 2011, 32, S257–S263 CrossRef CAS PubMed.
- R. Zmuidinaite, F. I. Sharara and R. K. Iles, Int. J. Mol. Sci., 2021, 22, 1–13 Search PubMed.
- L. Botros, D. Sakkas and E. Seli, Mol. Hum. Reprod., 2008, 14, 679–690 CrossRef CAS PubMed.
- M. G. Katz-Jaffe and S. McReynolds, in Fertility and Sterility, 2013, vol. 99, pp. 1073–1077 Search PubMed.
- A. N. Andersen, V. Goossens, A. P. Ferraretti, S. Bhattacharya, R. Felberbaum, J. de Mouzon and K. G. Nygren, Hum. Reprod., 2008, 23, 756–771 CrossRef PubMed.
- G. Paternot, A. M. Wetsels, F. Thonon, A. Vansteenbrugge, D. Willemen, J. Devroe, S. Debrock, T. M. D'Hooghe and C. Spiessens, Reprod. Biol. Endocrinol., 2011, 9, 1–5 CrossRef.
- P. L. Matson, Hum. Reprod., 1998, 13, 156–165 CrossRef PubMed.
- J. Harper, E. Jackson, K. Sermon, R. J. Aitken, S. Harbottle, E. Mocanu, T. Hardarson, R. Mathur, S. Viville, A. Vail and K. Lundin, Hum. Reprod., 2017, 32, 485–491 CrossRef PubMed.
- M. Lu, Y. Wen, Y. Liu, C. Ding, C. Zhou and Y. Xu, Fertil. Steril., 2020, 114, 801–808 CrossRef CAS PubMed.
- C. E. Gordon and C. Racowsky, Fertil. Steril., 2020, 1–2 Search PubMed.
- A. Penzias, K. Bendikson, S. Butts, C. Coutifaris, T. Falcone, G. Fossum, S. Gitlin, C. Gracia, K. Hansen, A. La Barbera, J. Mersereau, R. Odem, R. Paulson, S. Pfeifer, M. Pisarska, R. Rebar, R. Reindollar, M. Rosen, J. Sandlow, M. Vernon and E. Widra, Fertil. Steril., 2018, 109, 429–436 CrossRef PubMed.
- Z. Rosenwaks, A. H. Handyside, F. Fiorentino, N. Gleicher, R. J. Paulson, G. L. Schattman, R. T. Scott, M. C. Summers, N. R. Treff and K. Xu, Fertil. Steril., 2018, 110, 353–361 CrossRef PubMed.
- H. J. Leese, Reproduction, 2012, 143, 417–427 CAS.
- J. P. Renard, A. Philippon and Y. Menezo, Reproduction, 1980, 58, 161–164 CrossRef CAS PubMed.
- E. Seli, D. Sakkas, R. Scott, S. C. Kwok, S. M. Rosendahl and D. H. Burns, Fertil. Steril., 2007, 88, 1350–1357 CrossRef PubMed.
- D. Sakkas, Methods Mol. Biol., 2014, 1154, 533–540 CrossRef CAS PubMed.
- T. Bracewell-Milnes, S. Saso, H. Abdalla, D. Nikolau, J. Norman-Taylor, M. Johnson, E. Holmes and M. Y. Thum, Hum. Reprod. Update, 2017, 23, 723–736 CrossRef CAS PubMed.
- R. K. Iles, F. I. Sharara, R. Zmuidinaite, G. Abdo, S. Keshavarz and S. A. Butler, J. Assisted Reprod. Genet., 2019, 36, 1153–1160 CrossRef PubMed.
- A. Ahlström, M. Wikland, L. Rogberg, J. S. Barnett, M. Tucker and T. Hardarson, Reprod. BioMed. Online, 2011, 22, 477–484 CrossRef PubMed.
- T. Hardarson, A. Ahlstrm, L. Rogberg, L. Botros, T. Hillensj, G. Westlander, D. Sakkas and M. Wikland, Hum. Reprod., 2012, 27, 89–96 CrossRef CAS PubMed.
- I. Sfontouris, I. Zorzovilis, G. Lainas, T. Lainas, G. Petsas and D. Sakkas, J. Hum. Reprod. Sci., 2013, 6, 133 CrossRef PubMed.
- E. Seli, C. G. Vergouw, H. Morita, L. Botros, P. Roos, C. B. Lambalk, N. Yamashita, O. Kato and D. Sakkas, Fertil. Steril., 2010, 94, 535–542 CrossRef PubMed.
