Meta-analyses of host metagenomes from colorectal cancer patients reveal strong relationship between colorectal cancer-associated species†
Abstract
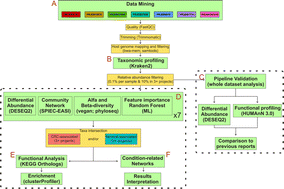
Colorectal cancer (CRC) is one of the most common types of cancer, with many studies associating its development with changes in the gut microbiota. Recent developments in sequencing technologies and subsequent meta-analyses of gut metagenome provided a better understanding of species related to CRC tumorigenesis. Still, the importance of high-importance taxonomic singletons (i.e. species highly associated with a given condition but observed only in the minority of datasets) and the species interactions and co-occurrence across cohorts need further exploration. It has been shown that the gut metagenome presents a high functional redundancy, meaning that species interactions could mitigate the absence of any given species. In a CRC framework, this implies that species co-occurrence could play a role in tumorigenesis, even if CRC-associated species show low abundance. We propose to evaluate the prevalence of microbial species in tumor by initially analyzing each dataset individually and subsequently intersecting the results for differentially abundant species between CRC and healthy samples. We then identify metabolic pathways from these species based on KEGG orthologs, highlighting metabolic pathways associated with CRC. Our results indicate seven species with high prevalence across all projects and with high association to CRC, including the genus Bacteroides, Enterocloster and Prevotella. Finally, we show that CRC is also characterized by the co-occurrence of species that do not present significant differential abundance, but have been described in the literature as potential CRC biomarkers. These results indicate that between-species interactions could also play a role in CRC tumorigenesis.
- This article is part of the themed collection: Celebrating Latin American Chemistry


 Please wait while we load your content...
Please wait while we load your content...