Real-time visualization of autophagy by monitoring the fluctuation of lysosomal pH with a ratiometric two-photon fluorescent probe†
Peng
Ning‡
a,
Liling
Hou‡
a,
Yan
Feng
 *a,
Guoyong
Xu
b,
Yuyuan
Bai
a,
Haizhu
Yu
a and
Xiangming
Meng
*a,
Guoyong
Xu
b,
Yuyuan
Bai
a,
Haizhu
Yu
a and
Xiangming
Meng
 *ab
*ab
aSchool of Chemistry and Chemical Engineering, Center for Atomic Engineering of Advanced Materials, and Anhui Province Key Laboratory of Chemistry for Inorganic/Organic Hybrid Functionalized Materials, Anhui University, Hefei, Anhui 230601, China. E-mail: fy70@163.com; mengxm@ahu.edu.cn; Fax: +86 551 63861467; Tel: +86 551 63861467
bInstitute of Physical Science and Information Technology, Anhui University, Hefei 230601, P. R. China
First published on 11th January 2019
Abstract
A benzimidazole-decorated two-photon fluorescent probe (Lyso-MPCB) based on the p-methoxyphenylacetylene-substituted carbazole was developed for detecting lysosomal pH with a double-channel signal, which can be used to visualize autophagy by real-time imaging the fluctuation of the pH in the lysosomes.
Autophagy is a lysosome-dependent degradation pathway for recycling misfolded proteins and dysfunctional organelles as well as providing nutrients during starvation.1 Several conventional methods have been used to monitor such metabolic processes, including transmission electron microscopy (TEM), western blotting (Atg8/LC3), and GFP-Atg8/LC3 fluorescence microscopy.2 Nevertheless, it's difficult to achieve real-time, rapid and low-cost visualization of autophagy in living cells by employing the above methods. Therefore, real-time detection of autophagy efficiently and economically is still a challenge.
In general, a lysosome and an autophagosome can be fused to generate an autolysosome during the membrane fusion process of autophagy.3 The microenvironments (including polarity, pH, and viscosity, etc.) would fluctuate owing to the difference between the lysosome and autophagosome in this process.4 Accordingly, it is feasible to monitor autophagy by detecting the microenvironmental variation, especially the pH, in a real-time fashion, which is the vital factor for the activity of acid hydrolases.5 To date, a few small-molecule fluorescent probes have been reported for autophagy detection by responding to the changes in pH value.6 However, the existing probes yield fluctuation of the emission intensity at a single particular wavelength. Such a turn-on type probe is liable to uneven probe loading and photobleaching. These limitations can be improved and simplified by developing one novel ratiometric pH fluorescent probe for autophagy detection. Ratiometric fluorescent probes can overcome several problems including local concentrations of probes, photobleaching and variations of the instrument, which can inhibit the quantitative and accurate determination of the analyte.7 In addition, our reported studies have evidenced that the two-photon (TP) fluorescence performance of probes could enhance the spatial resolution in autophagy detection.8 Therefore, it is of pressing need to design a TP ratiometric fluorescent probe to visualize autophagy by ratiometric imaging of the lysosomal pH fluctuation.
Here, we report a novel pH-sensitive TP ratiometric fluorescent probe (Lyso-MPCB) on the basis of p-methoxyphenylacetylene-substituted carbazole, an effective TP fluorophore reported in our previous work.8c The benzimidazole group in Lyso-MPCB was employed to respond to changes in pH with an obvious emission shift (65 nm). In addition, Lyso-MPCB was configured with the lysosome-locating group, morpholine.9 This probe can specifically respond to changes in lysosomal pH and has good biocompatibility. To our knowledge, Lyso-MPCB was the first ratiometric small-molecule TP fluorescent probe to visualize autophagy by real-time imaging of the lysosomal pH fluctuation (Scheme 1). The synthetic route and detailed characterizations of Lyso-MPCB are shown in the ESI† (Scheme S1 and Fig. S11–S21).