- C. G. Vergouw, D. C. Kieslinger, E. H. Kostelijk, L. L. Botros, R. Schats, P. G. Hompes, D. Sakkas and C. B. Lambalk, Hum. Reprod., 2012, 27, 2304–2311 CrossRef CAS PubMed.
- C. G. Vergouw, L. L. Botros, P. Roos, J. W. Lens, R. Schats, P. G. A. Hompes, D. H. Burns and C. B. Lambalk, Hum. Reprod., 2008, 23, 1499–1504 CrossRef CAS PubMed.
- D. Sakkas, L. Botros, M. Henson, K. Judge and P. Roos, in Practical Manual of In Vitro Fertilization, 2012, pp. 405–412 Search PubMed.
- D. R. Brison, K. Hollywood, R. Arnesen and R. Goodacre, Reprod. BioMed. Online, 2007, 15, 296–302 CrossRef PubMed.
- T. Grunert, R. Stephan, M. Ehling-Schulz and S. Johler, Food Control, 2016, 60, 361–364 CrossRef CAS.
- L. E. Rodriguez-Saona, M. M. Giusti and M. Shotts, in Advances in food authenticity testing, 2016, pp. 71–116 Search PubMed.
- G. Kosa, K. S. Vuoristo, S. J. Horn, B. Zimmermann, N. K. Afseth, A. Kohler and V. Shapaval, Appl. Microbiol. Biotechnol., 2018, 102, 4915–4925 CrossRef CAS PubMed.
- C. A. Sellick, R. Hansen, R. M. Jarvis, A. R. Maqsood, G. M. Stephens, A. J. Dickson and R. Goodacre, Biotechnol. Bioeng., 2010, 106, 432–442 CAS.
- C. Musmann, K. Joeris, S. Markert, D. Solle and T. Scheper, Eng. Life Sci., 2016, 16, 405–416 CrossRef CAS.
- T. Scholz, V. V. Lopes and C. R. C. Calado, Biotechnol. Bioeng., 2012, 109, 2279–2285 CrossRef CAS PubMed.
- M. Bağcıoğlu, M. Fricker, S. Johler and M. Ehling-Schulz, Front. Microbiol., 2019, 10, 1–10 CrossRef PubMed.
- S. Vogt, K. Löffler, A. G. Dinkelacker, B. Bader, I. B. Autenrieth, S. Peter and J. Liese, Front. Microbiol., 2019, 10, 1–11 CrossRef PubMed.
- J. Depciuch, I. Zawlik, M. Skrzypa, J. Pajak, N. Potocka, K. Łach, H. Bartosik-Psujek, A. Koziorowska, E. Kaznowska and J. Cebulski, J. Alzheimer’s Dis., 2019, 68, 281–293 CAS.
- A. Blat, E. Wiercigroch, M. Smeda, A. Wislocka, S. Chlopicki and K. Malek, J. Biophotonics, 2019, 12, 1–11 CrossRef PubMed.
- V. Untereiner, G. Dhruvananda Sockalingum, R. Garnotel, C. Gobinet, F. Ramaholimihaso, F. Ehrhard, M. D. Diebold and G. Thiéfin, J. Biophotonics, 2014, 7, 241–253 CrossRef CAS PubMed.
- M. Wenning and S. Scherer, Appl. Microbiol. Biotechnol., 2013, 97, 7111–7120 CrossRef CAS PubMed.
- J. Ollesch, M. Heinze, H. M. Heise, T. Behrens, T. Brüning and K. Gerwert, J. Biophotonics, 2014, 7, 210–221 CrossRef CAS PubMed.
- X. Zhang, G. Thiéfin, C. Gobinet, V. Untereiner, I. Taleb, B. Bernard-Chabert, A. Heurgué, C. Truntzer, P. Ducoroy, P. Hillon and G. D. Sockalingum, Transl. Res., 2013, 162, 279–286 CrossRef CAS PubMed.
- E. Peuchant, S. Richard-Harston, I. Bourdel-Marchasson, J.-F. Dartigues, L. Letenneur, P. Barberger-Gateau, S. Arnaud-Dabernat and J.-Y. Daniel, Transl. Res., 2008, 152, 103–112 CrossRef CAS PubMed.
- C. Lacombe, V. Untereiner, C. Gobinet, M. Zater, G. D. Sockalingum and R. Garnotel, Analyst, 2015, 140, 2280–2286 RSC.
