Functionalized nanomaterials targeting NLRP3 inflammasome driven immunomodulation: Friend or Foe
Kanika
and
Rehan
Khan
 *
*
Chemical Biology Unit, Institute of Nano Science and Technology, Knowledge City, Sector-81, 5 Sahibzada Ajit Singh Nagar, Punjab Pin – 140306, India. E-mail: rehankhan@inst.ac.in; Tel: +91 172-2210075
First published on 28th August 2023
Abstract
The advancement in drug delivery systems in recent times has significantly enhanced therapeutic effects by enabling site-specific targeting through nanocarriers. These nanocarriers serve as invaluable tools for pharmacotherapeutic advancements against various disorders that enhance the effectiveness of encapsulated drugs by reducing their toxicity and increasing the efficacy of less potent drugs, thereby improving the therapeutic index. Inflammasomes, protein complexes located in the activated immune cell cytoplasm, regulate the activation of caspases involved in inflammation. However, aberrant activation of inflammasomes can result in uncontrolled tissue responses, contributing to the development of various diseases. Therefore, achieving a precise balance between inflammasome inhibition and activation is crucial for effectively treating inflammatory disorders through targeted functionalized nanocarriers. Despite the wealth of available data on the relevance of functionalized nanocarriers in inflammatory disorders, the nanotechnological potential to modulate inflammasomes has not been adequately explored. In this comprehensive review, we highlight the latest research on the modulation of the inflammasome cascade, both upregulating and downregulating its function, using nanocarriers in the context of inflammatory disorders. The utilization of nanocarriers as a therapeutic strategy holds immense potential for researchers aiming to effectively target and modulate inflammasomes in the treatment of inflammatory disorders, thus improving disease severity outcomes.
1. Introduction
In the realm of pharmaceutical research, the application of nanotechnology has been substantially advanced in recent years, resulting in significant improvements. The use of nanomaterials for the prevention, diagnosis, treatment, examination and control of many diseases and disorders has made nanotechnology the most important Key Enabling Technology in the European Union. Drugs are delivered to the target cells by functionalised drug delivery nanocarriers that act specifically as nanomedicines through solubilization and active/passive diffusion by targeting any mechanistic pathway that would give a site specific therapeutic effect of the carrier that further would increase the efficacy of a drug that has less potency. Additionally, nanomaterial-based drug delivery carriers including exosomes, liposomes, metal-based nanoparticles, polymer-based nanoparticles, hydrogels, etc. are effective at treating inflammatory illnesses by taking advantage of various molecular signalling pathways.1,2Amphiphilic copolymer chains exhibit self-assembly in aqueous environments, yielding polymeric nanoparticles. Utilizing distinct hydrophilic and hydrophobic blocks with varying structural, length, and charge attributes, a diverse array of polymeric nanoparticles has been synthesized. Liposomes, small amphipathic vesicles composed of phospholipids, exhibit varying properties influenced by lipid composition, size, surface charge, and production methods.3 Exosomes, nanoscale extracellular vesicles, hold prominence in rapidly advancing research due to their role as messengers in diverse clinical disorders, pivotal for intercellular communication.4 Metal nanoparticles garner substantial interest owing to optical traits like surface plasmon resonance (SPR), enabling optical field modulation for potential biomedical applications. Their diminutive size facilitates passage through biological membranes often impervious to larger macromolecules.5 Amphiphilic monomers give rise to polymeric crosslinked networks, generating hydrogels with high water-holding capacity. Hydrogel formation involves supramolecular interactions like H-bonding, metal coordination, van der Waals forces, and electrostatic interactions, as well as polymeric bonding mechanisms.6
In accordance with various studies reviewed by Nel et al. (2009), it is evident that the interaction between nanoparticles (NPs) and cells, as well as tissues in vivo, is influenced by the specific proteins to which they can bind. These interactions play a crucial role in determining the fate and behavior of NPs within the biological environment. Understanding the protein–NP interactions is essential for predicting the potential bio-distribution, cellular uptake, and overall biocompatibility of nanoparticles, which are crucial factors for their safe and effective use in various biomedical applications.7
The inflammasome pathway represents a pivotal signaling cascade involving cytosolic protein complexes known as inflammasomes. These complexes, originally identified by Martinon and colleagues in 2002, play a crucial role in orchestrating the activation of potent inflammatory mediators.8 Being a critical part of the innate immune system, they get activated by stressors or cellular infections. Furthermore, they aid in alleviating the maturation, expression, and release of a wide range of cytokines that are proinflammatory such as pro-IL-18 and pro-IL-1β, which are mainly produced by the cleavage of caspase-1, resulting in a cascade of inflammatory responses.9,10 Aside from cytokine production, inflammasome activation can also cause pyroptosis, a type of inflammatory death of cells that helps prevent intracellular pathogen proliferation.11 NLRs, such as galectins, C-type lectins (CTLs), and Toll-like receptors (TLRs), play a crucial role in the innate immune response by detecting pathogenic microbes and other pathogens, forming inflammasomes. Inflammasomes consist of five PRRs, including NLRP4, NLRP3, NLRC1, AIM2, and pyrin. Additionally, NLRP6, NLRP7, NLRP12, and IFI16 from the NLR and PYHIN families can also form inflammasomes. However, the specific composition of these inflammasomes remains unknown12,13 according to recent data. NLRs possess a tripartite structure, consisting of a central NOD domain flanked by C-terminal leucine-rich repeats (LRRs) and N-terminal domains, either caspase recruitment domains (CARDs) or pyrin domains (PYDs).9,14 NLRs are classified and named based on their domain structure. They typically contain common domains, including the nucleotide-binding domain, the LRR domain (which aids in ligand binding), and a signaling domain. Notably, all NLRs, except for NLRP10, exhibit these common domains.15 The signaling region of NLRs facilitates the recruitment of caspase-1, either directly through a CARD domain or indirectly through a PYRIN domain, which can bind to the PYRIN-CARD adaptor ASC.
Among these inflammasomes, the NLRP3 inflammasome has been the subject of much research due to its potential role in various human disorders. The NLRP3 inflammasome's vital role lies in its ability to sense diverse danger signals from infections, the environment, and the host. However, numerous chronic diseases are closely associated with excessive and prolonged NLRP3 inflammasome activation. These encompass autoimmune conditions such as Alzheimer's disease, multiple sclerosis, rheumatoid arthritis, systemic lupus erythematosus, and autoinflammatory disorders like Cryopyrin-Associated Periodic Syndromes and Familial Cold Autoinflammatory Syndrome. A range of cardiovascular ailments driven by NLRP3 include gout, atherosclerosis, acute myocardial infarction, silicosis, and non-alcoholic fatty liver disease. Additionally, numerous skin disorders, including acne, atopic dermatitis, and vitiligo, manifest NLRP3-driven mechanisms. The underlying mechanism in these conditions involves upregulated expression of the NLRP3 gene, amplifying the activity of pro-inflammatory cytokines, thereby triggering inflammation.16
The NLRP3 inflammasome exhibits a dichotomous nature, acting as both a friend and a foe in the context of inflammation. The beneficial or pathogenic role of the NLRP3 inflammasome in inflammation remains a subject of debate. Our review disseminates the most recent findings on the nanotechnological approach towards the modulation of NLRP3 inflammasomes. Furthermore, it also includes an inflammatory cascade that affects the activation and assembly processes of NLRP3 signalling. By consolidating current knowledge, our review enhances the understanding of the intricate role of nanotechnological interventions in precisely modulating the NLRP3 inflammasome across diverse inflammatory contexts. This review will benefit researchers using nanocarriers as a therapeutic approach to target inflammasomes and treat inflammatory-based diseases.
2. NLRP3 inflammasome
2.1 Biological characteristics of the NLRP3 inflammasome
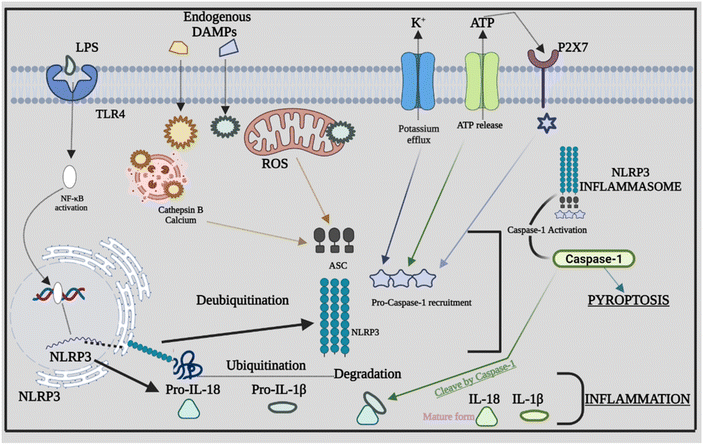
As discussed above there are many types of inflammasomes, but among them, the one that is the most characterized is the NLRP3 inflammasome, which aids in strategically targeting the inflammatory disease. The NLRP3 inflammasome mainly consists of a three-component protein assembly that includes procaspase-1, adaptor protein apoptosis-associated speck-like protein (ASC), and lastly, NLRP3 protein.17–19 The pyrin domain-containing protein 3 (NLRP3) of the NLR family is the receptor that helps in recognizing the pattern that is activated by numerous stimuli, including lipopolysaccharide (LPS) and ATP-rich, crystalline, high fat, and high sugar meals.20,21 The NACHT domain and LRRs have a relationship under normal conditions, which prevents the formation of inflammasomes by inhibiting the interaction between the ASC and NLRP3 protein.22 The structure of the NLRP3 protein is modified in response to PAMPs, DAMPs, and other exogenous invaders or environmental stress to enhance the interaction between PYDs in the ASC and NLRP3 protein. After the interaction of the NLRP3 protein with ASC, procaspase-1, another NLRP3 inflammasome component, binds to ASC through CARD that is part of procaspase-1 binding to the homologous area of ASC forming the NLRP3 inflammasome. The development of the protein complex causes the self-cleavage of procaspase-1, resulting in the active tetramer named caspase-1 p10/p20, which leads to the maturation of cytokines like IL-18 and IL-1β that are found to be pro-inflammatory in nature by converting their pro-forms into active forms, persuading inflammatory and immune responses.23–262.2 Signalling mechanism of the NLRP3 inflammasome
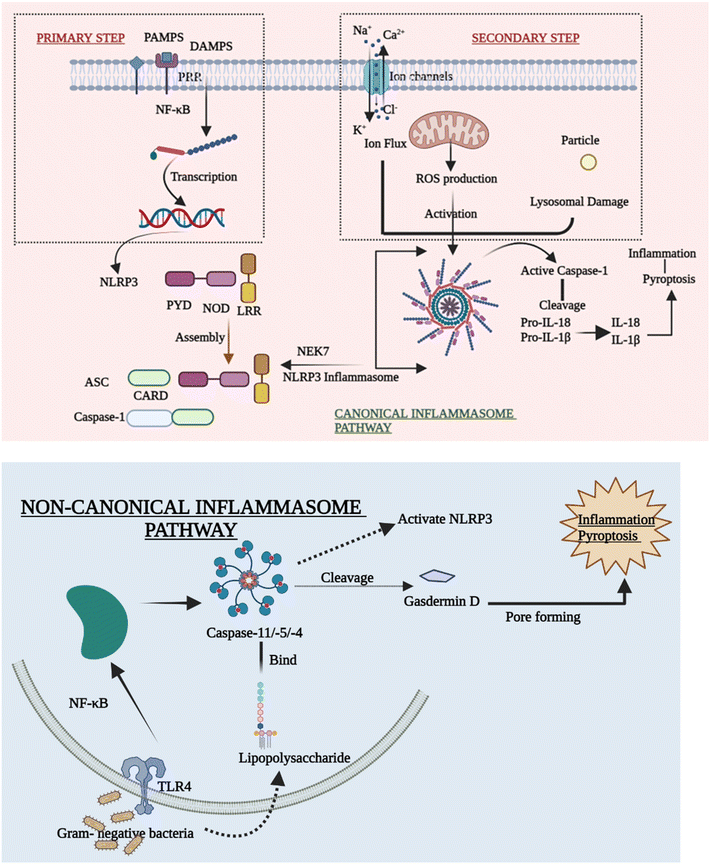
Despite mounting evidence of the NLRP3 inflammasome's critical involvement in the innate immune system, the methods by which NLRP3 reacts to numerous stimuli and events at both cellular and molecular levels to cause the NLRP3 inflammasome to assemble remain a mystery. The canonical and non-canonical inflammasome pathways are briefly outlined in order to explain how the activation of NLRP3 inflammasomes takes place.27(A) Priming step. Various priming stimuli activate the transcription factor NF-κB, which gives the initial signal for inflammasome activation, causing macrophages to upsurge NLRP3 and pro-IL-1β production. The existence of priming stimuli, on the other hand, is insufficient to activate the inflammasome. Although upregulating NLRP3 may increase inflammatory responses, current research has shown that the priming phase promotes NLRP3 inflammasome activation at the transcriptional level but is not required for activation of other inflammasomes.30,31
(B) Secondary step. Following the priming process, NLRP3 may be triggered by a diversity of chemicals and organisms, including bacteria. Due to differences in their structure and biology, NLRP3 cannot directly interact with these agonists. As a result, current research assumes that the NLRP3 inflammasome is activated by shared signaling events generated by NLRP3 agonists at the molecular and cellular levels, such as ROS generation, ionic flux, and lysosomal damage.32
(C) Ion flux. Ion mobilization in multiple intracellular signaling pathways has long been thought to occur when NLRP3 activators are stimulated. Even before inflammasomes were found, there was a strong link between IL-1β maturation and potassium (K+) efflux.27,33 Despite the difficulty in identifying the channel, the fluxion of K+ across the plasma membrane is widely accepted as a common cause of the activation of the inflammasome. Furthermore, excessive levels of extracellular K+ can prevent activation.34 Moreover, the inflow of Na+ and Ca2+,35 as well as the outflow of Cl−, is thought to be an upstream event in the activation of the NLRP3 inflammasome. They also take part in a number of signaling pathways that are implicated in inflammasome activation. Ion channels are future aims for anti-inflammatory treatment, despite the fact that the specific mechanism by which numerous ions coordinate in inflammasome activation is yet unknown.