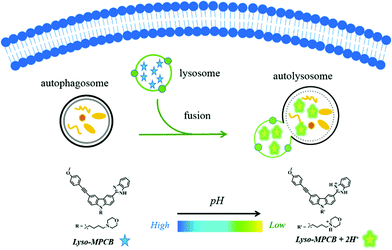 | ||
| Scheme 1 Illustration of real-time visualization of autophagy with a double-channel signal using probe Lyso-MPCB in cells. | ||
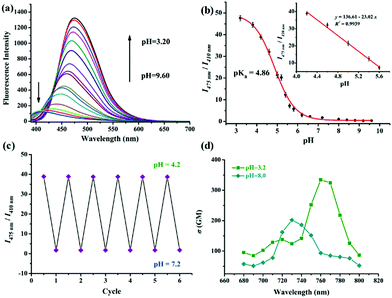
Firstly, the pH-responsive profile of Lyso-MPCB was studied spectroscopically in Britton–Robinson (B–R) buffer (10 μM, 30% DMSO, v/v). Under basic conditions (pH = 8.2), the UV-Vis spectrum showed the maximum absorption peak at 340 nm (Fig. S1a, ESI†). With the pH value decreasing from 8.2 to 3.2, the peak at 340 nm was reduced gradually along with the emergence of a new peak at 385 nm. The nonlinear pH titration curve was established by plotting the ratio (A340nm/A385nm) of absorbance against pH value from 3.2 to 8.2 according to the Henderson–Hasselbalch equation10 and good linearity was presented in a narrow acidic pH range (pH = 4.6–6.2) (Fig. S1b, ESI†). The isosbestic point at 370 nm indicated the protonation of Lyso-MPCB. As shown in Fig. 1a, the fluorescence emission spectra of Lyso-MPCB were tested in the pH range from 9.6 to 3.2. Under basic conditions (pH = 9.6), Lyso-MPCB exhibited weak blue fluorescence emission at 410 nm. When the pH decreased from 9.6 to 3.2, the evident green fluorescence emission at 475 nm was gradually increased with a large red-shift of 65 nm. The ratios (I475nm/I410nm) of fluorescence intensity displayed apparent enlargement from 0.34 to 47.62 (∼140-fold enhancements) along with the enlarged fluorescence quantum yield from 5.23% to 11.27%. These results indicated that Lyso-MPCB was highly sensitive to changes in pH in vitro. Moreover, the pKa value was obtained to be 4.86 fitted by the Henderson–Hasselbalch equation (eqn (S1), ESI†).5b,10 Such lower pKa value was suitable for determining the changes in pH in the lysosomal range (4.5–5.5).11
More importantly, the ratio signals (I475nm/I410nm) of Lyso-MPCB displayed a good linear relationship with pH values ranging from 4.2 to 5.6 (inset of Fig. 1b), which demonstrated that the probe can be an efficient tool for detecting the lysosomal pH change quantitatively.
1H NMR titration experiments using DCl (deuterium chloride) in DMSO-d6 were conducted to confirm the sensing mechanism of Lyso-MPCB for acid pH (Fig. S2, ESI†).12 Compared with H1–H4 of the benzimidazole moiety in Lyso-MPCB, the corresponding four protons (H1′–H4′) of Lyso-MPCB + 2H+ obviously shifted downfield due to the protonation of the nitrogen atom in the benzimidazole unit (H1 shifted 0.27 ppm, H4 shifted 0.15 ppm and H2, H3 shifted 0.35 ppm, respectively). Moreover, the time-dependent density functional theory (TD-DFT) calculations were employed to study the photophysical properties of Lyso-MPCB and its protonated configuration Lyso-MPCB + 2H+.13 As shown in Fig. S3 (ESI†), electrons were totally localized over the molecular skeleton at the ground and excited states of Lyso-MPCB, respectively. When probe Lyso-MPCB reacted with H+, electrons were changed to distribute on the p-methoxyphenylacetylene-substituted carbazole unit as the donor group at the ground state, while the protonated benzimidazole unit as the acceptor group at the excited state of Lyso-MPCB + 2H+, indicating an enhanced ICT process. The HOMO–LUMO gap value of Lyso-MPCB is larger than that of Lyso-MPCB + 2H+ (3.92 eV vs. 2.10 eV), which is consistent with the hypochromic shift (from 340 nm to 385 nm) of the absorption spectra. It's clear that the enhancement of the ICT process in Lyso-MPCB + 2H+ was caused by H+-binding of the ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N-atom in the benzimidazole group of Lyso-MPCB, showing that the probe is indeed pH-dependent.
