Differential roles of transcriptional and translational negative autoregulations in protein dynamics
Abstract
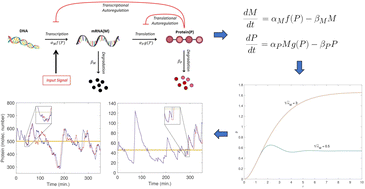
Cells continuously respond to stimuli to function properly by employing a wide variety of regulatory mechanisms that often involve protein up or down regulations. This study focuses on dynamics of a protein with negative autoregulations in E. coli, and assumes that the input signal up-regulates the protein, and then the protein down-regulates its own production via 2 distinct types of mechanisms. The mathematical models describe the dynamics of mRNA and protein for 3 scenarios: (i) a simplistic model with no regulation, (ii) a model with transcriptional negative autoregulation, and (iii) a model with translational negative autoregulation. Our analysis shows that the negative autoregulation models produce faster responses and quicker return times to the input signals compared to the model with no regulation, while the transcriptional autoregulation model is the only model capable of producing oscillatory dynamics. The stochastic simulations predict that the transcriptional autoregulation model is the noisiest followed by the simplistic model, and the translational autoregulation model has the least noise. The noise level depends on the strength of inhibition. Furthermore, the transcriptional autoregulation model filters out the noise in the input signal for longer periods of time, and this time increases as the strength of the feedback gets stronger.


 Please wait while we load your content...
Please wait while we load your content...