Parameterization and atomistic simulations of biomimetic membranes†
Abstract
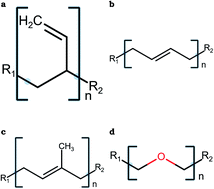
Biomimetic membranes, designed by combining proteins or protein-mimics with self-assembled block copolymers, are emerging as novel hybrid materials with applications in the next generation of sensing and separation devices. However, designing such membranes requires a fundamental understanding of the atomic-scale interactions between biological channel proteins and their non-native polymeric membrane environment as it affects their stability and function. In principle, all-atom molecular dynamics (MD) simulations are well-suited to probe the atomistic details of channel/membrane interactions, but the absence of interatomic potentials is a major limiting factor in conducting such simulations. To alleviate this, we have developed CHARMM force-field compatible parameters and conducted all-atom explicit-solvent MD simulations of biomimetic membranes composed of block copolymers of poly(butadiene), poly(isoprene), and poly(ethylene oxide). Consistent with scaling laws and literature data, we report measurements on several structural properties that inform on molecular-scale features of chain conformations. Finally, we report simulations of a synthetic transport channel in selected membranes and characterize its functional behavior by measuring the single-channel water permeability. We suggest that the interatomic potentials and membrane models reported here could be useful in studies of other proteins as well as for deriving potentials for coarse-grained models to permit future simulations of large-scale protein/polymer membranes.
- This article is part of the themed collection: Artificial Water Channels


 Please wait while we load your content...
Please wait while we load your content...