- D. A. Scott, D. E. Renaud, S. Krishnasamy, P. Meriç, N. Buduneli, Ş. Çetinkalp and K.-Z. Liu, Diabetol. Metab. Syndr., 2010, 2, 48 CrossRef PubMed.
- A. Bosch, A. Miñán, C. Vescina, J. Degrossi, B. Gatti, P. Montanaro, M. Messina, M. Franco, C. Vay, J. Schmitt, D. Naumann and O. Yantorno, J. Clin. Microbiol., 2008, 46, 2535–2546 CrossRef CAS PubMed.
- P. Lasch, M. Beekes, T. Udelhoven, M. Eiden, H. Fabian, W. Petrich, D. Naumann, R. Koch-institut, S. Kg and K. C. Strasse, Anal. Chem., 2002, 56, 6673–6678 Search PubMed.
- H. Fabian, P. Lasch and D. Naumann, J. Biomed. Opt., 2005, 10, 031103 CrossRef PubMed.
- G. Kosa, V. Shapaval, A. Kohler and B. Zimmermann, Microb. Cell Fact., 2017, 16, 1–11 CrossRef PubMed.
- D. I. Ellis, W. B. Dunn, J. L. Griffin, J. W. Allwood and R. Goodacre, Pharmacogenomics, 2007, 8, 1243–1266 CrossRef CAS PubMed.
- M. Cavelaars, J. Rousseau, C. Parlayan, S. de Ridder, A. Verburg, R. Ross, G. Visser, A. Rotte, R. Azevedo, J.-W. Boiten, G. A. Meijer, J. A. M. Belien and H. Verheul, J. Clin. Bioinf., 2015, 5, S2 CrossRef.
- G. Palermo, H. Joris, P. Devroey and A. C. Van Steirteghem, Lancet, 1992, 340, 17–18 CrossRef CAS.
- A. C. Van Steirteghem, Z. Nagy, H. Joris, J. Liu, C. Staessen, J. Smitz, A. Wisanto and P. Devroey, Hum. Reprod., 1993, 8, 1061–1066 CrossRef CAS PubMed.
- A. Penzias, K. Bendikson, S. Butts, C. Coutifaris, T. Falcone, G. Fossum, S. Gitlin, C. Gracia, K. Hansen, A. La Barbera, J. Mersereau, R. Odem, R. Paulson, S. Pfeifer, M. Pisarska, R. Rebar, R. Reindollar, M. Rosen, J. Sandlow and M. Vernon, Fertil. Steril., 2017, 107, 882–896 CrossRef PubMed.
- A. Bosch, M. A. Golowczyc, A. G. Abraham, G. L. Garrote, G. L. De Antoni and O. Yantorno, Int. J. Food Microbiol., 2006, 111, 280–287 CrossRef CAS PubMed.
- M. J. Baker, S. R. Hussainb, L. Lovergnea, V. Untereinerb, C. Hughese, R. A. Lukaszewskif, G. Thiéfinb and G. D. Sockalingum, Chem. Soc. Rev., 2016, 45, 1803–1818 RSC.
- P. Lasch and D. Naumann, Infrared Spectroscopy in Microbiology, 2015 Search PubMed.
- P. Lasch, Chemom. Intell. Lab. Syst., 2012, 117, 100–114 CrossRef CAS.
- V. Tafintseva, V. Shapaval, M. Smirnova and A. Kohler, J. Biophotonics, 2020, 13, 1–15 CrossRef PubMed.
- A. Kohler, U. Böcker, V. Shapaval, A. Forsmark, M. Andersson, J. Warringer, H. Martens, S. W. Omholt and A. Blomberg, PLoS One, 2015, 10, 1–22 Search PubMed.
- C. A. Rebuffo-Scheer, J. Dietrich, M. Wenning and S. Scherer, Anal. Bioanal. Chem., 2008, 390, 1629–1635 CrossRef CAS PubMed.
- D. Helm, H. Labischinski, G. Schallehn, D. Naumann, G. Schallehn and D. Naumann, J. Gen. Microbiol., 1991, 137, 69–79 CAS.
- S. Wold and M. Sjöström, in Chemometrics: Theory and Application, 1977, pp. 243–282 Search PubMed.
