Themed collection Physical Foundations of Protein Folding

Probing molecular kinetics with Markov models: metastable states, transition pathways and spectroscopic observables
Markov (state) models are useful for identifying metastable conformations from simulated data, to investigate ensembles of folding pathways and to link MD simulation with kinetic experiments.
Phys. Chem. Chem. Phys., 2011,13, 16912-16927
https://doi.org/10.1039/C1CP21258C
Diffusion models of protein folding
Descriptions of protein folding as diffusion along low-dimensional reaction coordinates capture the observed simulation dynamics and help interpret single-molecule experiments.

Phys. Chem. Chem. Phys., 2011,13, 16902-16911
https://doi.org/10.1039/C1CP21541H
Coarse-grained force field: general folding theory
UNRES force field is a tool with which to carry out ab initio simulations of protein folding.

Phys. Chem. Chem. Phys., 2011,13, 16890-16901
https://doi.org/10.1039/C1CP20752K
Protonation/deprotonation effects on the stability of the Trp-cage miniprotein
The effect of protonating the aspartic acid on the Trp-cage folding/unfolding equilibrium is studied by explicit solvent molecular dynamics simulations.
Phys. Chem. Chem. Phys., 2011,13, 17056-17063
https://doi.org/10.1039/C1CP21193E
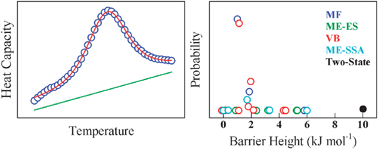
Estimation of protein folding free energy barriers from calorimetric data by multi-model Bayesian analysis
A global multi-model Bayesian analysis of scanning calorimetry data allows a high-sensitivity, robust and model-independent estimation of thermodynamic protein folding barriers.
Phys. Chem. Chem. Phys., 2011,13, 17064-17076
https://doi.org/10.1039/C1CP20156E
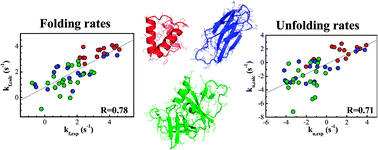
Integrated prediction of protein folding and unfolding rates from only size and structural class
A simple physics-based model that predicts protein folding and unfolding rates using size and structural class as only specific input.
Phys. Chem. Chem. Phys., 2011,13, 17030-17043
https://doi.org/10.1039/C1CP20402E
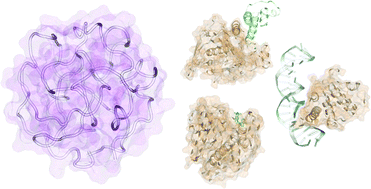
Why not consider a spherical protein ? Implications of backbone hydrogen bonding for protein structure and function
Comparing artificial (orange) and quasi-spherical (purple) protein models, hydrogen bonding leads to protein-like cavities for ligand-binding and interfaces for protein/DNA-binding.
Phys. Chem. Chem. Phys., 2011,13, 17044-17055
https://doi.org/10.1039/C1CP21140D
About this collection
We are delighted to present a high-profile themed collection on the Physical Foundations of Protein Folding. This themed collection, Guest Edited by Modesto Orozco (IRB Barcelona), contains a mix of exciting ‘Perspective’ review articles and cutting-edge research papers. Watch out for more PCCP articles on this theme which will be added to this collection throughout 2011.
This special themed collection emphasizes the recent advances from all three approaches in protein folding: experiment, theory and simulations. It also provides a platform for discussing interesting concepts in protein folding and the direction this field will take in the future.