(D) ROS production. Membrane damage, hypoxia, and other stressful situations have all been linked to the formation of reactive oxygen species (ROS), which has been considered another standard signal of NLRP3 inflammasome activation. Inflammatory disorders are related to disrupted mitochondrial ROS (mtROS) formation. The mitochondrial respiratory chain is inhibited, which in turn inhibits autophagy that causes the formation of ROS, which can activate the NLRP3 inflammasome. The fact that NLRP3 and ASC are both found in the mitochondria backs up this claim.36
(E) Lysosomal damage. Lysosomal damage, which is induced by the absorption of particles that are both endogenous and exogenous such as alum, calcium crystals,37 silica, and cholesterol crystals,38 is another primary source of inflammasome activation. After the particulate matter is endocytosed and lysosomal contents are spilled into the cytosol, lysosome disruption occurs as a crucial step in the secondary phase of NLRP3 inflammasome activation. Hornung39 showed this inflammatory process by stating that phagocytosis induced by crystal and lysosomal instability might stimulate the NLRP3 inflammasome.
3. Interplay between the NLRP3 inflammasome and factors affecting inflammation
Inflammasome components combine and oligomerize in response to infection and/or damage, causing procaspase-1 to auto-cleave into its active form. Proinflammatory cytokines are transformed into mature forms by activated caspase-1, which subsequently contribute to the inflammatory response.44 The sensor PRRs recognize the danger or stressor, which triggers the activation of NLRP3. PPRs may detect PAMPs such as bacterial secretion systems and parts of microbial cell walls and microbial nucleic acids.45,46 Furthermore, uric acid crystals and ATP that come under the category of DAMPs aid in triggering the PPRs. Being the two-stage process of its activation, the first step is the detection and production stage, which starts with TLRs recognizing DAMPs and PAMPs. TLRs identify numerous stress stimuli and trigger NF-κB signaling, which leads to increased creation of precursor proteins such as pro-IL-18, NLRP3 protein, and pro-IL-1β.47 The assembly and effector stage, which commences with the assembly of the NLRP3 inflammasome, is the second step. The mature complex is formed by procaspase-1, NLRP3 protein, and ASC, which changes the immature versions of IL-1β and IL-18 into their mature forms. The ensuing inflammatory response is mediated by IL-18 and IL-1β.48 Furthermore, other danger signals, including mitochondrial dysfunction (MtD). Nitric oxide (NO), calcium, and ROS can activate the NLRP3 inflammasome. ROS are produced in the cell by two sources: cytosolic ROS and mitochondria-derived ROS (mtROS). Following the identification of PAMPs from microorganisms or DAMPs, mtROS can operate as a second messenger to activate inflammasomes.49 The appropriate functionality of mitochondria is also required for the stimulation of the NLRP3 inflammasome. MtD can be influenced by a series of causes, including NO and Ca2+, which can trigger the NLRP3 inflammasome by releasing oxidized mitochondrial DNA (mtDNA) following TLR engagement.50 Several studies evaluated the different signals or stimuli that have been activating the NLRP3 inflammasomes. The NLRP3 inflammasome is involved in the development of atherosclerosis by driving the production of IL-1β. Through the NLRP3 inflammasome, hydroxyapatite crystals can activate IL-1β and increase its production in osteoarthritis, triggering inflammation and joint disorders. The NLRP3 inflammasome and ROS signaling may be activated by pulmonary macrophages which become active when endocytosis of silica and asbestos takes place and further cause silicosis and asbestosis.51 The specific signaling mechanism behind the cross-talk between ROS in NLRP3 remains unknown; rather, it has been recognized extensively. Furthermore, we explore the rapidly expanding cross-talks between the inflammasome and various factors that affect inflammation. Fig. 2 provides a comprehensive overview of NLRP3's involvement in inflammation, highlighting how the assembly of NLRP3 adapts in response to infections, giving rise to diverse PAMPs, DAMPs, and other activating factors.4. Cross-talk between the NLRP3 inflammasome, caspase-8 and caspase-11
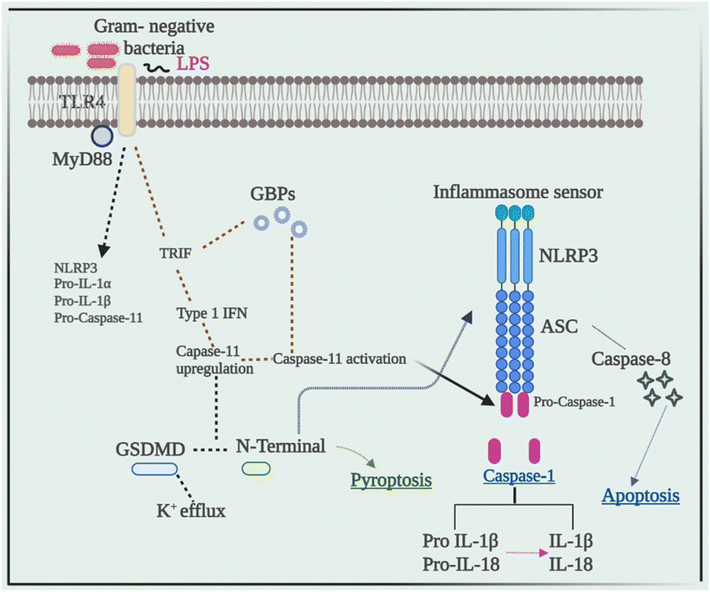
Current research has revealed that caspase 8 plays a crucial role in the activation of inflammasomes, cell death, and production of IL-1β in response to a variety of ligands. Another research showed that inhibiting caspase-8 decreased NLRP3 and AIM2-induced caspase-1 activation, indicating that caspase-8 plays a role in inflammasome activation.52 In response to AIM2 and NLRP3, recruitment of caspase-8 through ASC causes its activation, which leads to apoptosis. Although pyroptosis is found to be a process of rapid cell death, this work indicated that cells missing caspase-1 suffer apoptosis that will be via caspase-8 with slower kinetics.53Despite the fact that caspase-8 is a recognized predictor of cell survival or death via apoptosis, recent investigations have assigned it a critical role in inflammasome activation. Talking next about caspase 11, it was later discovered to be an independent inducer of pyroptosis, without the need for caspase 1 as well as the need for activation of NLRP3 through the CARD domain of Gram-negative bacteria.54 Caspase-11 was found to be serving as a sensor and a persuader of LPS-induced pyroptotic reactions, as well as aids in triggering the NLRP3 inflammasome's assembly. This differentiates between the bifunctional roles of activation of inflammasome pyroptosis that is triggered by caspase-11 activation, and on the other hand, processing of the cytokine that is triggered by NLRP3-mediated caspase-1 activation.55Fig. 3 elucidates the pivotal role of caspases in inflammation, illustrating their impact through the NLRP3 signaling pathway, ultimately culminating in the production of IL-1β, a potent proinflammatory cytokine, thus driving the inflammatory response.5. Cross-talk between the NLRP3 inflammasome and autophagy
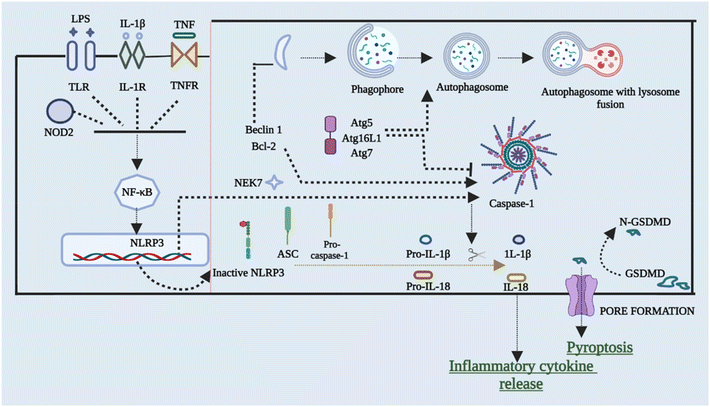
To restrict the infection and improve pathogen clearance, cells initiate autophagy in response to a pathogen. The relationship between autophagy, inflammasome activation, and cytokine processing was initially demonstrated by Saitoh et al.56 Autophagy also removes damaged organelles that are produced as a result of homeostatic circumstances, reducing the inflammasome response.57,58 Blocking autophagy enhances inflammasome activation by accumulating ROS-producing mitochondria. On the other hand, autophagy malfunction can result in disorders characterized by hyper inflammation and excessive activation of the NLRP3 inflammasome, making it a key regulator of inflammasomes. Enhanced ROS generation in mitochondria caused via suppression of complex I and III of mitochondria results in boosted activation of caspase 1 that is NLRP3-dependent and release of IL-1β in both macrophages and monocytes, according to Zhou et al.59 inflammasome activation is also elevated in cells that have missing components of autophagosome formation, such as beclin-1, ATG16L, and LC3B. Furthermore, IL-1β stimulates autophagy, implying a negative feedback loop for inflammasome activation.57,59,60 The NLRP3 inflammasome has been found to interrelate with the machinery of autophagy so that co-localization with autophagosomes takes place. This further leads to an increase in NLRP3 inflammasome activation when the autophagic route is inhibited. This shows that post-translational changes of NLRP3 (such as phosphorylation and ubiquitination) and subsequent autophagic clearance of inflammasome components after activation function as a negative-feedback loop to prevent an over-inflammatory response. Ubiquitination and phosphorylation changes of NLRP3 and clearance of autophagic components after it gets activated act as the negative-signal loop to avoid any response that would be over-inflammatory.61–64Fig. 4 briefly outlines the interplay between cellular autophagy initiation inhibition and the activation of NLRP3. This reciprocal relationship demonstrates the dual and contentious nature of the process.6. Cross-talk between NLRP3 inflammasomes and the gut microbiota
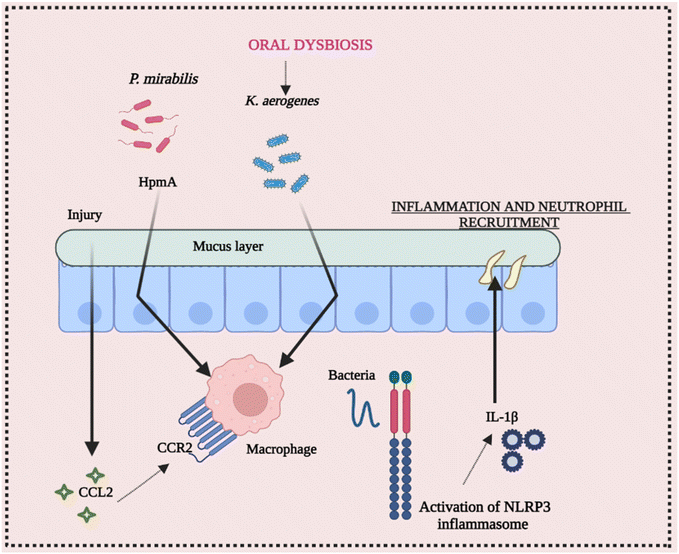
The ecology of the gut microbiota, which impacts the activation state of inflammasomes, may be affected by diet and aseptic procedures utilized by various facilities. Endogenous danger signals such as dietary cholesterol and deoxycholic acid activate the NLRP3 inflammasome, contributing to colorectal cancer (CRC) and high fat diet (HFD-related colonic inflammation. Inflammation is reduced when NLRP3 is activated by short-chain fatty acids or inhibited by omega-3 fatty acids.65–68 The NLRP3 inflammasome affects the gut microbiota, according to several studies. Animal models have been used to investigate the function of commensal microorganisms in NLRP3 activation in the etiology of inflammatory disorders. Skin commensal microorganisms activate the NLRP3 inflammasome in the CAPS mouse model Nlrp3R258W, which promotes cutaneous inflammation. Certain commensal pathobionts in the gut have been shown to be capable of activating the NLRP3 inflammasome in intestinal mononuclear phagocytes.69 The activation of NLRP3 by P. mirabilis is essential for the worsening of colitis produced by this bacterium's colonization. Kitamoto and his group found that ingested K. aerogenes enters the gastrointestinal system, and its ectopic gut colonization exacerbates intestinal inflammation in DSS-induced colitis and Il-10-deficient animals.70 As a result, growing evidence suggests that the NLRP3 inflammasome and downstream mediators like IL-18 and IL-1β play a vital role in the initiation and development of intestinal inflammation. Furthermore, pathobiont-refereed stimulation of the NLRP3 inflammasome may play a critical role in the onset or worsening of intestinal inflammation. These conclusions point to a dynamic and growing link between the daily routine diet, microbiota, and inflammasome activation, implying that dysbiosis affected by inflammasomes should be examined and interpreted with caution.71,72 Conclusively, Fig. 5 elucidates the intricate interplay between intestinal inflammation and NLRP3 signaling. It highlights the significant involvement of the gut microbiota in triggering inflammasome assembly, underscoring their role in this context.7. Cross-talk between NLRP3 inflammasome activation and bacterial infection
Gram-positive Listeria monocytogenes and Staphylococcus aureus, Gram-negative Shigella flexneri and Shigella sonnei, but not Gram-negative Francisella tularensis and Salmonella typhimurium, all stimulate NLRP3-dependent IL-1β production, according to previous research that was demonstrated to activate the inflammasome named NLRP3.73 The primary murine macrophages that have been LPS primed depict that S. sonnei promoted the production of IL-1β via P2X7 receptor-mediated efflux of potassium, ROS formation, acidification of lysosomes, and mitochondrial damage.74 In human monocytes and macrophages, live intracellular Mycobacterium tuberculosis infection resulted in NLRP3-dependent IL-1β production and pyroptosis.75,76 Infection of human macrophages with Salmonella typhimurium triggered the NLRP3 inflammasome in an indirect manner. Salmonella sequestration within the vesicles that contain Salmonella is caused by S. typhimurium infection, which finally gets burst by releasing its pathogens into the cytosol. In addition, the NLRP3 inhibitor only decreased the release of cytokine IL-1β and pyroptosis in macrophages when NLRC4 activity was inhibited simultaneously.778. NLRP3 inhibitors: promising therapeutic candidates
The oligomerization of NLRP3 and subsequent ASC alignment onto NLRP3 oligomers are essential steps in the formation of the NLRP3 inflammasome complex. Through the recruitment and oligomerization of ASC, the ATPase activity of NLRP3's NACHT domain drives the oligomerization of NLRP3, culminating in caspase 1 activation. In light of this, inhibitors specifically targeting NLRP3 within the inflammasome hold substantial promise as therapeutic targets.78(A) MCC950: MCC950, a potent compound, exhibits direct interaction with the Walker B motif within the NLRP3 NACHT domain. This interaction leads to the inhibition of NLRP3 inflammasome activation, concurrently impeding ATP hydrolysis, ASC oligomerization, and chloride ion export.79 The remarkable in vivo efficacy of MCC950 has been substantiated across diverse disease models. Notably, MCC950's inhibitory impact remains unaffected by NLRP3 inflammasome activation, NLRP3–NLRP3 interactions, calcium signaling, mitochondrial respiration, ROS generation, potassium efflux, or NEK7–NLRP3 interactions. The therapeutic utility of MCC950 in addressing inflammatory disorders and related complications has been firmly established.80
(B) OLT1177: OLT1177, an oral sulfonyl cyanide compound, exerts inhibitory effects on ASC oligomerization and reduces ATPase activity by directly binding to NLRP3. Moreover, OLT1177 disrupts the interaction between NLRP3 and ASC, effectively impeding the assembly of the NLRP3 inflammasome. Consequently, OLT1177 emerges as an orally administrable therapeutic agent, boasting notable benefits for inflammatory disorders attributed to its minimal toxicity and limited adverse effects.81,82
(C) INF39: INF39 stands out as a non-toxic, enduring, and selective inhibitor of the NLRP3 inflammasome, effectively impeding NLRP3 activation. INF39's anti-inflammatory effects predominantly stem from its capacity to hinder the NEK7–NLRP3 interaction. This initial inhibition subsequently halts the oligomerization process of NLRP3, further extending to the oligomerization of ASC, culminating in the prevention of speckle formation.83
(D) Tranilast: Tryptophan metabolite, tranilast, effectively hampers NLRP3 inflammasome activation. Notably, this inhibition occurs without compromising K+ efflux, ATPase role, or mitochondrial integrity. Tranilast's mechanism involves binding to the NACHT domain, consequently obstructing NLRP3 oligomerization and the ensuing assembly of the NLRP3 inflammasome. This interruption curbs caspase-1 activation and subsequent IL-1β synthesis.84
(E) CY-09: An alternate NLRP3 inhibitor engages in direct binding with the ATP binding motif within the NACHT domain of the NLRP3 assembly. Unlike its impact on the mitochondrial integrity, potassium efflux, or chloride ion efflux during NLRP3 activation, this inhibitor distinctly targets and suppresses NLRP3 ATPase activity.85
(F) JC124: JC124, a selective NLRP3 inflammasome inhibitor, centers its attention on ASC oligomerization within macrophages. This compound demonstrates not only activity but also constitutive expression of the active NLRP3 inflammasome. Across multiple investigations, JC124 exhibited its inhibitory prowess by strategically targeting oligomerization, thereby effectively combating inflammation.86
(G) 3,4-Methylenedioxy-β-nitrostyrene (MNS): MNS decreases the NLRP3 ATPase activity, resulting in the inhibition of NLRP3 inflammasome activation. Importantly, MNS achieves this inhibition without disrupting K+ efflux or impeding the activation of NLRC4 or AIM2 inflammasomes.87
Consequently, these inhibitors have been comprehensively documented for their direct interference with NLRP3 assembly, paving the way for the formulation of inflammation-targeted strategies utilizing them as potential anti-inflammatory drugs.