- Y. V. Zontov, O. Y. Rodionova, S. V. Kucheryavskiy and A. L. Pomerantsev, Chemom. Intell. Lab. Syst., 2017, 167, 23–28 CrossRef CAS.
- E. Seli, C. Robert and M. A. Sirard, Mol. Hum. Reprod., 2010, 16, 513–530 CrossRef CAS PubMed.
- A. Sakudo, Clin. Chim. Acta, 2016, 455, 181–188 CrossRef CAS PubMed.
- B. Liang, Y. Gao, J. Xu, Y. Song, L. Xuan, T. Shi, N. Wang, Z. Hou, Y. L. Zhao, W. E. Huang and Z. J. Chen, Fertil. Steril., 2019, 111, 753–762 CrossRef PubMed.
- D. K. Gardner, M. Meseguer, C. Rubio and N. R. Treff, Hum. Reprod. Update, 2015, 21, 727–747 CrossRef CAS PubMed.
- D. K. Gardner, P. Wela and P. L. Wale, Fertil. Steril., 2013, 99, 1062–1072 CrossRef CAS PubMed.
- D. R. Brison, F. D. Houghton, D. Falconer, S. A. Roberts, J. Hawkhead, P. G. Humpherson, B. A. Lieberman and H. J. Leese, Hum. Reprod., 2004, 19, 2319–2324 CrossRef CAS PubMed.
- F. D. D. Houghton, J. A. Hawkhead, P. G. Humpherson, J. E. Hogg, A. H. Balen, A. J. Rutherford and H. J. Leese, Hum. Reprod., 2003, 18, 1756–1757 CrossRef CAS.
- T. Velde and P. L. Pearson, Hum. Reprod. Update, 2002, 8, 141–154 CrossRef PubMed.
- D. F. Albertini, R. Anderson, S. Bhattacharya, J. L. H. Evers, D. J. Mclernon, S. Repping, E. Somigliana, D. T. Baird, P. G. Crosignani, K. Diedrich, R. G. Farquharson, K. Lundin, J. S. Tapanainen and A. Van Steirteghem, Hum. Reprod., 2017, 32, 1556–1559 CrossRef PubMed.
- R. J. Van Kooij, C. W. N. Looman, J. D. F. Habbema, M. Dorland and E. R. Te Velde, Fertil. Steril., 1996, 66, 769–775 CrossRef CAS PubMed.
- C.-C. Chuang, C.-D. Chen, K.-H. Chao, S.-U. Chen, H.-N. Ho and Y.-S. Yang, Fertil. Steril., 2003, 79, 63–68 CrossRef PubMed.
- K. Sharif, M. Elgendy, H. Lashen and M. Afnan, BJOG: Int. J. Obstet. Gynaecol., 1998, 105, 107–112 CrossRef CAS PubMed.
- S. L. Padilla and J. E. Garcia, Fertil. Steril., 1989, 52, 270–273 CrossRef CAS PubMed.
- B. J. Van Voorhis, N. Engl. J. Med., 2007, 356, 379–386 CrossRef CAS PubMed.
- P. K. Dubey, A. Tripathi and A. Ali, in Male Infertility: Understanding, Causes and Treatment, ed. R. Singh and K. Singh, Springer Singapore, Singapore, 2017, pp. 481–497 Search PubMed.
- R. J. Hart, Physiol. Rev., 2016, 96, 873–909 CrossRef CAS PubMed.
- G. A. N. Gowda, S. Zhang, H. Gu, V. Asiago, N. Shanaiah and D. Raftery, Expert Rev. Mol. Diagn., 2008, 8, 617–633 CrossRef CAS PubMed.
- K. Elder and B. Dale, In vitro Fertilization, Cambridge University Press, 4th edn, 2019 Search PubMed.
- V. Tafintseva, E. Vigneau, V. Shapaval, V. Cariou, E. M. Qannari and A. Kohler, J. Biophotonics, 2018, 11, 1–10 CrossRef PubMed.
- V. Tafintseva, V. Shapaval, U. Blazhko and A. Kohler, Chemom. Intell. Lab. Syst., 2021, 215, 1–9 CrossRef.
- A. Kohler, J. Solheim, V. Tafintseva, B. Zimmermann and S. Shapaval, Model-Based Pre-Processing in Vibrational Spectroscopy, in Comprehensive Chemometrics, ed. Brown, Tauler, Walczak, Elsevier, 2020, pp. 83–100 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an01191j |
| This journal is © The Royal Society of Chemistry 2021 |