Logic-based models in systems biology: a predictive and parameter-free network analysis method†
Abstract
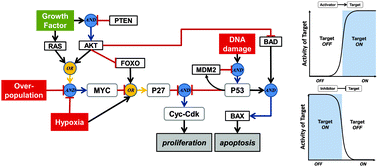
Highly complex molecular networks, which play fundamental roles in almost all cellular processes, are known to be dysregulated in a number of diseases, most notably in cancer. As a consequence, there is a critical need to develop practical methodologies for constructing and analysing molecular networks at a systems level. Mathematical models built with continuous differential equations are an ideal methodology because they can provide a detailed picture of a network's dynamics. To be predictive, however, differential equation models require that numerous parameters be known a priori and this information is almost never available. An alternative dynamical approach is the use of discrete logic-based models that can provide a good approximation of the qualitative behaviour of a biochemical system without the burden of a large parameter space. Despite their advantages, there remains significant resistance to the use of logic-based models in biology. Here, we address some common concerns and provide a brief tutorial on the use of logic-based models, which we motivate with biological examples.

 Please wait while we load your content...
Please wait while we load your content...