9. Nanotechnological approach towards the butterfly nature of the NLRP3 inflammasome
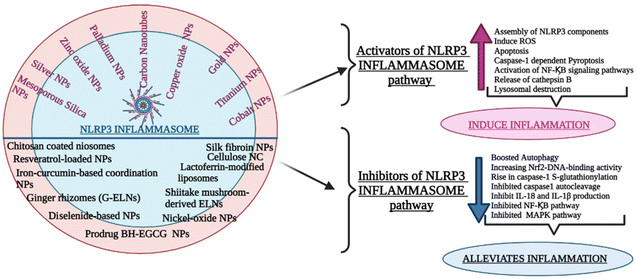
The NLRP3 inflammasome can impact various inflammatory disease pathogenesis and control the production of proinflammatory cytokines, including IL-1β and IL-18. Whether the NLRP3 inflammasome is beneficial or harmful to diseased patients remains a point of contention. Recently the growing body of studies examined the pathogenic and preventive functions of the NLRP3 inflammasome and the underlying mechanism of their regulation by different nanocarriers.9.1 Drug delivery nanocarriers targeting the upregulation of the signalling cascade of the NLRP3 inflammasome : pathogenic role
Nanoparticle-associated molecular patterns, mediated by synthetic materials like nanoparticles and nanomaterials, have lately been postulated as NLRP3 inflammasome danger signals. These nanoparticles are absorbed into immune cells, which activates them by causing tissue damage. In skin keratinocytes, monocytes, and hepatoma cells, many metal oxide, and metal-based nanoparticles have been shown to initiate the NLRP3 inflammasome. Particulate stimulants activate the NLRP3 inflammasome, which has been linked to the pathophysiology of a diversity of inflammatory disorders.(A) Palladium and silver nanoparticles. Metal nanoparticles are frequently exposed to oral epithelial keratinocytes. In addition, Eri et al. discovered that palladium and silver nanoparticles diminish the number of acidic organelles and cathepsin B expression in oral keratinocytes, indicating that NLRP3 inflammasome stimulation by metal-based nanoparticles is linked to lysosomal injury.88 In summary, ingestion of metal-based nanoparticles into human oral keratinocytes caused autophagic-lysosomal related dysfunctions and activated the specific inflammasome that is NLRP3. Metal nanoparticles might be integrated into oral keratinocytes and trigger the NLRP3 inflammasome, as well as immune cells, according to Eri's findings.
According to recent research, depending on the nanoparticle surface charge and size, modification of surface-produced nanoparticles activated the NLRP3 inflammasome and boosted the production of antibodies. Improved uptake of inflammasome activators, more ROS production, repressed degradation of inflammasomes due to the cooperative activity of lysosomes, and other dangerous signal releases that are presently anticipated mechanisms of nanoparticle-induced activation of NLRP3.89–92
Another relevant study on silver nanoparticles elucidated that AgNPs are thought to cause cytotoxicity by triggering apoptosis and non-apoptotic cell death, according to growing research.93 They discovered that particle sizes that include 10, 50, and 100 nm of AgNPs cause cytotoxicity in liver cells, which is facilitated via AgNP-induced lysosomal membrane permeabilization and inflammasome-dependent stimulation of caspase-1.94 This proinflammatory protease governs cell death. AgNPs activated caspase-1 in HepG2 cells, and inhibiting the autophagy-lysosomal pathway exacerbates NLRP3 inflammasome-dependent caspase-1 activation, according to their findings. AgNPs can promote autophagy and caspase-1 activation even at low, non-cytotoxic concentrations, according to their conclusions.95 The findings of Mishra and his group have shown that limiting lysosomal acidification or inhibiting siRNA-mediated autophagosome formation exacerbates activation of AgNP-induced caspase-1 and the death of the cell. The results show that autophagy may have a protective function in AgNP-induced activation of inflammasomes at sub-cytotoxic concentrations. Caspase-1 activation and cell death are exacerbated when AgNP-induced autophagy is blocked. AgNPs cause autophagy and permeabilization of the lysosomal membrane, resulting in caspase-1 activation via the NLRP3 inflammasome.96
(B) Gold nanoparticles. Gold (Au) nanoparticles activate innate immune signaling pathways in a size-dependent manner, according to this study. The NLRP3 inflammasome is preferentially activated by ultrasmall Au nanoparticles (10 nm) for maturation of caspase-1 and interleukin-1 generation, whereas the NF-κB signaling pathway is activated by more significant Au nanoparticles (>10 nm). Au nanoparticles (Au 4.5) that are ultrasmall (4.5 nm) trigger the NLRP3 inflammasome by directly penetrating into the cytoplasm of the cell, promoting strong ROS production and targeting protein-LC3 (microtubule-associated protein 1-light chain 3) in an endocytic independent manner for proteasomal degradation. Autophagy relies on LC3, which is needed for limiting the activation of NLRP3 and plays an important role in inflammasome negative regulation.97
(C) Silica dioxide and titanium dioxide nanoparticles. Previously reported literature showed that both in in vitro and in vivo cases, titanium dioxide (TiO2) nanoparticles cause the development of ROS along with inflammation. In the presence of titanium dioxide, the pyrin domain of the NLRP3 inflammasome, a nucleotide receptor, is activated, which was already known.92,98 Ruiz and his group revealed that the NLRP3 inflammasome was implicated in the worsening of acute colitis when TiO2 nanoparticles were given orally. Crystals were found to be accumulated in the spleens of mice that had been administered with TiO2 nanoparticles. TiO2 particles were taken up by macrophages and human intestinal epithelial cells (IECs) in vitro, resulting in the assembly of NLRP3-ASC-caspase-1, then the cleavage of caspase-1, and finally the release of proinflammatory interleukins 18 and 1β.99
Another group, Baron's, recently showed that nanoparticles of SiO2 (nano-SiO2 and TiO2 (nano-TiO2) were detected by NLRP3 and caused the secretion of mature IL-1β, similar to asbestos or silica in the environment. However, its mechanism of activation of inflammasome is still unclear. Furthermore, they found that inflammasome activation requires the active release of ATP, ADP, and adenosine receptor signaling. P2Y1, P2Y2, A2A, and A2B receptor expression in NLRP3 was significantly increased by nano-SiO2 or nano-TiO2. However, P2X7 receptor expression was significantly decreased.100,101 Surprisingly, increased ATP and ADP hydrolysis boost IL-1β production in response to nanoparticles, whereas adenosine degradation or specific A2A or A2B receptor blockade suppresses it. Nanoparticles stimulate the NLRP3 inflammasome via activating PLC-InsP3 and/or inhibiting adenylate cyclase (ADCY)-cAMP pathways downstream of these receptors, according to their findings.102 By blocking adenosine kinase activity, nucleotide receptors, and nucleoside transporters, their results may lead to novel therapeutic methods for controlling chronic inflammation. These findings offer fresh insights into the processes of NLRP3 inflammasome activation as well as novel anti-inflammatory treatment methods.103
Kim and his group reported another relevant study on the same metal nanoparticle named TiO2 NPs that have been demonstrated to cause oxidative stress and inflammation in HepG2 and Caco-2 cells in previous research. In addition to in the respiratory system, TiO2 NPs have also been demonstrated to induce ROS, apoptosis, and inflammation.104 Kim et al. stated that TiO2 NPs had been shown to trigger the inflammasome pathway, causing lung tissue injury. This pathway's activation damages tissue and leads to the development of liver and intestinal disorders. The primary component of this route is the NLR family pyrin domain containing 3 (NLRP3), which triggers caspase-1. When this caspase is activated, pro-IL-1β is activated, resulting in tissue damage and inflammation, and it also becomes a part of the pathogenesis of various disorders like chronic liver disease. The findings of this investigation revealed that the administration of TiO2 NPs causes inflammation, oxidative stress, and apoptosis in the liver and intestine in a dose-dependent way, which was substantially accompanied by histological alterations in the liver and intestine.105
(D) Silica nanoparticles. Some metal nanoparticles are found to be crucial for human applications. Silica nanoparticles (SiNPs) have various human applications as they can induce multiple immune responses.106The findings of Gomez et al. showed that both 200 nm and 12 nm SiNPs cause PBMCs to produce IL-1β and IL-6, consistent with earlier research. The main results show that SiNPs cause inflammasome components (NLRP3, ASC, Casp-1), as well as IL-1b, IL-6, and IL-18, to be expressed in PBMCs and neutrophils in a dose-dependent way.