N-atom in the benzimidazole group of Lyso-MPCB, showing that the probe is indeed pH-dependent.
In addition, the reversible response to pH between 4.2 and 7.2 of Lyso-MPCB was conducted in B–R buffer solution (with 30% DMSO) (Fig. 1c). The results distinctly showed that the process was fully reversible for at least 6 cycles, which indicated that Lyso-MPCB was sensitive and stable for pH detection between the protonation and deprotonation processes. Two-photon (TP) absorption properties of unprotonated and protonated Lyso-MPCB were carried out (Fig. 1d).14 The maximum TP absorption cross-section value (δ) was 202 GM at 730 nm for the former and 340 GM at 760 nm for the latter, which attested that the protonated probe possessed a better two-photon absorption performance than that of the unprotonated one due to the enhanced ICT. Besides, Lyso-MPCB displayed no response to other analytes (amino acids and ions), showing that it's an excellent pH-dependent TP fluorescent probe with high selectivity (Fig. S4, ESI†). The time-dependent fluorescence response results of Lyso-MPCB in B–R buffer (pH = 5.0, with 30% DMSO) indicated that the ratios were stabilized at 2 min. These data fitted well to a monoexponential model and a first-order kinetic rate constant can be recovered as 0.0307 s−1 (R2 = 0.99835) in this kinetic experiment (Fig. S5, ESI†).15
The cytotoxicity of probe Lyso-MPCB was tested by MTT assay before biological imaging, and the results indicated that the cell survival was as high as 89% even when the concentration of Lyso-MPCB reached 15 μM (Fig. S6, ESI†). Therefore, the probe was capable of biological imaging in living MCF-7 cells as expected. Its specificity of localizing lysosomes was carried out in MCF-7 cells via colocalization fluorescence imaging technology. As shown in Fig. S7 (ESI†), probe Lyso-MPCB and LysoTracker Red overlapped very well, and the Pearson's colocalization coefficient (R) was found to reach 0.935. On the contrary, the Pearson's colocalization coefficient between Lyso-MPCB and MitoTracker Red was only 0.336 in MCF-7 cells. Hence, these results demonstrated that our probe could localize to the lysosome perfectly and could be utilized as a reliable tool for further investigations in lysosomal regions.
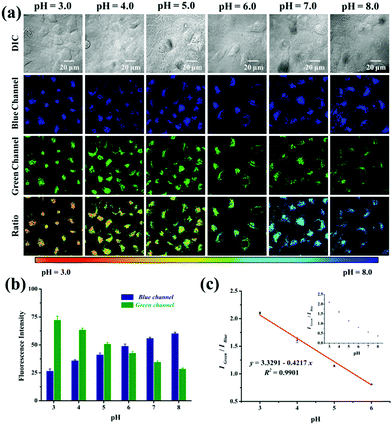
Subsequently, we verified the capability of probe Lyso-MPCB to quantify the intracellular pH in a ratiometric manner in living cells. The intracellular pH of MCF-7 cells was well-distributed by using the high K+ buffer (0.5 mM MgSO4, 1 mM CaCl2, 1 mM NaH2PO4, 5 mM glucose, 30 mM NaCl, 120 mM KCl, and 20 mM HEPES) with 10 mM nigericin at different pH values (3.0–8.0).5c,9a The blue channel exhibited a continual increase along with a gradual decrease in the green fluorescence when the intracellular pH values enhanced from 3.0 to 8.0 (Fig. 2a). Similarly, obvious remarkable pseudocolor changes were observed in the pH range 3.0–8.0. Meanwhile, the ratio images (Igreen/Iblue) displayed a pH-dependent characteristic greatly from pH 3.0 to 8.0. When images were quantified using Image J software, the blue fluorescence intensity showed enhanced features along with reduced green fluorescence intensity (Fig. 2b). The fluorescence intensity ratio (Igreen/Iblue) in cells also exhibited a good linear calibration curve to pH (pH = 3.0–6.0, Fig. 2c). It is just within the scope of the lysosomal pH value, which can help us to analyze the pH changes of lysosomes quantitatively. Taken together, Lyso-MPCB showed a wonderful capability for monitoring lysosomal pH value changes quantitatively.
