Dual mechanism of ionic liquid-induced protein unfolding†
Abstract
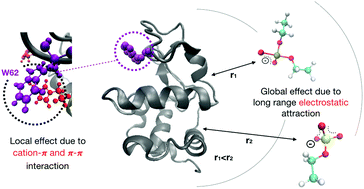
Ionic liquids (ILs) are gaining attention as protein stabilizers and refolding additives. However, varying degrees of success with this approach motivates the need to better understand fundamental IL–protein interactions. A combination of experiment and simulation is used to investigate the thermal unfolding of lysozyme in the presence of two imidazolium-based ILs (1-ethyl-3-methylimidazolium ethylsulfate, [EMIM][EtSO4] and 1-ethyl-3-methylimidazolium diethylphosphate, [EMIM][Et2PO4]). Both ILs reduce lysozyme melting temperature Tm, but more gradually than strong denaturants. [EMIM][Et2PO4] lowers lysozyme Tm more readily than [EMIM][EtSO4], as well as requiring less energy to unfold the protein, as determined by the calorimetric enthalpy ΔH. Intrinsic fluorescence measurements indicate that both ILs bind to tryptophan residues in a dynamic mode, and furthermore, molecular dynamics simulations show a high density of [EMIM]+ near lysozyme's Trp62 residue. For both ILs approximately half of the [EMIM]+ cations near Trp62 show perfect alignment of their respective rings. The [EMIM]+ cations, having a “local” effect in binding to tryptophan, likely perturb a critically important Arg-Trp-Arg bridge through favorable π–π and cation–π interactions. Simulations show that the anions, [EtSO4]− and [Et2PO4]−, interact in a “global” manner with lysozyme, due to this protein's strong net positive charge. The anions also determine the local distribution of ions surrounding the protein. [Et2PO4]− is found to have a closer first coordination shell around the protein and stronger Coulomb interactions with lysozyme than [EtSO4]−, which could explain why the former anion is more destabilizing. Patching of ILs to the protein surface is also observed, suggesting there is no universal IL solvent for proteins, and highlighting the complexity of the IL–protein environment.


 Please wait while we load your content...
Please wait while we load your content...