In summary, the findings of Gomez and the group showed that SiNPs stimulate the creation of proinflammatory cytokines in a dose-dependent manner and that this stimulation occurs regardless of the NP size. Although results imply that the NLRP3 inflammasome is involved in the generation and release of IL-1β, both 200 nm and 12 nm silica nanoparticles are used at low doses and show mild inflammation action; hence, this might be highly valuable for biological applications.107
In addition to this study, in the year 2021, Yang Song and his group revealed that silica nanoparticles (SiO2 NPs) have been reported to enter the bloodstream via inhalation and cause inflammation of vascular endothelial cells. They discovered that amorphous SiO2 NPs cause signal axis modulation in human umbilical vein endothelial cells (HUVECs), which leads to proinflammatory responses. HUVECs exposed to SiO2 NPs produce increased expression of inflammatory cell factors.108
Furthermore, recently they found that amorphous SiO2 NPs produce excessive ROS that activates the NLRP3 inflammasome, speeds up the lysine acetylation of HMGB1, enhances its translocation, and release from the nucleus to the cytoplasm, and ultimately causes inflammatory injury in HUVECs via TLR4/MyD88/NF-κB signaling pathways.108
Later, increased medicinal uses of mesoporous silica nanoparticles (MSNs) have sparked interest in their toxicological consequences; nonetheless, MSN toxicity is still unclear, as are the underlying processes.109 MSNs exhibited dose- and time-dependent cytotoxicity in hepatic L02 cells. NLRP3 inflammasomes in hepatocytes were then activated by MSNs, causing caspase-1-dependent pyroptosis, a new model of cell death.110,111 Furthermore, tests revealed that MSNs increased the production of ROS in mitochondria and that a ROS scavenger may reduce MSN-induced NLRP3 inflammasomes and pyroptosis in the liver. The findings of Zhang and his group revealed that MSNs generated hepatocyte pyroptosis and inflammation of the liver by activating NLRP3 inflammasomes, which was driven by MSN-induced ROS production. Their group research also revealed new information on MSN hepatotoxicity and the processes that underpin it, as well as a viable strategy for improving MSN biosafety.112
Something had to be done to lower the toxicity of metal nanoparticles. Another group thought of surface modification of nanoparticles that became an emerging method for “masking” the harmful effects of nanoparticles, and it has been found to minimize non-specifically that further reducing its aggregation.113 Surface modified silica nanoparticle (SiO2 NPs) lowers their possibility for activation of inflammasome and its cytotoxicity, according to in vitro investigation.114,115
Furthermore, the previous group elucidated that surface-modified SiO2 NPs reduce the immunomodulatory and proinflammatory effects in the murine ovalbumin (OVA)-induced allergic airway inflammation model.116 Viviana et al. studies proved various studies regarding the impact of surface-modified Si nanoparticles, both in vitro and in vivo based studies. It has been found that surface-modified SiO2 NPs with the combination of both amino (–NH2) group and phosphonate (–P) significantly lessen the activation of inflammasome in murine bone marrow-derived dendritic cells (BMDCs), according to their findings rather than plain SiO2 NPs. Plain SiO2 NPs were not found to activate the inflammasome in non-sensitized mice (NS) but in sensitized (S) animals; when administered with PEGylated (-PEG), SiO2-NPs showed higher IL-1β and caspase-1 mRNA expression. Furthermore, phosphonate and amino modification, but not PEGylation, significantly lessen the expression of genes that are inflammatory in nature in the lungs of both S and NS mice when given SiO2 plain NPs intratracheally.117 The majority of the investigations have concentrated on discovering and reporting inflammasome-activating nanoparticles such as silver, SiO2, mesoporous silica, DOTAP, iron oxide, carbon black, and TiO2 particles.103,118–120
(E) Zinc oxide nanoparticles. Several in vitro and in vivo investigations have found that zinc-oxide nanoparticle (ZnO-NPs) exposure has a high risk of harm, particularly oxidative stress and lung inflammation. Liang and his group found that in A549 cells, generation of ROS, activation of NLRP3 inflammasome, and activation of NF-κB pathway were persuaded by ZnO-NPs. The ROS scavenger NAC might prevent ZnO-NP-induced NF-κB and NLRP3 inflammasome activation. BAY11-7082, the NF-κB inhibitor, had little effect on ROS generation but reduced NLRP3 inflammasome activation produced by ZnO-NPs. The NLRP3 inflammasome was primarily responsible and was helping in regulating the production of IL-18 and IL-1β by ZnO-NPs. Hence, this can be summarised that ROS generation was induced after exposure to ZnO-NPs which further led to activate NF-κB helping in regulating the activation of inflammasomes in A549 cells.121
(F) Copper oxide nanoparticles. Another metal nanoparticle copper oxide nanoparticles (CuONPs) treatment triggered interleukin 1-mediated inflammation in J774A.1 macrophages for the first time via the NOD-, LRR-, and pyrin domain of NLRP3 inflammasome.122,123 Tao et al. found that activation of NLRP3 inflammasome by CuONPs includes a two-fold process. CuONPs produced lysosomal destruction, as well as the release of cathepsin B, which facilitated the activation of NLRP3 inflammasomes directly. CuONPs may release copper ions after deposition in lysosomes, owing to the acidic environment. As a result, the free copper ions significantly increased cellular oxidative stress and facilitated NLRP3 inflammasome activation.124Furthermore, CuONPs exposure might stimulate J774A.1 macrophages to produce pro-IL-1β via the toll-like receptor 4 (TLR4) signal pathway, which is dependent on myeloid differentiation factor 88 (MyD88), consequently activating nuclear transcription factor kappa B (NF-κB) and hence affecting inflammatory pathway.125,126
(G) Cobalt nanoparticles. Many nanomaterials, including silver nanoparticles, amino-modified poly (phenylene glycol), and quantum dots nanoparticles, have been demonstrated in previous research to activate NLRP3 inflammation and cause the production of the inflammatory cytokine IL-1β.103,127
Feng and his group have used cobalt nanoparticles and revealed that nano-Co exposure to human fetal hepatocytes L02 resulted in dose- and time-dependent cytotoxicity. These nanoparticles might have caused activation of NLRP3 inflammasome in hepatocytes as they stated that the increase of NLRP3, caspase-1 p20, and IL-1β expression was seen when hepatocytes were exposed to 7.5 g mL−1 nano-Co for 24 hours. Nano-Co exposure at 7.5 or 10 g mL−1 resulted in the generation of IL-1β and IL-18.128
Furthermore, the Sisi group proposed that nano-Co-induced enhancement of caspase-1 p20, IL-1, and IL-18 was dramatically inhibited by NLRP3 siRNA treatment. Nano-Co-induced cytotoxicity was likewise considerably decreased by NLRP3 siRNA treatment. These findings suggested that nano-Co exposure might activate the NLRP3 inflammasome in hepatocytes, resulting in hepatocyte injury.129
(H) Nickel nanoparticle. Concerns concerning nickel oxide nanoparticles’ (NiONPs) are increasing as manufacturing and application technology improve.130,131 Cao and his group found that both in vivo and in vitro, NiONP exposure caused prolonged lung inflammation and NLRP3 inflammasome activation. Inflammatory cell infiltration, alveolar proteinosis, and cytokine production were all seen after NiONP exposure. The NiONPs significantly increased Nlrp3 expression, which went together with overexpression of the active form of caspase-1 (p20) and IL-1β secretion in the in vivo study.132
Furthermore, siRNA-mediated Nlrp3 knockdown in macrophages inhibited NiONP-induced release of cytokine and activity of caspase-1 proved with in vitro based study. Their group findings imply that the NLRP3 inflammasome participated in NiONPs-induced inflammation of the lung, and they provide novel ways to counteract NiONPs-induced pulmonary toxicity.132
Yang et al. proved that CNTs activate the NLRP3 inflammasome, and this property is demonstrated for the first time in carbon nano-onions (CNOs) that were spherical in the shape of 6 nm size. Purified CNTs and CNOs with benzoic acid functionalization, on the other hand, have dramatically reduced inflammatory characteristics. Following injection into mice, there was a significant reduction in the recruitment of immune cells, as well as a reduction in the release of the inflammatory cytokine IL-1β. Overall, their findings show that carbon NPs’ inflammatory qualities are strongly reliant on their physicochemical features and that chemical surface functionalization may significantly reduce these properties.135
In contrast, a thorough examination of the nanoparticle-associated molecular patterns (NAMPs) that cause it has received little attention.102,135 Both core and surface hydrophobicity of nanoparticles have been found to alter the permeability of the membrane and endo-lysosomal rupture (NLRP3 signal 2), which makes them promising research prospects.140–143 Dipika et al. evaluated four distinct series of nanoparticles, including core stiffness, surface hydrophobicity, surface charge, and core hydrophobicity, to represent various NAMPs and their effect on NLRP3 activation. They also discovered that core hydrophobicity is a new NAMP that regulates NLRP3 activation via many signaling pathways, including calcium flux-mitochondrial ROS production array and lysosomal rupture-cathepsin B. In comparison to surface charge, core stiffness, and surface hydrophobicity, core hydrophobicity dramatically stimulates and positively correlates with the assembly of NLRP3 in this study.144
These all discussed were the latest studies that have been explored in the previous years that aid in targeting the NLRP3 inflammasome.
9.2 Drug delivery nanocarriers targeting the downregulation of signalling cascade of NLRP3 inflammasome : Protective Role
(A) Zinc oxide nanoparticle. Other research contradicted the above-mentioned conclusions. The most generally found metal oxide nanoparticles are zinc oxide nanoparticles (ZnO NPs) that are broadly employed in a variety of applications due to their unique physical and chemical characteristics, which allow them to interact with cellular macromolecules and produce a variety of therapeutic effects.145 ZnO NPs also have a variety of medicinal uses, including anti-inflammatory, bioimaging anticancer, and antibacterial. ZnO NPs have been proposed as a potential anti-diabetic drug.146 A recent study found that giving ZnO NPs to streptozotocin (STZ) rats for six weeks relieves renal histological abnormalities, reduces renal hypertrophy and improves renal functions, as measured by creatinine clearance urine albumin, and creatinine.147
An existing study found that the STZ-induced DN model is well inhibited by ZnO NPs not only by monitoring advanced glycation end products and renal functions, but also by a variety of other beneficial effects such as by enhancing Nrf2-DNA-binding activity inhibiting TXNIP (thioredoxin-interacting protein) expression, which leads to suppression of oxidative stress and impeding inflammatory response by reducing activation of NLRP3.
As a consequence, ZnO NPs should be evaluated as a prospective healing agent for DN treatment.148
(B) Silver nanoparticles. Another study states that Jabir et al. revealed the green synthesis of silver nanoparticles (Ag-NPs) by using the extract of Annona muricata. Meanwhile, in both in vivo and in vitro models, the AgNPs significantly boosted autophagy and decreased IL-1β and NLRP3 levels. The IL-1β secretion was decreased, whereas the breakdown of NLRP3 inflammasome was increased. These proven data suggest that AgNPs have an anti-proliferative effect on THP-1 and AMJ-13 cell lines via inducing apoptosis through mitochondrial damage and activation of the p53 protein pathway. Furthermore, AgNP-induced autophagy decreased IL-1β and NLRP3 inflammasome activation.149 Overall, this research implies that AgNPs might be a promising cancer treatment alternative to anti-inflammatory drugs by promoting autophagy and affecting the NLRP3 inflammasome activation.
The novel finding states that an obligate Gram-negative anaerobic bacterium Porphyromonas gingivalis (P. gingivalis) was the most common bacteria in periodontal disease and has been linked to the activation of the NLRP3 inflammasome.153 The Naip subfamily proteins of the nod-like receptor family identify the secreting and flagellin system subparts of pathogens in inflammasomes.154 As a result, inhibiting the NLRP3 inflammasome offers a novel therapeutic technique for periodontitis treatment.155 Ionic gelation was employed to create complex nanoparticles (CS/CMCS-NPs), including carboxymethyl chitosan (CMCS) and chitosan (CS), that form a polyelectrolyte complex which was then used as a doxycycline carrier (Dox: CS/CMCS-NPs). Dox: CS/CMCS-NPs displayed an ordered morphology and high cytocompatibility, according to the findings. In comparison with the control group, Dox: CS/CMCS-NPs significantly suppressed P. gingivalis. In HGFs (human gingival fibroblasts), Dox: CS/CMCS-NPs efficiently reduced the gene and protein levels of the NLRP3 inflammasome and IL-1β. Their research offers a novel technique for using Dox in the therapeutic treatment of periodontal disease, as well as a new avenue for understanding the mechanical action of Dox: CS/CMCS-NPs and other drug-carrying based nanoparticles.156
Inflammatory bowel disease (IBD) is a gastrointestinal inflammatory illness that is persistent and idiopathic.157 In all of the previously reported studies, P. Diez-Echave et al. postulated that silk fibroin nanoparticles would efficiently distribute quercetin to injured intestinal cells, hence increasing quercetin intestinal wound healing potential. In addition, they examine quercetin's anti-inflammatory effects in the intestine when it's loaded in silk fibroin nanoparticles in the DSS mouse colitis model.158 The elevated expression of Nlrp3 was linked to colonic inflammation, which was considerably reduced when colitis mice were given quercetin-loaded silk fibroin nanoparticles (QSFN). Quercetin has been demonstrated to reduce NLRP3 inflammasome activation in several experimental conditions in previous in vitro and in vivo research.159,160 However, in the current work, quercetin treatment in colitis mice only demonstrated a significant trend to the lower colonic expression of Nlrp3 when loaded in nanoparticles, suggesting that quercetin needed to be incorporated into SFN to have an anti-inflammatory impact in the intestine.161
Seven enlisted different mushroom species were analyzed, and the author found that only shiitake mushroom-derived ELNs (S-ELNs) significantly reduced the activation of NLRP3 inflammasome by inhibiting the production of inflammasomes in primary macrophages. Protein levels of inflammatory gene IL-1β and interleukin (IL)-6 secretion were all inhibited by S-ELNs. Interestingly, mice were protected from acute liver injury by pre-treatment with S-ELNs caused by GalN/LPS. As a result, S-ELNs, which have been discovered as potent novel NLRP3 inflammasome inhibitors, constitute a promising class of medicines with the potential to treat FHF.169
Another interesting, relevant study elucidated that the exosome-like nanoparticles have been strongly identified from various food-borne exosome-like nanoparticles that prevent NLRP3 inflammasome activity.170–173 From them the uniquely studied ELN to which macrophages readily absorbed is the ELNs from ginger rhizomes (G-ELNs) because it was comprised of RNAs, lipid, and proteins. G-ELN therapy inhibited autocleavage of caspase1, production of IL-18 and 1β, and pyroptotic cell death, all of which are downstream of inflammasome activation. G-ELNs inhibited NLRP3 inflammasome assembly and speck production in apoptotic speck proteins containing a caspase recruitment domain (ASC). Furthermore, It has also been found that the inhibitory action identified was due to the lipids in G-ELNs.174 Hence, Chen and his group's findings revealed that G-ELNs are new potent inhibitors of NLRP3 inflammasome formation and activation.