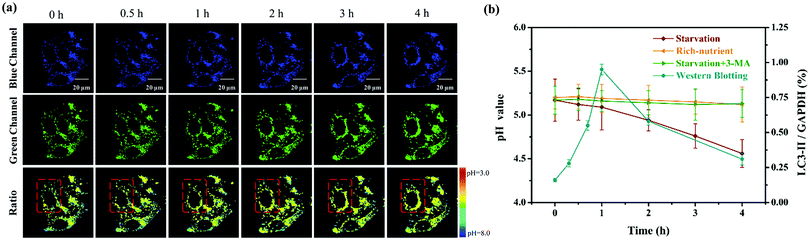
Considering that the microenvironmental pH of cells would fluctuate during the autophagy process, we monitored the real-time changes of the lysosomal pH values with Lyso-MPCB in this lysosomes-related physiological pathway. The autophagy process here was stimulated under starvation conditions. In the beginning, the MCF-7 cells, co-stained with Lyso-MPCB (10 μM) were cultured in a medium without nutrients, Hank's Balanced Salt Solution (HBSS).16 Then, the TP fluorescence images in the blue and green channel, including the ratio image analysis, were all acquired within 4 hours (Fig. 3a). In the first 1 hour, the fluorescence intensities both in the blue and green channels displayed no obvious change, as well as the ratio analysis. In particular, we chose a single cell in the red dashed box as the representative case for observing the variations of parameters within 4 hours. In the later 3 hours, the distribution range of the pseudocolor gradually became larger in the single cell. Moreover, the average level of pseudocolor also became deeper. These results demonstrated that the lysosomes in this cell were migrated step by step and the pH values in the lysosomal regions were decreased continuously in the later 3 h. In addition, the western blotting results (as shown in Fig. S8 (ESI†) and Fig. 3b, blue line) supplied accurate evidence that the membrane fusion procedure between the lysosomes and autophagosomes occurred after incubating cells with HBSS for 1 hour, which was in conformity with the TEM results in our reported work.8b Moreover, the specific pH values at different times of autophagy can be calculated intuitively and accurately, and the value was reduced from 5.2 to 4.6 (Fig. 3b, red line) in accordance with the analysis of Fig. 3a. Correspondingly, the pH values remained almost constant both under “rich-nutrient” conditions (Fig. S9 (ESI†) and Fig. 3b, orange line) and autophagy-inhibited conditions (HBSS with 3-methyladenine (3-MA) for inhibiting autophagy) (Fig. S10 (ESI†) and Fig. 3b, green line). These consequences demonstrated that Lyso-MPCB can be utilized as an available TP fluorescent probe for visualizing the autophagy process in real-time via quantifying the undulation of pH values with ratio signals.
In summary, a novel ratiometric TP fluorescent probe (Lyso-MPCB) was developed for detecting lysosomal pH changes. The probe displayed a dual-color response to pH with high sensitivity and selectivity, and the ratio values of the fluorescence intensities at 475 nm and 410 nm showed a great linear relationship with pH changes in solution as well as in living cells. More importantly, Lyso-MPCB can be used as an effective ratiometric fluorescence imaging tool for real-time visualization of autophagy by monitoring the lysosomal pH fluctuation.
