Informatics analysis of capillary electropherograms of autologously doped and undoped blood†
Abstract
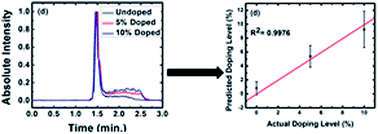
An ‘Autologous Blood Transfusion’ (ABT) is the reinjection of blood previously taken from an athlete to increase its oxygen transport capabilities. Despite the World Anti-Doping Agency's ban on such practices, ABT abuse continues. Autologous blood doping (ABD) is challenging to detect because of the similarities between an individual's doped and undoped blood. Recently, Harrison et al. reported that high-speed capillary electrophoresis may identify ABD. In their work, first order derivatives of the electropherograms were used to identify doping. However, this method suffered from false negatives due to the subjective nature of the analysis. Here, we provide an informatics analysis of the data from this study, contrasting the results of traditional statistical methods and less traditional mathematical techniques. First, three well-known multivariate statistical tools: cluster analysis, principal component analysis (PCA), and partial least squares (PLS) are applied to develop calibrations and/or group electropherograms of undoped (0%) and doped (5% and 10%) blood samples. (These doping levels were chosen due to the low physiological effect of doping below 5%, with 10% corresponding to the approximate ‘gain’ derived from the transfusion of a single unit of blood into an adult.) Different preprocessing and variable selection methods were considered. Due to variation in the electropherograms and the limited sample size, these methods were inadequate. We next considered four less commonly used mathematical/informatics tools: pattern recognition entropy (PRE), the Euclidean distance between vectors, a peak fitting/integration method, and the second moment (SM). Each of these techniques showed some ability to differentiate between the 0, 5, and 10% doped samples. We then evaluated the prediction capabilities of inverse least squares (ILS) models based on these summary statistics. An ILS calibration based on PRE, the Euclidean distance, and peak fitting/integration proved more successful than the PLS model at predicting levels of blood doping from the corresponding electropherograms; the ILS model distinguished between doped (5% and 10%) and undoped (0%) blood. This methodology may be applicable to other challenging informatics problems like determining risk factors for genetically linked diseases, robust pattern finding in peak-like data such as ChIP-seq, or other genomic sequencing for understanding the 3D genome.
- This article is part of the themed collection: Analytical Methods Recent HOT articles


 Please wait while we load your content...
Please wait while we load your content...