Another plant-derived drug also worked as an inhibitor, as in the case of chronic kidney disease (CKD), which is marked by a progressive loss of renal function that is irreversible, further marked by a lower glomerular filtration rate. The key therapeutic objective for reducing renal injury and slowing the course of CKD is to inhibit renal inflammation.179–181 Inflammasome activation has been linked to injury and inflammation in a variety of kidney disorders, according to previous research studies.89,182,183 Resveratrol – loaded nanoparticles (Res NPs) coupled with an antibody named KIM-1 were employed in this study as a unique way to increase resveratrol pharmacokinetic properties, as well as its bioavailability and targetability. Resveratrol NPs may help patients with CKD by enhancing autophagy and inhibiting the NLRP3 inflammasome. Res NPs also triggered autophagy in kidney cells by boosting AMPK and hindering the Akt/mTOR signaling pathways. Kidney injury molecule-1 (KIM-1-Res NPs) improved CKD in an in vivo trial. These data also imply that Res NPs could be useful in treating this disease and as a possible pharmacological agent for disorders involving inflammasome.184
The newest and most interesting inhibitor of NLRP3 inflammasome is dopamine which also acts as curing therapy for liver disease.188 Inflammasome, a newly discovered and intriguing target for successful therapy of immunity-associated disorders such as liver disease.189 This study shows that a diselenide-based nanodrug can be used to treat ALF (acute liver failure) by suppressing the NLRP3 inflammasome and improving liver regeneration. In conclusion, Zhan and his group developed a diselenide-based nanosystem that has been synthesized by linking two dopa molecules by the covalent bond that results in stable nanoparticles when seen in aqueous media for the treatment of ALF, which is the first attempt to use a nano-prodrug generated by molecular diselenide in the treatment of liver disease. This method can not only prevent additional liver injury by suppressing the production of the NLRP3 inflammasome but also accelerate liver regeneration by downregulating 15-PGDH (prostaglandin-degrading enzyme 15), resulting in enhanced therapeutic effectiveness for ALF.190
In 2023, Ravi et al. and their team developed pH-responsive nanomicelles for ischemic stroke therapy. They incorporated MCC950, an NLRP3 inhibitor, into transferrin-conjugated nanomicelles to facilitate blood–brain barrier penetration. The study demonstrated that nanomicelle treatment notably reduces NLRP3 inflammasome biomarker levels, which were elevated in the right vitelline artery (RVA) of I/R-induced chick embryos, the MCAO rat model, and oxygen-glucose deprived (OGD) SH-SY5Y cells. A key finding is the significant mitigation of oxidative and inflammatory stress through pharmacological NLRP3 suppression by MCC950, observed in in vitro, in ovo, and in vivo contexts.195
(A) Top of form. Therefore, all the above-cited group of studies gives an overview of the interplay between NLRP3 inflammasome and various other nanoparticles affecting its mechanism of signaling and elucidating their potential as therapeutic targets. Fig. 6 provides a comprehensive overview of diverse drug delivery carriers employed for targeting the inflammatory pathway within different disease models via the NLRP3 signaling pathway.
10. Conclusion and future perspectives
Inflammasome biology has advanced rapidly in recent years, surpassing the fundamental notion established in the early 2000s. The molecular basis of ligand recognition has been discovered for numerous receptors. The finding and characterization of the non-canonical inflammasome pathway and pyrin inflammasome have generated new paradigms for how the innate immune system recognizes microbial invaders.196 In addition, numerous vital actors that influence downstream signaling have been discovered to affect the molecular processes that drive the oligomerization of receptors, complex building, and signal propagation within that complex. As a critical node for immunological detection within the innate immune system, NLRP3 activation or inhibition may be therapeutically valuable in some cases. Abnormal NLRP3 activation is at the root of the harmful inflammation that underpins many degenerative diseases linked to ageing and lifestyle. Small compounds that suppress NLRP3 activation have been shown to reduce inflammasome signaling in mouse models of different inflammatory illnesses in recent research (Table 1). The novel approaches that outlined the various nanoparticle targeting strategies emphasized in this review provide a road map for creating targeted therapies by altering inflammasome signaling. This might pave the way for inflammasome signaling targeting in specified inflammatory illnesses.| Delivery carrier | Signalling pathway | In vivo and in vitro studies | Activation/inhibition | Ref. |
|---|---|---|---|---|
| Silver and palladium NPs | Decrease cathepsin B expression and the quantity of acidic organelles via lysosomal injury | Human oral keratinocytes | Activation of NLRP3 | 88 |
| Gold NPs | Ultrasmall Au nanoparticles – robust ROS production and targeting autophagy protein-LC3 for proteasomal degradation in an endocytic/phagocytic-independent manner | Mouse bone marrow-derived dendritic cells | Activation of NLRP3 | 97 |
| Titanium dioxide NPs | Cause assembly of NLRP3-ASC-caspase-1, caspase-1 cleavage, and the release of NLRP3-associated interleukin (IL)-1β and IL-18 | In vitro: human intestinal epithelial cells and macrophages | Activation of NLRP3 | 99 |
| In vivo-acute colitis | ||||
| Nano-TiO2 and nano-SiO2NPs | P2Y1, P2Y2, A2A, or A2B receptor expression in NLRP3 increased via activating PLC-InsP3 and/or inhibiting adenylate cyclase (ADCY)-cAMP pathways | In vitro: murine bone-marrow-derived macrophages | Activation of NLRP3 | 103 |
| Titanium dioxide NPs | Induce ROS, apoptosis, caspase-1 activation, and inflammation | In vivo: liver and intestinal disorders | Activation of NLRP3 | 105 |
| Silica NPs | Stimulate the production of inflammasome components (NLRP3, ASC, Casp-1), as well as IL-1b, IL-6, and IL-18 | PBMC and neutrophils in a dose dependent manner | Activation of NLRP3 | 107 |
| Silica NPs | Produce excessive ROS that activate the NLRP3 inflammasome via TLR4/MyD88/NF-κB signaling pathways, and speed up the lysine acetylation of HMGB1 | Human umbilical vein endothelial cells (HUVECs) | Activation of NLRP3 | 108 |
| Mesoporous silica nanoparticles (MSNs) | Activation of NLRP receptor by excessive production of ROS and leading to caspase-1-dependent pyroptosis | Hepatic L02 cell and liver inflammation | Activation of NLRP3 | 112 |
| Zinc-oxide NPs | Generation of ROS, activation of the NLRP3 inflammasome via activation of the NF-κB pathway | A549 cells | Activation of NLRP3 | 121 |
| Copper oxide NPs | Liberated copper ions, increase oxidative stress, lysosomal destruction, release of cathepsin B, and activation of NLRP3 via MyD88 and NF-κB pathways | J774A.1 macrophages | Activation of NLRP3 | 124 |
| Cobalt NPs | Increase of NLRP3, caspase-1 p20, and IL-1β expression | Human fetal hepatocytes L02 | Activation of NLRP3 | 128 |
| Silver NPs | Cause autophagy and permeabilization of the lysosomal membrane, resulting in caspase-1 activation via the NLRP3 inflammasomes | HepG2 cells | Activation of NLRP3 | 107 |
| Nickel-oxide NPs | Increase NLRP3 expression, overexpression of the active form of caspase-1 (p20), and IL-1β secretion | In vivo: lung inflammatory model | Activation of NLRP3 | 132 |
| Chitosan NPs | Cathepsin B mediated activation of NLRP3 via the lysosomal rupture activation process | In vitro: murine BMDCs | Activation of NLRP3 | 139 |
| In vivo: peritoneal macrophages and in vitro | ||||
| Zinc oxide NPs | Increasing Nrf2-DNA-binding activity and downregulating TXNIP gene expression, which leads to oxidative stress suppression and inhibiting the inflammatory response by attenuating NLRP3 inflammasome activation | STZ-induced DN model | Inhibition of NLRP3 | 148 |
| Silver NPs | Boosted autophagy and decreased IL-1β and NLRP3 levels via promoting autophagy | THP-1 and AMJ-13 cells lines | Inhibition of NLRP3 | 149 |
| Cellulose nanocrystals-grafted poly-N-aminoethylmethacrylamide | The highest drop in NLRP3 and rise in caspase-1 S-glutathionylation, prolonged complex formation, and reduced pro-IL-1β processing and release of its active state | Mouse macrophage-like cells J774A.1 | Inhibition of NLRP3 | 152 |
| CS/CMCS-NPs | Reduced protein levels of the NLRP3 inflammasome and IL-1β | Primary human gingival fibroblast cell line | Inhibition of NLRP3 | 156 |
| Iron-curcumin-based coordination NPs | Activating the nuclear factor erythroid 2 related factor-2 (Nrf2) and inhibiting nod-like receptor protein-3 (NLRP3) inflammasome activation | In vivo: rat osteoarthritis model | Inhibition of NLRP3 | 166 |
| Shiitake mushroom-derived ELNs (S-ELNs) | Inhibit the production of inflammasomes, interleukin IL-6 secretion, and protein and mRNA levels of IL-1β | Fulminant hepatic failure primary macrophages | Inhibition of NLRP3 | 169 |
| Ginger rhizomes (G-ELNs) | Inhibited caspase1 autocleavage, interleukin (IL)-1 and IL-18 production, and pyroptotic cell death, all of which are downstream of inflammasome activation | Bone marrow-derived macrophages (BMDMs) from C57BL/6J mice | Inhibition of NLRP3 | 174 |
| Chitosan coated niosomes | Reduced IL-1β levels by downregulating protein expression of components of the inflammasome that include caspase-1, NLRP3, and adaptor protein components | In vivo: DSS-induced colitis model | Inhibition of NLRP3 | 159 |
| Resveratrol-loaded nanoparticles (Res NPs) coupled with KIM-1 antibody | Triggered autophagy in kidney cells by boosting AMPK and inhibiting the Akt/mTOR signaling pathway | In vivo: chronic kidney disease mouse model | Inhibition of NLRP3 | 184 |
| In vitro: HK-2 cell line | ||||
| Prodrug BH-EGCG NPs | Inhibiting the NF-κB pathway and suppressing NLRP3 inflammasome activation | CAR-induced hind paw edema and ConA-induced autoimmune hepatitis mouse models | Inhibition of NLRP3 | 157 |
| Silk fibroin NPs | Lower the colonic expression of Nlrp3 when loaded in nanoparticles | In vivo: DSS mouse colitis model | Inhibition of NLRP3 | 161 |
| Lactoferrin-modified liposomes | Inhibited the MAPK/NF-κB pathway and lowered the levels of inflammatory cytokines and ROS | In vitro: colonic macrophages | Inhibition of NLRP3 | 193 |
| Diselenide-based nanodrug | Suppressing the production of the NLRP3 inflammasome but accelerating liver regeneration by downregulating 15-PGDH | In vitro: NCTC-1469 cells | Inhibition of NLRP3 | 190 |
| In vivo: ALF mouse model |
Author contributions
Dr Rehan Khan conceived the idea and the outline of this manuscript and edited it. Kanika wrote the manuscript, drew the figures, organized the table and revised the manuscript.Abbreviations
| NLRP3 | NOD-, LRR- and pyrin domain-containing 3 |
| IL-1β | Interleukin-1β |
| TLRs | Toll-like receptors |
| CTLs | C-type lectins |
| NLRs | Nod like receptors |
| PRRs | Pattern recognition receptors |
| AIM2 | Absent in melanoma 2 |
| CARD | Caspase recruitment domain |
| PYDs | Pyrin domain |
| LRRs | leucine-rich repeats |
| ASC | Associated speck-like protein |
| LPS | Lipopolysaccharide |
| PAMPs | Pathogen-associated molecular patterns |
| DAMPs | Danger-associated molecular patterns |
| IL-18 | Interleukin IL-18 |
| ATP | Adenosine triphosphate |
| NEK7 | Serine/threonine-protein kinase |
| ROS | Reactive oxygen species |
| mtROS | Mitochondrial ROS |
| IBD | Inflammatory bowel disease |
| MtD | Mitochondrial dysfunction |
| NO | Nitric oxide |
| mtDNA | Mitochondrial DNA |
| ATG16L | Autophagy related protein 16 L |
| CRC | Colorectal cancer |
| HFD | High fat diet |
| CAPS | Cryopyrin-associated periodic fever syndrome |
| DSS | Dextran sulfate sodium |
| DCs | Dendritic cells |
| Au | Gold |
| LC3 | Light chain 3 |
| IECs | Intestinal epithelial cells |
| TiO2 | Titanium dioxide |
| Ag-NPs | Silver nanoparticles |
| MSNs | Mesoporous silica nanoparticles |
| CuONPs | Copper oxide nanoparticles |
| MyD88 | Myeloid differentiation factor 88 |
| NF-κB | Nuclear transcription factor kappa B |
| ADP | Adenosine diphosphate |
| SiO2 | Silica dioxide |
| ADC | Adenylate cyclase |
| ALF | Acute liver failure |
| 15-PGDH | Prostaglandin-degrading enzyme 15 |
| SiNPs | Silica nanoparticles |
| HUVECs | Human umbilical vein endothelial cells |
| HMGB1 | High-mobility group box 1 |
| P. gingivalis | Porphyromonas gingivalis |
| CS | Chitosan |
| CMCS | Carboxymethyl chitosan |
| Dox: CS | Doxycycline carrier |
| HGHs | Human gingival fibroblasts |
| CNOs | Carbon nano-onions |
| CNTs | Carbon nanotubes |
| ZnO NPs | Zinc oxide nanoparticles |
| STZ | Streptozotocin |
| AGEs | Advanced glycation end products |
| DN | Diabetic nephropathy |
| TXNIP | Thioredoxin-interacting protein |
| CNPs | Chitosan nanoparticles |
| IFA | Incomplete Freund's adjuvant |
| NiONPs | Nickel oxide nanoparticles |
| OVA | Ovalbumin |
| BMDCs | Bone marrow-derived dendritic cells |
| NS | Non-sensitized mice |
| RA | Rosmarinic acid |
| HO-1 | Heme oxygenase-1 |
| ELNs | Exosome-like nanoparticles |
| S-ELNs | Shiitake mushroom-derived ELNs |
| FHF | Fulminant hepatic failure |
| EGCG | Epigallocatechin-3-gallate |
| CNCs | Cellulose nanocrystals |
| ECM | Extracellular matrix |
| PGE2 | Prostaglandin E2 |
| MMPs | Matrix metalloproteinases |
| Fe-Cur NPs | Iron-curcumin-based coordination nanoparticles |
| Nrf2 | Nuclear factor-erythroid 2 related factor-2 |
| CKD | Chronic kidney disease |
| IBD | Inflammatory bowel disease |
| QSFN | Quercetin silk-fibroin nanoparticle |
| LF-Lipo | Lactoferrin-modified liposomes |
| NAMPs | Nanoparticle-associated molecular patterns |
| CNC-AEMA2 | Cellulose nanocrystal-grafted poly-N-aminoethylmethacrylamide |
| CH-Al NPs | Chitosan-aluminum nanoparticles |
Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This work was supported by the Department of Science and Technology (DST), SERB, with Grant No. CRG/2019/004018.References
- S. Soares, J. Sousa, A. Pais and C. Vitorino, Front. Chem..