This work was supported by the National Natural Science Foundation of China (21778001 and 21372005), the Natural Science Foundation of Anhui Province (1608085MB39 and 1808085MB47) and the Open fund for Discipline Construction in Institute of Physical Science and Information Technology of Anhui University.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) J. J. Lum, R. J. DeBerardinis and C. B. Thompson, Nat. Rev. Mol. Cell Biol., 2005, 6, 439–448 CrossRef CAS PubMed; (b) N. J. Dolman, K. M. Chambers, B. Mandavilli, R. H. Batchelor and M. S. Janes, Autophagy, 2013, 9, 1653–1662 CrossRef CAS PubMed.
- (a) N. Sun, D. Malide, L. Jie, I. I. Rovira, C. A. Combs and T. Finkel, Nat. Protoc., 2017, 12, 1576–1586 CrossRef CAS PubMed; (b) Y. X. Lin, Y. Wang and H. Wang, Small, 2017, 13, 1700996 CrossRef PubMed; (c) A. Kuma, M. Matsui and N. Mizushima, Autophagy, 2007, 3, 323–328 CrossRef CAS PubMed.
- J. J. Diao, R. Liu, Y. G. Rong, M. L. Zhao, J. Zhang, Y. Lai, Q. J. Zhou, L. M. Wilz, J. X. Li, S. Vivona, R. A. Pfuetzner, A. T. Brunger and Q. Zhong, Nature, 2015, 520, 563–566 CrossRef CAS PubMed.
- (a) M. B. Azad, Y. Chen, E. S. Henson, J. Cizeau, E. McmillanWard, S. J. Israels and S. B. Gibson, Autophagy, 2008, 4, 195–204 CrossRef CAS PubMed; (b) G. Maulucci, M. Chiarpotto, M. Papi, D. Samengo, G. Pani and M. DeSpirito, Autophagy, 2015, 11, 1905–1916 CrossRef CAS PubMed.
- (a) Y. K. Yue, F. J. Huo, S. Y. Lee, C. X. Yin and J. Yoon, Analyst, 2017, 142, 30–41 RSC; (b) H. J. Kim, C. H. Heo and H. M. Kim, J. Am. Chem. Soc., 2013, 135, 17969–17977 CrossRef CAS PubMed; (c) L. L. Wu, Y. Wang, T. D. James, N. Q. Jia and C. S. Huang, Chem. Commun., 2018, 54, 5518–5521 RSC; (d) Y. B. Zhang, S. A. Xia, M. X. Fang, W. F. Mazi, Y. B. Zeng, T. Johnston, A. Pap, R. L. Luck and H. Y. Liu, Chem. Commun., 2018, 54, 7625–7628 RSC.
- (a) M. H. Lee, N. Park, C. Yi, J. H. Han, J. H. Hong, K. P. Kim, D. H. Kang, J. L. Sessler, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2014, 136, 14136–14142 CrossRef CAS PubMed; (b) H. Iwashita, S. Torii, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, S. Shimizu and K. Okuma, ACS Chem. Biol., 2017, 12, 2546–2551 CrossRef CAS PubMed; (c) L. Q. Niu, J. Huang, Z. J. Yan, Y. H. Men, Y. Luo, X. M. Zhou, J. M. Wang and J. H. Wang, Spectrochim. Acta, Part A, 2019, 207, 123–131 CrossRef CAS PubMed; (d) Y. Liu, J. Zhou, L. J. Wang, X. X. Hu, X. J. Liu, M. R. Liu, Z. H. Cao, D. H. Shangguan and W. H. Tan, J. Am. Chem. Soc., 2016, 138, 12368–12374 CrossRef CAS PubMed; (e) Y. C. Liu, L. L. Teng, L. L. Chen, H. C. Ma, H. W. Liu and X. B. Zhang, Chem. Sci., 2018, 9, 5347–5353 RSC; (f) H. Iwashita, H. T. Sakurai, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, K. Okuma, S. Shimizu and Y. Ueno, FEBS Lett., 2018, 592, 559–567 CrossRef CAS PubMed.
- (a) M. H. Lee, J. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 4185–4191 RSC; (b) M. A. Haidekker and E. A. Theodorakis, J. Mater. Chem. C, 2016, 4, 2707–2718 RSC.
- (a) L. L. Hou, P. Ning, Y. Feng, Y. Q. Ding, L. Bai, L. Li, H. Z. Yu and X. M. Meng, Anal. Chem., 2018, 90, 7122–7126 CrossRef CAS PubMed; (b) J. C. Jiang, X. H. Tian, C. Z. Xu, S. X. Wang, Y. Feng, M. Chen, H. Z. Yu, M. Z. Zhu and X. M. Meng, Chem. Commun., 2017, 53, 3645–3648 RSC; (c) W. J. Wang, P. Ning, Q. Wang, W. Zhang, J. Jiang, Y. Feng and X. M. Meng, J. Mater. Chem. B, 2018, 6, 1764–1770 RSC.
- (a) Y. Wen, W. J. Zhang, T. Liu, F. J. Huo and C. X. Yin, Anal. Chem., 2017, 89, 11869–11874 CrossRef CAS PubMed; (b) Q. Q. Wan, S. M. Chen, W. Shi, L. H. Li and H. M. Ma, Angew. Chem., Int. Ed., 2014, 53, 10916–10920 CrossRef CAS PubMed.
- (a) X. Luo, H. T. Yang, H. L. Wang, Z. W. Ye, Z. N. Zhou, L. Y. Gu, J. Q. Chen, Y. Xiao, X. W. Liang, X. H. Qian and Y. Y. Yang, Anal. Chem., 2018, 90, 5803–5809 CrossRef CAS PubMed; (b) D. Yoshihara, T. Noguchi, B. Roy, J. Sakamoto, T. Yamamoto and S. Shinkai, Chem. – Eur. J., 2017, 23, 17663–17666 CrossRef CAS PubMed; (c) M. J. Chang, K. Kim, K. S. Park, J. S. Kang, C. S. Lim, H. M. Kim, C. Kang and M. H. Lee, Chem. Commun., 2018, 54, 13531–13534 RSC.
- (a) J. T. Hou, W. X. Ren, K. Li, J. Seo, A. Sharma, Q. Yu and J. S. Kim, Chem. Soc. Rev., 2017, 46, 2076–2090 RSC; (b) F. Galindo, M. I. Burguete, L. Vigara, S. V. Luis, N. Kabir, J. Gavrilovic and D. A. Russell, Angew. Chem., Int. Ed., 2005, 44, 6504–6508 CrossRef CAS PubMed.
- G. P. Li, D. J. Zhu, L. Xue and H. Jiang, Org. Lett., 2013, 15, 5020–5023 CrossRef CAS PubMed.
- (a) A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS PubMed; (b) L. Yuan, W. Y. Lin, J. Z. Song and Y. T. Yang, Chem. Commun., 2011, 47, 12691–12693 RSC.
- C. Xu and W. W. Webb, J. Opt. Soc. Am. B, 1996, 13, 481–491 CrossRef CAS.
- (a) Z. X. Liu, X. Zhou, Y. Miao, Y. Hu, N. Y. Kwon, X. Wu and J. Y. Yoon, Angew. Chem., Int. Ed., 2017, 56, 5812–5816 CrossRef CAS PubMed; (b) L. P. Qiu, T. Zhang, J. H. Jiang, C. C. Wu, G. Z. Zhu, M. X. You, X. G. Chen, L. Q. Zhang, C. Cui, R. Q. Yu and W. H. Tan, J. Am. Chem. Soc., 2014, 136, 13090–13093 CrossRef CAS PubMed.
- E. Desideri, G. Filomeni and M. R. Ciriolo, Autophagy, 2012, 8, 1769–1781 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section, UV-Vis spectra, 1H-NMR titration, selectivity experiment, theoretical calculations, cytotoxicity assay, colocalization experiments, western blotting, HRMS and NMR spectra. See DOI: 10.1039/c8cc09517e |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |