High-throughput ultra-sensitive discrimination of single nucleotide polymorphism via click chemical ligation†
Abstract
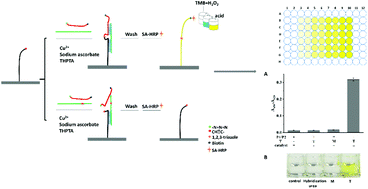
Single nucleotide polymorphisms (SNPs) have been proven to be important biomarkers for disease diagnosis, prognosis and disease pathogenesis. Here, taking the advantages of a self-assembled oligonucleotide sandwich structure and robust chemical reactions, we have developed a simple, high-throughput and effective colorimetric analytical technique termed CuAAC-based ligation-assisted assays (CuAAC-LA) for SNP detection using a DNA-BIND 96-well plate. With the 5′-azide and 3′-alkyne groups labelled on two oligonucleotide probes, the target DNA can direct a Cu(I)-catalyzed alkyne–azide cycloaddition (CuAAC) click reaction. Since the small difference in duplex stability caused by a single-nucleotide mismatch was amplified by the steric effects of these reactive groups for the ligation reaction of an unstable duplex, CuAAC-LA exhibited an ultra-sensitive discrimination ability for a mutant type target in the presence of large amounts of wild type targets. As low as 0.05% SNP could be clearly detected, which was better than most previously reported methods by various DNA ligases, indicating that a simple and rapid synthetic method i.e., the DNA template-directed click reaction held the potential to replace the ligase for SNP detection.


 Please wait while we load your content...
Please wait while we load your content...