- J. Jeevanandam, A. Barhoum, Y. S. Chan, A. Dufresne and M. K. Danquah, Beilstein J. Nanotechnol., 2018, 9, 1050–1074 CrossRef CAS PubMed.
- A. P. Singh, A. Biswas, A. Shukla and P. Maiti, Signal Transduction Targeted Ther., 2019, 4, 1–21 CrossRef CAS PubMed.
- R. Kar, R. Dhar, S. Mukherjee, S. Nag, S. Gorai, N. Mukerjee, D. Mukherjee, R. Vatsa, M. Chandrakanth Jadhav, A. Ghosh, A. Devi, A. Krishnan and N. D. Thorat, ACS Biomater. Sci. Eng., 2023, 9, 577–594 CrossRef CAS PubMed.
- Review on metal nanoparticles as nanocarriers: current challenges and perspectives in drug delivery systems | SpringerLink, https://link.springer.com/article/10.1007/s42247-021-00335-x, (accessed August 21, 2023).
- Umesh, S. Sarkar, S. Bera, P. Moitra and S. Bhattacharya, Mater. Today Chem., 2023, 30, 101554 CrossRef CAS.
- Understanding biophysicochemical interactions at the nano–bio interface | Nature Materials, https://www.nature.com/articles/nmat2442, (accessed July 20, 2023).
- M. Fabio, K. Burns and J. Tschopp, Mol. Cell, 2002, 10(2), 417–426 Search PubMed.
- K. Schroder and J. Tschopp, Cell, 2010, 140, 821–832 CrossRef CAS PubMed.
- H. Yaribeygi, N. Katsiki, A. E. Butler and A. Sahebkar, Drug Discovery Today, 2019, 24, 256–262 CrossRef PubMed.
- E. A. Miao, J. V. Rajan and A. Aderem, Immunol. Rev., 2011, 243, 206–214 CrossRef CAS PubMed.
- M. Lamkanfi and V. M. Dixit, Cell, 2014, 157, 1013–1022 CrossRef CAS PubMed.
- S. M. Man and T.-D. Kanneganti, Immunol. Rev., 2015, 265, 6–21 CrossRef CAS PubMed.
- A. Grebe, F. Hoss and E. Latz, Circ. Res., 2018, 122, 1722–1740 CrossRef CAS PubMed.
- J. P.-Y. Ting, R. C. Lovering, E. S. Alnemri, J. Bertin, J. M. Boss, B. K. Davis, R. A. Flavell, S. E. Girardin, A. Godzik, J. A. Harton, H. M. Hoffman, J.-P. Hugot, N. Inohara, A. Mackenzie, L. J. Maltais, G. Nunez, Y. Ogura, L. A. Otten, D. Philpott, J. C. Reed, W. Reith, S. Schreiber, V. Steimle and P. A. Ward, Immunity, 2008, 28, 285–287 CrossRef CAS PubMed.
- J. K. Seok, H. C. Kang, Y.-Y. Cho, H. S. Lee and J. Y. Lee, Arch. Pharmacal Res., 2021, 44, 16–35 CrossRef CAS PubMed.
- J. K. Kim, H. S. Jin, H.-W. Suh and E.-K. Jo, Immunol. Cell Biol., 2017, 95, 584–592 CrossRef CAS PubMed.
- Q. Liu, D. Zhang, D. Hu, X. Zhou and Y. Zhou, Mol. Immunol., 2018, 103, 115–124 CrossRef CAS PubMed.
- S. T. Sarvestani and J. L. McAuley, Antiviral Res., 2017, 148, 32–42 CrossRef CAS PubMed.
- X. Cai, H. Xu and Z. J. Chen, Cold Spring Harbor Perspect. Biol., 2017, 9, a023580 CrossRef PubMed.
- V. Singh, B. S. Yeoh, R. E. Walker, X. Xiao, P. Saha, R. M. Golonka, J. Cai, A. C. A. Bretin, X. Cheng, Q. Liu, M. D. Flythe, B. Chassaing, G. C. Shearer, A. D. Patterson, A. T. Gewirtz and M. Vijay-Kumar, Gut, 2019, 68, 1801–1812 CrossRef CAS PubMed.
- M. Inoue and M. L. Shinohara, Autoimmune Dis., 2013, 2013, 859145 Search PubMed.
- K. Shirasuna, T. Karasawa and M. Takahashi, J. Cell Physiol., 2019, 234, 5436–5450 CrossRef CAS PubMed.
- S. B. Willingham, I. C. Allen, D. T. Bergstralh, W. J. Brickey, M. T.-H. Huang, D. J. Taxman, J. A. Duncan and J. P.-Y. Ting, J. Immunol., 2009, 183, 2008–2015 CrossRef CAS PubMed.
- M. Groslambert and B. F. Py, J. Inflammation Res., 2018, 11, 359–374 CrossRef CAS PubMed.
- I. Jorgensen and E. A. Miao, Immunol. Rev., 2015, 265, 130–142 CrossRef CAS PubMed.
- Y. He, H. Hara and G. Núñez, Trends Biochem. Sci., 2016, 41, 1012–1021 CrossRef CAS PubMed.
- NEK7 interacts with NLRP3 to modulate the pyroptosis in inflammatory bowel disease via NF-κB signaling | Cell Death & Disease, https://www.nature.com/articles/s41419-019-2157-1, (accessed July 13, 2022).
- Y. He, M. Y. Zeng, D. Yang, B. Motro and G. Núñez, Nature, 2016, 530, 354–357 CrossRef CAS PubMed.
- J. Harder, L. Franchi, R. Muñoz-Planillo, J.-H. Park, T. Reimer and G. Núñez, J. Immunol., 2009, 183, 5823–5829 CrossRef CAS PubMed.
- F. G. Bauernfeind, G. Horvath, A. Stutz, E. S. Alnemri, K. MacDonald, D. Speert, T. Fernandes-Alnemri, J. Wu, B. G. Monks, K. A. Fitzgerald, V. Hornung and E. Latz, J. Immunol., 2009, 183, 787–791 CrossRef CAS PubMed.
- N. Kelley, D. Jeltema, Y. Duan and Y. He, Int. J. Mol. Sci., 2019, 20, E3328 CrossRef PubMed.
- D. Cheneval, P. Ramage, T. Kastelic, T. Szelestenyi, H. Niggli, R. Hemmig, M. Bachmann and A. MacKenzie, J. Biol. Chem., 1998, 273, 17846–17851 CrossRef CAS PubMed.
- R. Muñoz-Planillo, P. Kuffa, G. Martínez-Colón, B. L. Smith, T. M. Rajendiran and G. Núñez, Immunity, 2013, 38, 1142–1153 CrossRef PubMed.
- G.-S. Lee, N. Subramanian, A. I. Kim, I. Aksentijevich, R. Goldbach-Mansky, D. B. Sacks, R. N. Germain, D. L. Kastner and J. J. Chae, Nature, 2012, 492, 123–127 CrossRef CAS PubMed.
- S. J. Forrester, D. S. Kikuchi, M. S. Hernandes, Q. Xu and K. K. Griendling, Circ. Res., 2018, 122, 877–902 CrossRef CAS PubMed.
- Y. Yang, H. Wang, M. Kouadir, H. Song and F. Shi, Cell Death Dis., 2019, 10, 1–11 Search PubMed.
- A. R. Tall and M. Westerterp, J. Lipid Res., 2019, 60, 721–727 CrossRef CAS PubMed.
- V. Hornung, F. Bauernfeind, A. Halle, E. O. Samstad, H. Kono, K. L. Rock, K. A. Fitzgerald and E. Latz, Nat. Immunol., 2008, 9, 847–856 CrossRef CAS PubMed.
- N. Kayagaki, S. Warming, M. Lamkanfi, L. Vande Walle, S. Louie, J. Dong, K. Newton, Y. Qu, J. Liu, S. Heldens, J. Zhang, W. P. Lee, M. Roose-Girma and V. M. Dixit, Nature, 2011, 479, 117–121 CrossRef CAS PubMed.
- J. Yang, Y. Zhao and F. Shao, Curr. Opin. Immunol., 2015, 32, 78–83 CrossRef CAS PubMed.
- S. K. Vanaja, A. J. Russo, B. Behl, I. Banerjee, M. Yankova, S. D. Deshmukh and V. A. K. Rathinam, Cell, 2016, 165, 1106–1119 CrossRef CAS PubMed.
- G. Li, S. Wang and Z. Fan, Front. Med., 2022, 8, 750731 CrossRef PubMed.
- R. C. Rai, Life Sci., 2020, 240, 117084 CrossRef CAS PubMed.
- P. Broz and V. M. Dixit, Nat. Rev. Immunol., 2016, 16, 407–420 CrossRef CAS PubMed.
- L. Franchi, R. Muñoz-Planillo and G. Núñez, Nat. Immunol., 2012, 13, 325–332 CrossRef CAS PubMed.
- E. Tourkochristou, I. Aggeletopoulou, C. Konstantakis and C. Triantos, World J. Gastroenterol., 2019, 25, 4796–4804 CrossRef CAS PubMed.
- K. V. Swanson, M. Deng and J. P.-Y. Ting, Nat. Rev. Immunol., 2019, 19, 477–489 CrossRef CAS PubMed.
- Y. Yang, A. V. Bazhin, J. Werner and S. Karakhanova, Int. Rev. Immunol., 2013, 32, 249–270 CrossRef CAS PubMed.
- Z. Zhong, S. Liang, E. Sanchez-Lopez, F. He, S. Shalapour, X.-J. Lin, J. Wong, S. Ding, E. Seki, B. Schnabl, A. L. Hevener, H. B. Greenberg, T. Kisseleva and M. Karin, Nature, 2018, 560, 198–203 CrossRef CAS PubMed.
- V. Hornung, F. Bauernfeind, A. Halle, E. Samstad, H. Kono, K. Rock, K. Fitzgerald and E. Latz, Nat. Immunol., 2008, 9, 847–856 CrossRef CAS PubMed.
- Y.-H. Wu, W.-C. Kuo, Y.-J. Wu, K.-T. Yang, S.-T. Chen, S.-T. Jiang, C. Gordy, Y.-W. He and M.-Z. Lai, Cell Death Differ., 2014, 21, 451–461 CrossRef CAS PubMed.
- V. Sagulenko, S. J. Thygesen, D. P. Sester, A. Idris, J. A. Cridland, P. R. Vajjhala, T. L. Roberts, K. Schroder, J. E. Vince, J. M. Hill, J. Silke and K. J. Stacey, Cell Death Differ., 2013, 20, 1149–1160 CrossRef CAS PubMed.
- J. Shi, Y. Zhao, Y. Wang, W. Gao, J. Ding, P. Li, L. Hu and F. Shao, Nature, 2014, 514, 187–192 CrossRef CAS PubMed.
- D. Sharma and T.-D. Kanneganti, J. Cell Biol., 2016, 213, 617–629 CrossRef CAS PubMed.
- T. Saitoh, N. Fujita, M. H. Jang, S. Uematsu, B.-G. Yang, T. Satoh, H. Omori, T. Noda, N. Yamamoto, M. Komatsu, K. Tanaka, T. Kawai, T. Tsujimura, O. Takeuchi, T. Yoshimori and S. Akira, Nature, 2008, 456, 264–268 CrossRef CAS PubMed.
- M. S. Jabir, N. D. Ritchie, D. Li, H. K. Bayes, P. Tourlomousis, D. Puleston, A. Lupton, L. Hopkins, A. K. Simon, C. Bryant and T. J. Evans, Cell Host Microbe, 2014, 15, 214–227 CrossRef CAS.
- C.-S. Shi, K. Shenderov, N.-N. Huang, J. Kabat, M. Abu-Asab, K. A. Fitzgerald, A. Sher and J. H. Kehrl, Nat. Immunol., 2012, 13, 255–263 CrossRef CAS PubMed.
- R. Zhou, A. S. Yazdi, P. Menu and J. Tschopp, Nature, 2011, 469, 221–225 CrossRef CAS PubMed.
- C. Peral de Castro, S. A. Jones, C. Ní Cheallaigh, C. A. Hearnden, L. Williams, J. Winter, E. C. Lavelle, K. H. G. Mills and J. Harris, J. Immunol., 2012, 189, 4144–4153 CrossRef CAS.
- URB597 protects against NLRP3 inflammasome activation by inhibiting autophagy dysfunction in a rat model of chronic cerebral hypoperfusion | Journal of Neuroinflammation | Full Text, https://jneuroinflammation.biomedcentral.com/articles/10.1186/s12974-019-1668-0, (accessed July 14, 2022).
- S. Mehto, K. K. Jena, P. Nath, S. Chauhan, S. P. Kolapalli, S. K. Das, P. K. Sahoo, A. Jain, G. A. Taylor and S. Chauhan, Mol. Cell, 2019, 73, 429–445 CrossRef CAS.
- M. R. Spalinger, S. Lang, C. Gottier, X. Dai, D. J. Rawlings, A. C. Chan, G. Rogler and M. Scharl, Autophagy, 2017, 13, 1590–1601 CrossRef CAS.
- T. Kimura, A. Jain, S. W. Choi, M. A. Mandell, T. Johansen and V. Deretic, Autophagy, 2017, 13, 989–990 CrossRef CAS PubMed.
- Q. Du, Q. Wang, H. Fan, J. Wang, X. Liu, H. Wang, Y. Wang and R. Hu, Biochem. Pharmacol., 2016, 105, 42–54 CrossRef CAS PubMed.
- Dietary cholesterol promotes AOM-induced colorectal cancer through activating the NLRP3 inflammasome – PubMed, https://pubmed.ncbi.nlm.nih.gov/26921636/, (accessed July 15, 2022).
- L. Macia, J. Tan, A. T. Vieira, K. Leach, D. Stanley, S. Luong, M. Maruya, C. Ian McKenzie, A. Hijikata, C. Wong, L. Binge, A. N. Thorburn, N. Chevalier, C. Ang, E. Marino, R. Robert, S. Offermanns, M. M. Teixeira, R. J. Moore, R. A. Flavell, S. Fagarasan and C. R. Mackay, Nat. Commun., 2015, 6, 6734 CrossRef CAS PubMed.
- Y. Yan, W. Jiang, T. Spinetti, A. Tardivel, R. Castillo, C. Bourquin, G. Guarda, Z. Tian, J. Tschopp and R. Zhou, Immunity, 2013, 38, 1154–1163 CrossRef CAS PubMed.
- Y. Nakamura, L. Franchi, N. Kambe, G. Meng, W. Strober and G. Núñez, Immunity, 2012, 37, 85–95 CrossRef CAS PubMed.
- S. Kitamoto, H. Nagao-Kitamoto, Y. Jiao, M. G. Gillilland, A. Hayashi, J. Imai, K. Sugihara, M. Miyoshi, J. C. Brazil, P. Kuffa, B. D. Hill, S. M. Rizvi, F. Wen, S. Bishu, N. Inohara, K. A. Eaton, A. Nusrat, Y. L. Lei, W. V. Giannobile and N. Kamada, Cell, 2020, 182, 447–462 CrossRef CAS PubMed.
- C. Bauer, P. Duewell, C. Mayer, H. A. Lehr, K. A. Fitzgerald, M. Dauer, J. Tschopp, S. Endres, E. Latz and M. Schnurr, Gut, 2010, 59, 1192–1199 CrossRef CAS PubMed.
- S.-U. Seo, N. Kamada, R. Muñoz-Planillo, Y.-G. Kim, D. Kim, Y. Koizumi, M. Hasegawa, S. D. Himpsl, H. P. Browne, T. D. Lawley, H. L. T. Mobley, N. Inohara and G. Núñez, Immunity, 2015, 42, 744–755 CrossRef CAS PubMed.
- S. Rühl and P. Broz, Eur. J. Immunol., 2015, 45, 2927–2936 CrossRef PubMed.
- L.-H. Li, T.-L. Chen, H.-W. Chiu, C.-H. Hsu, C.-C. Wang, T.-T. Tai, T.-C. Ju, F.-H. Chen, O. V. Chernikov, W.-C. Tsai and K.-F. Hua, Front. Immunol., 2020, 11, 1115 CrossRef CAS.
- K. S. Beckwith, M. S. Beckwith, S. Ullmann, R. S. Sætra, H. Kim, A. Marstad, S. E. Åsberg, T. A. Strand, M. Haug, M. Niederweis, H. A. Stenmark and T. H. Flo, Nat. Commun., 2020, 11, 2270 CrossRef CAS.
- Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome – PubMed, https://pubmed.ncbi.nlm.nih.gov/20148899/, (accessed July 15, 2022).
- D. Bierschenk, M. Monteleone, F. Moghaddas, P. J. Baker, S. L. Masters, D. Boucher and K. Schroder, J. Leukocyte Biol., 2019, 105, 401–410 CrossRef CAS.
- B. K. Davis, H. Wen and J. P.-Y. Ting, Annu. Rev. Immunol., 2011, 29, 707–735 CrossRef CAS.
- D. Wu, Y. Chen, Y. Sun, Q. Gao, H. Li, Z. Yang, Y. Wang, X. Jiang and B. Yu, Inflammation, 2020, 43, 17–23 CrossRef CAS.
- A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases | Nature Medicine, https://www.nature.com/articles/nm.3806, (accessed August 21, 2023).
- An update on the regulatory mechanisms of NLRP3 inflammasome activation | Cellular & Molecular Immunology, https://www.nature.com/articles/s41423-021-00670-3, (accessed August 21, 2023).
- C. Marchetti, B. Swartzwelter, M. I. Koenders, T. Azam, I. W. Tengesdal, N. Powers, D. M. de Graaf, C. A. Dinarello and L. A. B. Joosten, Arthritis Res. Ther., 2018, 20, 169 CrossRef PubMed.
- Y. Shi, Q. Lv, M. Zheng, H. Sun and F. Shi, Int. Immunopharmacol., 2021, 92, 107358 CrossRef CAS PubMed.
- Y. Huang, H. Jiang, Y. Chen, X. Wang, Y. Yang, J. Tao, X. Deng, G. Liang, H. Zhang, W. Jiang and R. Zhou, EMBO Mol. Med., 2018, 10, e8689 CrossRef PubMed.
- Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders – PubMed, https://pubmed.ncbi.nlm.nih.gov/29021150/, (accessed August 21, 2023).
- C. Marchetti, S. Toldo, J. Chojnacki, E. Mezzaroma, K. Liu, F. N. Salloum, A. Nordio, S. Carbone, A. G. Mauro, A. Das, A. A. Zalavadia, M. S. Halquist, M. Federici, B. W. Van Tassell, S. Zhang and A. Abbate, J. Cardiovasc. Pharmacol., 2015, 66, 1–8 CrossRef CAS PubMed.
- Y. He, S. Varadarajan, R. Muñoz-Planillo, A. Burberry, Y. Nakamura and G. Núñez, J. Biol. Chem., 2014, 289, 1142–1150 CrossRef CAS PubMed.
- E. Sasabe, A. Tomomura, N. Kitamura and T. Yamamoto, Toxicol. in Vitro, 2020, 62, 104663 CrossRef PubMed.
- H. Guo, J. B. Callaway and J. P.-Y. Ting, Nat. Med., 2015, 21, 677–687 CrossRef CAS PubMed.
- N. R. G. Assis, A. J. Caires, B. C. Figueiredo, S. B. Morais, F. S. Mambelli, F. V. Marinho, L. O. Ladeira and S. C. Oliveira, J. Controlled Release, 2018, 275, 40–52 CrossRef CAS PubMed.
- Phagolysosome acidification is required for silica and engineered nanoparticle-induced lysosome membrane permeabilization and resultant NLRP3 inflammasome activity – PubMed, https://pubmed.ncbi.nlm.nih.gov/28126413/, (accessed July 15, 2022).
- A. S. Yazdi, G. Guarda, N. Riteau, S. K. Drexler, A. Tardivel, I. Couillin and J. Tschopp, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 19449–19454 CrossRef CAS PubMed.
- R. Roy, S. K. Singh, L. K. S. Chauhan, M. Das, A. Tripathi and P. D. Dwivedi, Toxicol. Lett., 2014, 227, 29–40 CrossRef CAS PubMed.
- C. A. Austin, T. H. Umbreit, K. M. Brown, D. S. Barber, B. J. Dair, S. Francke-Carroll, A. Feswick, M. A. Saint-Louis, H. Hikawa, K. N. Siebein and P. L. Goering, Nanotoxicology, 2012, 6, 912–922 CrossRef CAS PubMed.
- S. Knasmüller, W. Parzefall, R. Sanyal, S. Ecker, C. Schwab, M. Uhl, V. Mersch-Sundermann, G. Williamson, G. Hietsch, T. Langer, F. Darroudi and A. T. Natarajan, Mutat. Res., 1998, 402, 185–202 CrossRef PubMed.
- Silver nanoparticle-induced autophagic-lysosomal disruption and NLRP3-inflammasome activation in HepG2 cells is size-dependent – PubMed, https://pubmed.ncbi.nlm.nih.gov/26801583/, (accessed July 16, 2022).
- Cell-penetrating nanoparticles activate the inflammasome to enhance antibody production by targeting microtubule-associated protein 1-light chain 3 for degradation | ACS Nano, https://pubs.acs.org/doi/10.1021/acsnano.0c00962, (accessed July 15, 2022).
- Titanium dioxide nanoparticles induce DNA damage and genetic instability in vivo in mice – PubMed, https://pubmed.ncbi.nlm.nih.gov/19887611/, (accessed July 15, 2022).
- P. A. Ruiz, B. Morón, H. M. Becker, S. Lang, K. Atrott, M. R. Spalinger, M. Scharl, K. A. Wojtal, A. Fischbeck-Terhalle, I. Frey-Wagner, M. Hausmann, T. Kraemer and G. Rogler, Gut, 2017, 66, 1216–1224 CrossRef CAS PubMed.
- L. Antonioli, P. Pacher, E. S. Vizi and G. Haskó, Trends Mol. Med., 2013, 19, 355–367 CrossRef CAS PubMed.
- H. K. Eltzschig, M. V. Sitkovsky and S. C. Robson, N. Engl. J. Med., 2012, 367, 2322–2333 CrossRef CAS PubMed.
- C.-X. Sun, H. W. Young, J. G. Molina, J. B. Volmer, J. Schnermann and M. R. Blackburn, J. Clin. Invest., 2005, 115, 35–43 CrossRef CAS PubMed.
- L. Baron, A. Gombault, M. Fanny, B. Villeret, F. Savigny, N. Guillou, C. Panek, M. Le Bert, V. Lagente, F. Rassendren, N. Riteau and I. Couillin, Cell Death Dis., 2015, 6, e1629–e1629 CrossRef CAS PubMed.
- S. M. Duan, Y. L. Zhang and Y. Wang, Beijing Daxue Xuebao, Yixueban, 2018, 50, 395–400 CAS.
- E. Abbasi-Oshaghi, F. Mirzaei and M. Pourjafar, Int. J. Nanomed., 2019, 14, 1919–1936 CrossRef CAS PubMed.
- G. Bhakta, R. K. Sharma, N. Gupta, S. Cool, V. Nurcombe and A. Maitra, Nanomedicine, 2011, 7, 472–479 CrossRef CAS PubMed.
- D. M. Gómez, S. Urcuqui-Inchima and J. C. Hernandez, Innate Immun., 2017, 23, 697–708 CrossRef PubMed.
- X. Liu, B. Lu, J. Fu, X. Zhu, E. Song and Y. Song, J. Hazard. Mater., 2021, 404, 124050 CrossRef CAS PubMed.
- J.-H. Park, L. Gu, G. von Maltzahn, E. Ruoslahti, S. N. Bhatia and M. J. Sailor, Nat. Mater., 2009, 8, 331–336 CrossRef CAS PubMed.
- W. Chen, X. Zhang, J. Fan, W. Zai, J. Luan, Y. Li, S. Wang, Q. Chen, Y. Wang, Y. Liang and D. Ju, Theranostics, 2017, 7, 4135–4148 CrossRef CAS PubMed.
- R. Zhou, A. S. Yazdi, P. Menu and J. Tschopp, Nature, 2011, 469, 221–225 CrossRef CAS PubMed.
- X. Zhang, J. Luan, W. Chen, J. Fan, Y. Nan, Y. Wang, Y. Liang, G. Meng and D. Ju, Nanoscale, 2018, 10, 9141–9152 RSC.
- Surface modification of silica nanoparticles to reduce aggregation and nonspecific binding | Langmuir, https://pubs.acs.org/doi/10.1021/la052797j, (accessed July 16, 2022).
- A. Lankoff, M. Arabski, A. Wegierek-Ciuk, M. Kruszewski, H. Lisowska, A. Banasik-Nowak, K. Rozga-Wijas, M. Wojewodzka and S. Slomkowski, Nanotoxicology, 2013, 7, 235–250 CrossRef CAS PubMed.
- T. Morishige, Y. Yoshioka, H. Inakura, A. Tanabe, X. Yao, S. Narimatsu, Y. Monobe, T. Imazawa, S. Tsunoda, Y. Tsutsumi, Y. Mukai, N. Okada and S. Nakagawa, Biomaterials, 2010, 31, 6833–6842 CrossRef CAS PubMed.
- Surface modifications of silica nanoparticles are crucial for their inert versus proinflammatory and immunomodulatory properties – PubMed, https://pubmed.ncbi.nlm.nih.gov/24940059/, (accessed July 16, 2022).
- V. Marzaioli, C. J. Groß, I. Weichenmeier, C. B. Schmidt-Weber, J. Gutermuth, O. Groß and F. Alessandrini, Nanomaterials, 2017, 7, 355 CrossRef PubMed.
- K. Shirasuna, T. Karasawa and M. Takahashi, J. Cell Physiol., 2019, 234, 5436–5450 CrossRef CAS PubMed.
- J. Palomäki, E. Välimäki, J. Sund, M. Vippola, P. A. Clausen, K. A. Jensen, K. Savolainen, S. Matikainen and H. Alenius, ACS Nano, 2011, 5, 6861–6870 CrossRef PubMed.
- A. Harizaj, F. V. Hauwermeiren, S. Stremersch, R. D. Rycke, H. D. Keersmaecker, T. Brans, J. C. Fraire, K. Grauwen, S. C. D. Smedt, I. Lentacker, M. Lamkanfi and K. Braeckmans, Nanoscale, 2021, 13, 6592–6604 RSC.
- X. Liang, D. Zhang, W. Liu, Y. Yan, F. Zhou, W. Wu and Z. Yan, Toxicol. Ind. Health, 2017, 10, 737–745 CrossRef PubMed.
- C. Bartolucci, A. Antonacci, F. Arduini, D. Moscone, L. Fraceto, E. Campos, R. Attaallah, A. Amine, C. Zanardi, L. M. Cubillana-Aguilera, J. M. Palacios Santander and V. Scognamiglio, TrAC, Trends Anal. Chem., 2020, 125, 115840 CrossRef CAS.
- J.-P. Piret, D. Jacques, J.-N. Audinot, J. Mejia, E. Boilan, F. Noël, M. Fransolet, C. Demazy, S. Lucas, C. Saout and O. Toussaint, Nanoscale, 2012, 4, 7168–7184 RSC.
- X. Tao, X. Wan, D. Wu, E. Song and Y. Song, J. Hazard. Mater., 2021, 411, 125134 CrossRef CAS PubMed.
- K. Schroder and J. Tschopp, Cell, 2010, 140, 821–832 CrossRef CAS PubMed.
- T. Kawai and S. Akira, Nat. Immunol., 2010, 11, 373–384 CrossRef CAS PubMed.
- B. Sun, X. Wang, Z. Ji, M. Wang, Y.-P. Liao, C. H. Chang, R. Li, H. Zhang, A. E. Nel and T. Xia, Small, 2015, 11, 2087–2097 CrossRef CAS PubMed.
- S. Feng, Z. Zhang, Y. Mo, R. Tong, Z. Zhong, Z. Chen, D. He, R. Wan, M. Gao, Y. Mo, Q. Zhang and Y. Huang, Toxicol. in Vitro, 2020, 69, 104967 CrossRef CAS PubMed.
- L. Feng, Y. Zhang, M. Jiang, Y. Mo, R. Wan, Z. Jia, D. J. Tollerud, X. Zhang and Q. Zhang, Environ. Toxicol., 2015, 30, 490–499 CrossRef CAS PubMed.
- M. A. Siddiqui, M. Ahamed, J. Ahmad, M. A. Majeed Khan, J. Musarrat, A. A. Al-Khedhairy and S. A. Alrokayan, Food Chem. Toxicol., 2012, 50, 641–647 CrossRef CAS PubMed.
- Y. Mu, D. Jia, Y. He, Y. Miao and H.-L. Wu, Biosens. Bioelectron., 2011, 26, 2948–2952 CrossRef CAS PubMed.
- Z. Cao, Y. Fang, Y. Lu, F. Qian, Q. Ma, M. He, H. Pi, Z. Yu and Z. Zhou, Int. J. Nanomed., 2016, 11, 3331–3346 CrossRef CAS PubMed.
- Carbon nano-onions for imaging the life cycle of Drosophila melanogaster – PubMed, https://pubmed.ncbi.nlm.nih.gov/22012886/, (accessed July 16, 2022).
- Curling and closure of graphitic networks under electron-beam irradiation | Nature, https://www.nature.com/articles/359707a0, (accessed July 16, 2022).
- M. Yang, K. Flavin, I. Kopf, G. Radics, C. H. A. Hearnden, G. J. McManus, B. Moran, A. Villalta-Cerdas, L. A. Echegoyen, S. Giordani and E. C. Lavelle, Small, 2013, 9, 4194–4206 CrossRef CAS PubMed.
- S. B. Rizwan, B. J. Boyd, T. Rades and S. Hook, Expert Opin. Drug Delivery, 2010, 7, 1133–1144 CrossRef CAS PubMed.
- S. Gordon, E. Teichmann, K. Young, K. Finnie, T. Rades and S. Hook, Eur. J. Pharm. Sci., 2010, 41, 360–368 CrossRef CAS PubMed.
- B. Y. Chua, M. Al Kobaisi, W. Zeng, D. Mainwaring and D. C. Jackson, Mol. Pharm., 2012, 9, 81–90 CrossRef CAS PubMed.
- S. Neumann, K. Burkert, R. Kemp, T. Rades, P. Rod Dunbar and S. Hook, Immunol. Cell Biol., 2014, 92, 535–542 CrossRef CAS PubMed.
- J. J. Baljon, A. Dandy, L. Wang-Bishop, M. Wehbe, M. E. Jacobson and J. T. Wilson, Biomater. Sci., 2019, 7, 1888–1897 RSC.
- F. Shima, T. Akagi and M. Akashi, Biomater. Sci., 2014, 2, 1419–1425 RSC.
- Core role of hydrophobic core of polymeric nanomicelle in endosomal escape of siRNA | Nano Letters, https://pubs.acs.org/doi/full/10.1021/acs.nanolett.0c04468, (accessed July 16, 2022).
- Penetration of nanoparticles across a lipid bilayer: effects of particle stiffness and surface hydrophobicity – Nanoscale (RSC Publishing), https://pubs.rsc.org/en/content/articlelanding/2019/nr/c8nr09381d, (accessed July 16, 2022).
- D. Nandi, M. Shivrayan, J. Gao, J. Krishna, R. Das, B. Liu, S. Thayumanavan and A. Kulkarni, ACS Appl. Mater. Interfaces, 2021, 13, 45300–45314 CrossRef CAS PubMed.
- T. G. Smijs and S. Pavel, Nanotechnol., Sci. Appl., 2011, 4, 95–112 CrossRef CAS PubMed.
- The advancing of zinc oxide nanoparticles for biomedical applications, https://www.hindawi.com/journals/bca/2018/1062562/, (accessed July 16, 2022).
- Crossing biological barriers by engineered nanoparticles – PubMed, https://pubmed.ncbi.nlm.nih.gov/32223181/, (accessed July 16, 2022).
- S. R. Abd El-Khalik, E. Nasif, H. M. Arakeep and H. Rabah, Biol. Trace Elem. Res., 2022, 200, 1677–1687 CrossRef CAS PubMed.
- M. S. Jabir, Y. M. Saleh, G. M. Sulaiman, N. Y. Yaseen, U. I. Sahib, Y. H. Dewir, M. S. Alwahibi and D. A. Soliman, Nanomaterials, 2021, 11, 384 CrossRef CAS PubMed.
- Multitargeting by curcumin as revealed by molecular interaction studies – PubMed, https://pubmed.ncbi.nlm.nih.gov/21979811/, (accessed July 16, 2022).
- A. Shehzad and Y. S. Lee, BioFactors, 2013, 39, 27–36 CrossRef CAS PubMed.
- A. Guglielmo, A. Sabra, M. Elbery, M. M. Cerveira, F. Ghenov, R. Sunasee and K. Ckless, Chem.-Biol. Interact., 2017, 274, 1–12 CrossRef CAS PubMed.
- I. Olsen and Ö. Yilmaz, J. Oral Microbiol., 2016, 8, 30385 CrossRef PubMed.
- D. Lian, L. Dai, Z. Xie, X. Zhou, X. Liu, Y. Zhang, Y. Huang and Y. Chen, Mol. Immunol., 2018, 103, 209–219 CrossRef CAS PubMed.
- S. Xu, Q. Zhou, C. Fan, H. Zhao, Y. Wang, X. Qiu, K. Yang and Q. Ji, Arch. Oral Biol., 2019, 107, 104514 CrossRef CAS PubMed.
- The effect of doxycycline-containing chitosan/carboxymethyl chitosan nanoparticles on NLRP3 inflammasome in periodontal disease – PubMed, https://pubmed.ncbi.nlm.nih.gov/32241426/, (accessed July 16, 2022).
- L. Sun, J. Ouyang, Z. Zeng, C. Zeng, Y. Ma, F. Zeng and S. Wu, Bioact. Mater., 2022, 10, 79–92 CrossRef CAS PubMed.
- S. Yigit, N. S. Hallaj, J. L. Sugarman, L. C. Chong, S. E. Roman, L. M. Abu-Taleb, R. E. Goodman, P. E. Johnson and A. M. Behrens, Food Chem. Toxicol., 2021, 151, 112117 CrossRef CAS PubMed.
- Y. Xue, M. Du and M.-J. Zhu, Free Radicals Biol. Med., 2017, 108, 760–769 CrossRef CAS PubMed.
- Quercetin inhibits inflammasome activation by interfering with ASC oligomerization and prevents interleukin-1 mediated mouse vasculitis – PubMed, https://pubmed.ncbi.nlm.nih.gov/28148962/, (accessed July 17, 2022).
- P. Diez-Echave, A. J. Ruiz-Malagón, J. A. Molina-Tijeras, L. Hidalgo-García, T. Vezza, L. Cenis-Cifuentes, M. J. Rodríguez-Sojo, J. L. Cenis, M. E. Rodríguez-Cabezas, A. Rodríguez-Nogales, J. Gálvez and A. A. Lozano-Pérez, Int. J. Pharm., 2021, 606, 120935 CrossRef CAS PubMed.
- R. F. Loeser, J. A. Collins and B. O. Diekman, Nat. Rev. Rheumatol., 2016, 12, 412–420 CrossRef CAS PubMed.
- The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis – PubMed, https://pubmed.ncbi.nlm.nih.gov/24876674/, (accessed July 16, 2022).
- R. R. Bannuru, M. C. Osani, F. Al-Eid and C. Wang, Semin. Arthritis Rheum., 2018, 48, 416–429 CrossRef CAS PubMed.
- C. Kang, E. Jung, H. Hyeon, S. Seon and D. Lee, Nanomedicine, 2020, 23, 102104 CrossRef CAS PubMed.
- Natural product curcumin-based coordination nanoparticles for treating osteoarthritis via targeting Nrf2 and blocking NLRP3 inflammasome | SpringerLink, https://link.springer.com/article/10.1007/s12274-021-3864-3, (accessed July 16, 2022).
- Milk-derived exosomes and metabolic regulation – PubMed, https://pubmed.ncbi.nlm.nih.gov/30285461/, (accessed July 16, 2022).
- Plant-derived exosomal microRNAs shape the gut microbiota – PubMed, https://pubmed.ncbi.nlm.nih.gov/30449315/, (accessed July 16, 2022).
- B. Liu, Y. Lu, X. Chen, P. G. Muthuraj, X. Li, M. Pattabiraman, J. Zempleni, S. D. Kachman, S. K. Natarajan and J. Yu, Nutrients, 2020, 12, 477 CrossRef CAS PubMed.
- D. Fujita, T. Arai, H. Komori, Y. Shirasaki, T. Wakayama, T. Nakanishi and I. Tamai, Mol. Pharm., 2018, 15, 5772–5780 CrossRef CAS PubMed.
- Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles – PubMed, https://pubmed.ncbi.nlm.nih.gov/24842810/, (accessed July 17, 2022).
- Z. Zhao, S. Yu, M. Li, X. Gui and P. Li, J. Agric. Food Chem., 2018, 66, 2749–2757 CrossRef CAS PubMed.
- Plant-derived exosomal microRNAs shape the gut microbiota – PubMed, https://pubmed.ncbi.nlm.nih.gov/30449315/, (accessed July 17, 2022).
- Exosome-like nanoparticles from ginger rhizomes inhibited NLRP3 inflammasome activation | molecular pharmaceutics, https://pubs.acs.org/doi/10.1021/acs.molpharmaceut.9b00246, (accessed July 17, 2022).
- C. Luo, L. Zou, H. Sun, J. Peng, C. Gao, L. Bao, R. Ji, Y. Jin and S. Sun, Front. Pharmacol..
- M. K. Swamy, U. R. Sinniah and A. Ghasemzadeh, Appl. Microbiol. Biotechnol., 2018, 102, 7775–7793 CrossRef CAS PubMed.
- M. Ghasemzadeh Rahbardar and H. Hosseinzadeh, Naunyn-Schmiedeberg's Arch. Pharmacol., 2020, 393, 1779–1795 CrossRef CAS PubMed.
- S. Marinho, M. Illanes, J. Ávila-Román, V. Motilva and E. Talero, Biomolecules, 2021, 11, 162 CrossRef CAS PubMed.
- R. Guiteras, M. Flaquer and J. M. Cruzado, Clin. Kidney J., 2016, 9, 765–771 CrossRef CAS PubMed.
- Monitoring of inflammation in patients on dialysis: forewarned is forearmed – PubMed, https://pubmed.ncbi.nlm.nih.gov/21358695/, (accessed July 17, 2022).
- A.-E. Declèves and K. Sharma, Nat. Rev. Nephrol., 2014, 10, 257–267 CrossRef PubMed.
- The inflammasomes in kidney disease – PubMed, https://pubmed.ncbi.nlm.nih.gov/21566058/, (accessed July 17, 2022).
- The NLRP3 inflammasome in kidney disease and autoimmunity – PubMed, https://pubmed.ncbi.nlm.nih.gov/27011059/, (accessed July 17, 2022).
- Y.-F. Lin, Y.-H. Lee, Y.-H. Hsu, Y.-J. Chen, Y.-F. Lin, F.-Y. Cheng and H.-W. Chiu, Nanomedicine, 2017, 12, 2741–2756 CrossRef CAS PubMed.
- J.-J. Jhang, C.-C. Lu and G.-C. Yen, Mol. Nutr. Food Res., 2016, 60, 2297–2303 CrossRef CAS PubMed.
- U. E. N. C. for E. Assessment, Green tea polyphenols and epigallocatechin-3-gallate protect against perfluorodecanoic acid induced liver damage and inflammation in mice by inhibiting NLRP3 inflammasome activation, https://hero.epa.gov/hero/index.cfm/reference/details/reference_id/6323927, (accessed July 17, 2022).
- Molecular mechanisms of epigallocatechin-3-gallate for prevention of chronic kidney disease and renal fibrosis: preclinical evidence – PMC, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6752729/, (accessed July 17, 2022).
- Y. Yan, W. Jiang, L. Liu, X. Wang, C. Ding, Z. Tian and R. Zhou, Cell, 2015, 160, 62–73 CrossRef CAS PubMed.
- B. L. Woolbright and H. Jaeschke, J. Hepatol., 2017, 66, 836–848 CrossRef CAS PubMed.
- C. Zhan, G. Lin, Y. Huang, Z. Wang, F. Zeng and S. Wu, Biomaterials, 2021, 268, 120573 CrossRef CAS PubMed.
- X. Yu, G. Yang, H. Jiang, S. Lin, Y. Liu, X. Zhang, H. Zeng, Z. Su, S. Huang, L. Shen and X. Zhang, Exp. Ther. Med., 2017, 14, 1184–1192 CrossRef CAS PubMed.
- Macrophages in inflammation – PubMed, https://pubmed.ncbi.nlm.nih.gov/16101534/, (accessed July 17, 2022).
- Y. Zhao, Y. Yang, J. Zhang, R. Wang, B. Cheng, D. Kalambhe, Y. Wang, Z. Gu, D. Chen, B. Wang and Y. Huang, Acta Pharm. Sin. B, 2020, 10, 1966–1976 CrossRef CAS PubMed.
- L. Sun, J. Ouyang, F. Zeng and S. Wu, Biomaterials, 2022, 283, 121468 CrossRef CAS PubMed.
- R. Prakash, A. Vyawahare, R. Sakla, N. Kumari, A. Kumar, M. M. Ansari, Kanika, C. Jori, A. Waseem, A. J. Siddiqui, M. A. Khan, A. A. B. Robertson, R. Khan and S. S. Raza, ACS Nano, 2023, 17, 8680–8693 CrossRef CAS PubMed.
- Y. Song, Y. Zhao, Y. Ma, Z. Wang, L. Rong, B. Wang and N. Zhang, Cytokine Growth Factor Rev., 2021, 60, 61–75 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |